- *Corresponding Author:
- P. Khanal
Department of Pharmacology and Toxicology, KLE College of Pharmacy Belagavi, KLE Academy of Higher Education and Research (KAHER), Belagavi-590 010, India
E-mail: pukarkhanal58@gmail.com
| Date of Submission | 03 August 2018 |
| Date of Revision | 13 November 2018 |
| Date of Acceptance | 20 March 2019 |
| Indian J Pharm Sci 2019;81(3):550-555 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The current study was aimed to screen borapetoside C, cordifolioside A, and magnofl orine against targets that are involved in the pathogenesis of type 2 diabetes mellitus. All active compounds were retrieved from PubChem database and protein molecules were downloaded from Protein Data Bank. Each protein was prepared using Discovery studio, and the binding pocket was predicted using castP. Each ligand was docked against the target proteins using AutoDock 4.0 Drug-likeness, toxicity and probable antidiabetic activity of each compound were also predicted. Among the three phytoconstituents, magnofl orine scored highest druglikeness score and also predicted to be a potent antidiabetic molecule. Comparisons were made with two clinically approved antidiabetic drugs i.e. sitagliptin and repaglinide.
Keywords
Borapetoside C, cordifolioside A, docking, in silico, magnoflorine, Tinospora
Diabetes mellitus (DM) is a chronic metabolic disorder due to defective insulin secretion or resistance, or a combination of both that leads to various health complications[1]. According to a global report of World Health Organization (WHO) 2017, the prevalence of diabetes in adults aged 20-79 y was estimated to be 8.8 % in 2015 and expected to rise to 10.4 % in 2040[2]. Synthetic molecules are prescribed for the pharmacotherapy of type 2 diabetes mellitus (T2DM) but they possess well-known side