- *Corresponding Author:
- R. V. Hosur
UM-DAE Centre for Excellence in Basic Sciences, Mumbai University Campus, Kalina, Santa Cruz-400 098, Mumbai, India
E-mail: hosur@tifr.res.in
| Date of Submission | 02 July 2018 |
| Date of Revision | 21 October 2018 |
| Date of Acceptance | 11 March 2019 |
| Indian J Pharm Sci 2019;81(3):538-544 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Inactivation of antibiotics by enzymatic acetylation is considered as one of the major resistance mechanisms. Transfer of acetyl group to aminoglycoside antibiotics involved formation of a ternary complex. The acetyl- CoA-Hpa2-polyamine/antibiotic ternary complex was studied using in silico and mass spectrometric methods. Molecular interactions involved in the formation and stabilization of the ternary complex were analysed. To identify the amino acids necessary for stabilization of the ternary complex and to study the overall protein thermodynamic stability, FoldX algorithm was used. Results attested that Hpa2 has capacity to form stable ternary complex with acetyl-CoA and polyamines (spermine, spermidine, putrescine) or aminoglycoside antibiotics (kanamycin, streptomycin). This is the fi rst experimental report showing acetyl-CoA-Hpa2- kanamycin ternary complex and formation of this ternary complex is important for its function.
Keywords
Acinetobacter baumannii, drug resistance, polyamine, kanamycin, streptomycin
Introduction
Overuse of antibiotics hastens the development of resistance in microbes and the enormously rising antibiotic resistance is a serious public health concern [1]. To slow down the progress of resistance, it is necessary to understand the processes responsible for its development and expansion. Resistance is a natural unstoppable process and in bacteria it is the outcome of innate and/or acquired traits [2]. Controlling an innate process is more difficult than controlling the acquired trait. This concern has led to increased interest in studying the cellular processes, pathways, and enzymes involved in the development of resistance. Proper understanding of how a particular organelle or pathway or enzyme is evolving itself to contribute to the development of resistance is of immense value. Also, this information is useful in developing strategies to counter the resistance development and spread.
This study deals with one of the most important mechanisms responsible for the development of resistance, which is enzymatic inactivation of an antibiotic by adding a functional group [3,4]. Hpa2, an acetyltransferase enzyme of Acinetobacter baumannii, which acylates substrates including protein, peptides and polyamines is chosen for the ternary state characterization. Acylation involves formation of acetyl-CoA-Hpa2-substrate ternary complex. Here, FoldX was used to evaluate the amino acids and their interactions necessary for stabilization of the ternary complex. MALDI experiment data corroborate obtained in silico conclusions.
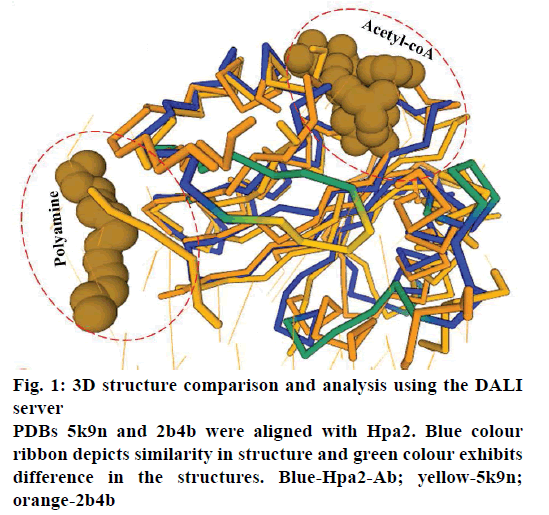
Hpa2 model was generated using modeller and model parameters were optimized by the RepairPDB command of FoldX [5]. Then, a dimer model was generated using HADDOCK [6] data driven docking, and the model was subjected to substrate-structure based data-driven docking [7]. Ternary complex (acetyl-CoA-Hpa2- substrate) was generated by HADDOCK data driven docking. Residues involved in the direct interaction with acetyl-CoA were used to generate the binary complex. Information about the residues involved in the formation of ternary complex was obtained by pair wise sequence alignment with homologous proteins using DALI server [8]. List of these residue are given in the Table 1.
| PDB id | Z-score | R.M.S.D (Å) | Residues crafting polyamine/antibiotics binding site |
|---|---|---|---|
| 1s3zvs. Hpa2 model | 11.5 | ~2.5Å | E28, Q33, D82, W84, L148, E152 and W154 E27, D34, D38, Y107, V108, Y124, Q125, E126, I129 and H131 |
| 2b4bvs. Hpa2 model |
12.5 | ~1.3Å | D34, W36, D37, D38, F39, R68, W124, Q125 and D126 E27, D34, D38, Y107, V108, Y124, Q125, E126, I129 and H131 |
Table 1: Substrate binding site analysis of PDBS 5K9N, 2B4B and 1S3Z, comparison with model HPA2 using DALI server
Binary complex was subjected for docking with polyamines such as spermine, spermidine, and putrescine and aminoglycoside antibiotics that include kanamycin and streptomycin. Accurate calculation of in silico binding affinity and selection of nearnative pose from the best-scoring clusters are most challenging tasks in computer-aided structure-based drug designing [9]. Discrimination of active pose from inactive ones requires proper analysis of clusters and prior information obtained from low resolution experiments [10]. Here, experimental information was used for grid mapping and data driven docking. Binding pose with strong binding affinity from the largest cluster was considered for the interaction analysis. Binding data obtained after docking study is depicted in the Table 2. Analysis of binding data with Hpa2 alone suggested that kanamycin and streptomycin exhibited high affinity for Hpa2. Analysis of docked poses of kanamycin and streptomycin exhibited consistency in the binding poses clustering. Higher value of binding energy and inconsistency in binding poses generally happens due to poor performance of force field. Clustering of binding poses discriminates active binders from inactive ones. Similar to kanamycin and streptomycin, homogeneity in the clustering of polyamine-docked poses attested that polyamines are true substrates of Hpa2. However, lower binding affinity obtained for polyamines is due to poor performance by the force field [11,12]. Thermodynamic parameters obtained using FoldX are given in Table 3. Analysis of these parameters indicated that binding of kanamycin and streptomycin is derived by favourable ΔGH−bond, ΔGvdw and ΔGelect terms. However, binding of polyamine was supported by ΔGvdw and ΔGelect terms only, this in line with the docking data (Table 2).
| Model Hpa2-Acetyl-CoA binary complex | Binding Energy (kcal/mol) | Number of conformations in the selected cluster (out of 1000) | Residues making H-bond with substrate | Non-bonded Interactions |
|---|---|---|---|---|
| Kanamycin | −8.274 | 727 | 10 | 56 |
| Streptomycin | −7.183 | 683 | 6 | 48 |
| Spermine | −5.132 | 816 | 0 | 44 |
| Spermidine | −5.014 | 799 | 0 | 37 |
| Putrescine | −4.139 | 959 | 0 | 29 |
Table 2: Features of HPA2-acetyl-COA binary complex docked with the polyamines and antibiotics
| Hpa2 model | ΔGtotal (kcal/mol) | ΔGH−bond (kcal/mol) | ΔGvdw (kcal/mol) | ΔGelect (kcal/mol) |
|---|---|---|---|---|
| Kanamycin | −7.76 | −1.96 | −4.26 | −0.79 |
| Streptomycin | −7.01 | −1.68 | −3.06 | −0.66 |
| Spermine | −6.60 | 0.0 | −3.84 | −0.56 |
| Spermidine | −6.19 | 0.0 | −3.06 | −0.46 |
| Putrescine | −5.08 | 0.0 | −1.68 | −0.39 |
Table 3: Detailed energy contributions from the favourable thermodynamic terms for HPA2 ternary complex using FOLDX
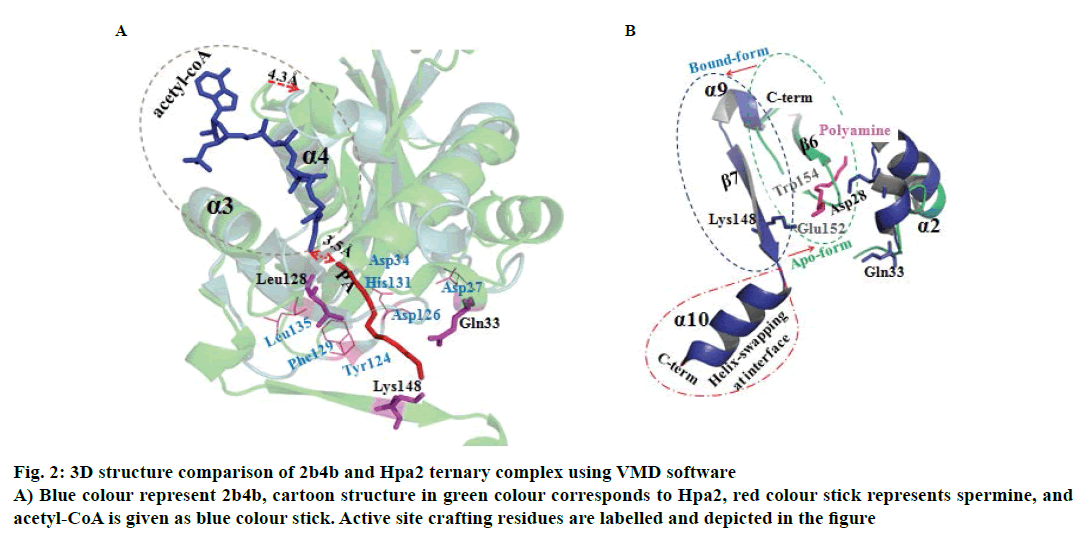
It is well established that GNAT superfamily members exhibit high structural conservation at acetyl-CoA binding site, however, regions responsible for the recognition of their diverse substrates might be different [13,14]. Three dimensional structural alignment and comparison of Hpa2 with its homologous protein using DALI server designated that binding interactions and residues involved in acetyl-CoA binding were almost identical; however, substrate binding site differed slightly. PDB of polyamine N-acetyltransferase 5k9n and 2b4b (rmsd ~2.5Å and Z-score ~11.5) showed considerable structural similarity with Hpa2 (fig. 1). Structural superposition of 5k9n, 2b4b and Hpa2 depicted similar observations as known for the other GNAT members, i.e., cofactor binding site was identical, however, polyamines binding site was not the same (fig. 2A). Also, binding of polyamines imposed large conformational changes as depicted in fig. 2B, polyamine binding led to opening of the C-terminal. In the unbound form, the C-terminal beta sheet-β8 and the loop preceding β8 folded back toward the acetyl-CoA binding site; however, in the bound form it extended outwardly. Residues involved in recognition of substrate polyamines acetylating 2b4b protein (E28, Q33, D82, W84, L148, E152 and W154) and Hpa2 (E27, D34, D38, Y107, V108, Y124, Q125, E126, I129 and H131) were similar in nature, i.e., mainly consisted of acidic and aromatic residues. Mutational study on 2b4b protein performed by Bewley and coworker [14] concluded that mutation of glutamic acid (E28) led to reduced activity by 55 %. Presence of E27 at the same site and its direct involvement in binding with spermine, spermidine and putrescine attested to the polyamine acetylation capacity of Hpa2. Binding sites of acetyl-CoA and polyamines are located side by side, and the distance between the acetyl-CoA and polyamines is ~4 Å, which suggests 1:1 ratio of binding. Polyamine binding site is crafted by the residues located at interface of dimer, due to which polyamine binding leads to conformational changes, and this reorganization would have an effect on the oligomeric state of the Hpa2.
Figure 2 : 3D structure comparison of 2b4b and Hpa2 ternary complex using VMD software
A) Blue colour represent 2b4b, cartoon structure in green colour corresponds to Hpa2, red colour stick represents spermine, and
acetyl-CoA is given as blue colour stick. Active site crafting residues are labelled and depicted in the figure
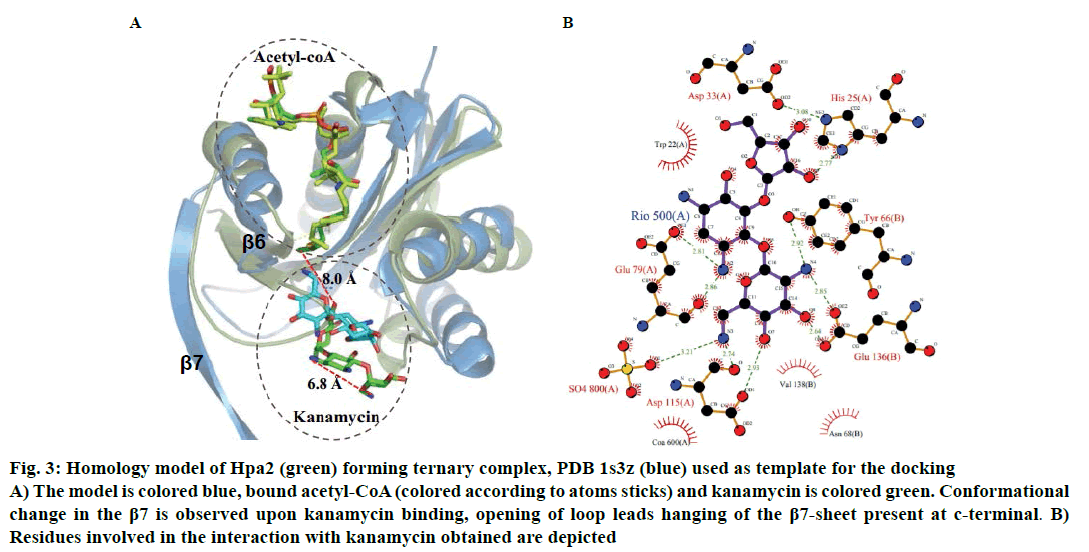
Binding of kanamycin and streptomycin was also carried out with binary complex using data driven docking and structural comparison with the homologous proteins was performed using the DALI server. Though Hpa2 exhibited very less sequence similarity (~27 %) with 1s3z, significant structural similarity was obtained (rmsd ~1.3 Å) for 143 of the 153 amino acids of (Z score ~12.5) AAC(6)-Iy (pdb-1s5k) (fig. 3A). AAC(6)-Iy enzyme possessed an extra helix between β1β2 sheets. AAC(6)-Iy catalysed the regioselective acetylation of aminoglycosides. In silico binding and thermodynamic data (Tables 2 and 3) of Hpa2 aminoglycoside complexation suggested that substrate recognition was derived by favourable interactions formed by acidic and aromatic residues (D34, W36, D37, D38, F39, R68, W124, Q125 and D126) fig. 3B. Regioselective acetylation and required chemistry for acetyl group transfer from acetyl-CoA needs ~3Å distance between 6’-amine and sulphur atom of bound acetyl-CoA. However, a distance of 8.0 Å was observed between the docked kanamycin and acetyl-CoA, which suggests possibility of 2:1 binding. In silico results suggested similar binding mode as well as interaction residues for all aminoglycoside antibiotics, however, why Hpa2 selectively binds with kanamycin and streptomycin remains unanswered. High resolution experiments such as NMR and crystallography characterization are needed to validate the exact mechanism.
Figure 3: Homology model of Hpa2 (green) forming ternary complex, PDB 1s3z (blue) used as template for the docking
A) The model is colored blue, bound acetyl-CoA (colored according to atoms sticks) and kanamycin is colored green. Conformational
change in the β7 is observed upon kanamycin binding, opening of loop leads hanging of the β7-sheet present at c-terminal. B)
Residues involved in the interaction with kanamycin obtained are depicted
In silico results were corroborated by MALDI data. For MALDI experiment all three polyamines and kanamycin and streptomycin were incubated with the Hpa2-acetyl-CoA binary complex. After overnight incubation, the samples were subjected to MALDI experiments using a Bruker ultrafleXtreme (Bruker Daltonics) mass spectrometer. For MALDI experiment, 100 μM of Hpa2, 100 μM of acetyl-CoA and 200 μM of substrate, were incubated overnight at 4°. Next day, 1 μl of acetyl-CoA-Hpa2-substrate ternary complex was taken and 1 μl of matrix α-cyano-4-hydroxycinnamic acid was added, mixed and 0.5 μl of this mixture was spotted on a well of the MALDI target plate. After complete solvent evaporation, sample was irradiated with laser for ionisation and then detected by using a time-of-flight mass spectrometer. Spectrum was recorded in reflector positive mode and the obtained MALDI data is discussed in the following section.
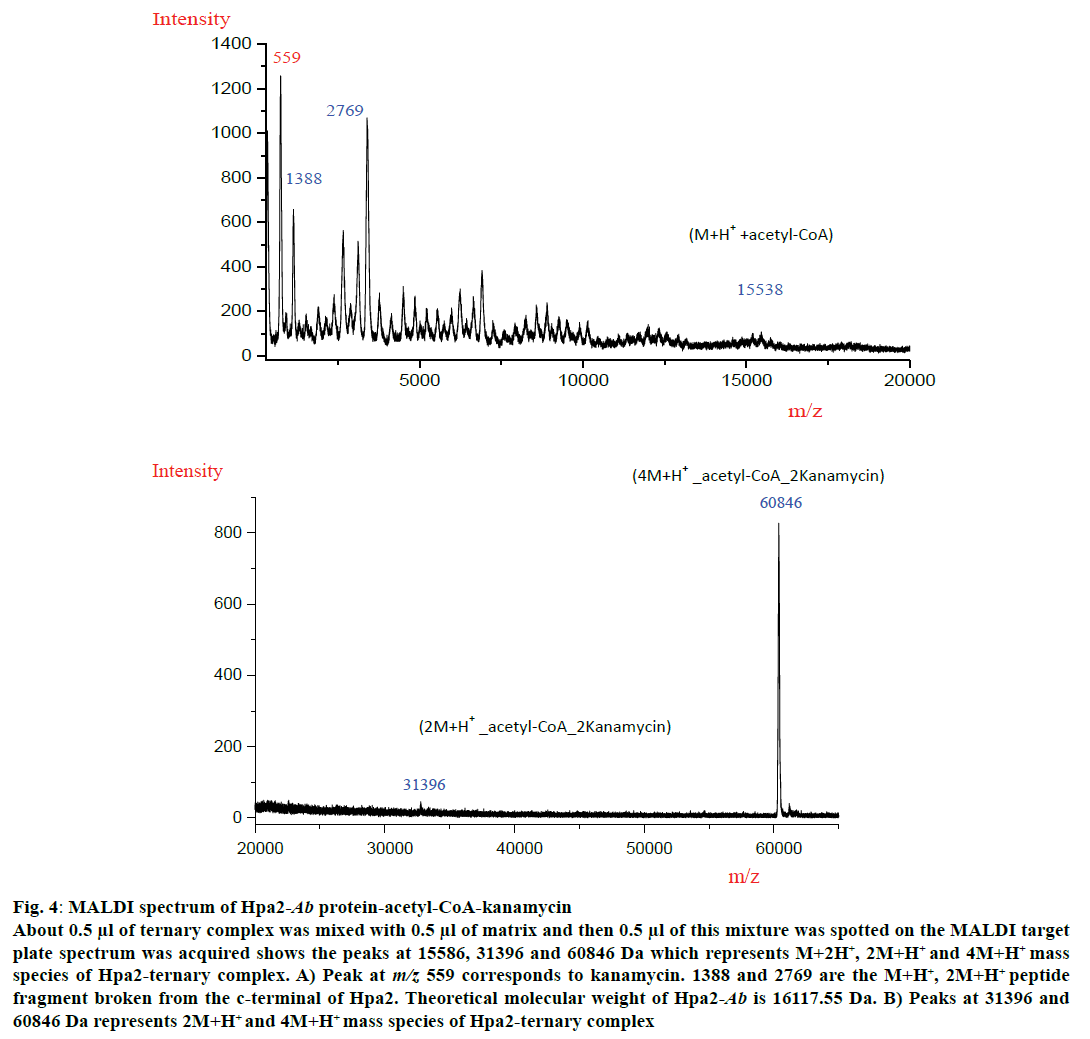
MALDI experiment was performed with all three polyamines and antibiotics but a ternary complex could be obtained with kanamycin (fig. 4). MALDI spectra analysis showed that Hpa2-Ab-acetyl-CoA formed complex with kanamycin as depicted in fig. 4A and 4B. For Hpa2-Ab-acetyl-CoA-kanamycin ternary complex, MALDI peaks were obtained at m/z values 559, 1388, 2769, 15586, 31396 and 60846 Da. Peak at m/z 559 represented kanamycin peak and 1388, 2769 peaks might correspond to the broken peptide fragment from the C-terminal. In silico analysis of ternary complexes indicate that formation of a ternary complex is assisted by a lot of conformational changes, especially near the C-terminal. The C-terminal beta-sheet (β7) unfolded to facilitate antibiotic binding, and the loop connecting this beta-sheet interacted with the bound kanamycin, however, β7-sheet hung outward as depicted in the fig. 3B. This C-terminal hanging beta-sheet (β7) might get chopped off during experiment. This is 13 residues (GIPHIDMTLALSL) long and theoretical molecular weight of this fragment is 1380.67 Da. It is reasonable to conclude that the peak obtained at m/z 1388 corresponded to M+H+ species of this peptide fragment, and peak at m/z 2769 corresponded to (2M+H+) species of this fragment. Peak obtained at m/z 15586 corresponded to (M+H+)+acetyl-CoA complex excluding the molecular weight of peptide fragment. Peaks obtained at m/z 31396 and 60846 corresponded to (2M+H+)+acetyl-CoA+2kanamycin and (4M+H+)+acetyl-CoA+2kanamycin ternary complex, respectively (fig. 4A). MALDI data suggested that binding of kanamycin induced conformational changes, which resulted in oligomerization of this protein.
Figure 4: MALDI spectrum of Hpa2-Ab protein-acetyl-CoA-kanamycin
About 0.5 μl of ternary complex was mixed with 0.5 μl of matrix and then 0.5 μl of this mixture was spotted on the MALDI target
plate spectrum was acquired shows the peaks at 15586, 31396 and 60846 Da which represents M+2H+, 2M+H+ and 4M+H+ mass
species of Hpa2-ternary complex. A) Peak at m/z 559 corresponds to kanamycin. 1388 and 2769 are the M+H+, 2M+H+ peptide
fragment broken from the c-terminal of Hpa2. Theoretical molecular weight of Hpa2-Ab is 16117.55 Da. B) Peaks at 31396 and
60846 Da represents 2M+H+ and 4M+H+ mass species of Hpa2-ternary complex
Aminoglycoside acetyltransferase enzyme showed considerable sequence similarity and high structural similarity to (AAC(6)-Iy), which exhibited histone acetylation capacity due to which they are considered as the evolutionary progenitor of Hpa2 [15]. However, no experimental reports are available on aminoglycoside acetylation capacity of Hpa2. In this investigation, from in silico and MALADI data it can be concluded that formation of Hpa2-acetly-CoA-kanamycin ternary complex might be associated with the kanamycin acetylation capacity of Hpa2.
Acknowledgements:
JST thanks Department of Atomic Energy for postdoctoral research fellowships.
Conflicts of interest:
The authors declare that there is no conflict of interest.
References
- Mak JK, Kim MJ, Pham J, Tapsall J, White PA. Antibiotic resistance determinants in nosocomial strains of multidrug-resistant Acinetobacter baumannii. J Antimicrob Chemother 2009;63:47-54.
- Spellberg B, Bonomo RA. Combination Therapy for Extreme Drug Resistant (XDR) Acinetobacter baumannii: Ready for Prime-Time? Crit Care Med 2015;43:1332-4.
- Hughes D, Anderson DI. Evolutionary consequences of drug resistance: shared principles across diverse targets and organisms. Nat Rev Genet 2015;16:459-71.
- Vetting MW, Magnet S, Nieves E, Roderick SL, Blanchard JS. A bacterial acetyltransferase capable of regioselective N-acetylation of antibiotics and histones. Chemist Biol 2004;11:565-73.
- Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: An online force field. Nucleic Acids Res 2005;33:W382-8.
- de Vries SJ, van Dijk M, Bonvin AM. The HADDOCK web server for data-driven biomolecular docking. Nat Protoc 2010;5:883-97.
- Tomar JS, Peddinti RK. A. baumannii histone acetyl transferases Hpa2: optimization of homology modeling, analysis of protein–protein interaction and virtual screening. J Biomol Struct Dyn 2017;35:1115-26.
- Holm L, Laakso LM. Dali server update. Nucleic Acids Res 2016;44:W351-5.
- Bharatham N, Finch KE, Min J, Mayasundari A, Dyer MA, Guy RK, et al. Performance of a docking/molecular dynamics protocol for virtual screening of nutlin-class inhibitors of Mdmx. J Mol Graph Model 2017;74:54-60.
- Entezari HY, Sereshti H, Saboury AA, Ghasemi J, Amirmostofian M, Supuran CT. 3D QSAR studies, pharmacophore modeling, and virtual screening of diarylpyrazole-benzenesulfonamide derivatives as a template to obtain new inhibitors, using human carbonic anhydrase II as a model protein. J Enzyme Inhib Med Chem 2017;32:688-700.
- Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 2006;65:712-25.
- Bharatham N, Bharatham K, Shelat AA, Bashford D. Ligand binding mode prediction by docking: mdm2/mdmx inhibitors as a case study. J Chem Inf Model 2014;54:648-59.
- Angus-Hill ML, Dutnall RN, Tafrov ST, Sternglanz S, Ramakrishnan V. Crystal structure of the histone acetyltransferase Hpa2: A tetrameric member of the Gcn5-related N-acetyltransferase superfamily. J Mol Biol 1999;294:1311-25.
- Bewley MC, Graziano V, Jiang J, Matz E, Studier FW, Pegg AP, et al. Structures of wild-type and mutant human spermidine/spermine N1-acetyltransferase, a potential therapeutic drug target. Proc Natl Acad Sci U S A 2005;103:2063-8.
- Filippova EV, Weigand S, Osipiuk J, Kiryukhina O, Joachimiak A, Anderson WF. Substrate-Induced Allosteric Change in the Quaternary Structure of the Spermidine N-Acetyltransferase SpeG. J Mol Biol 2015;427:3538-53.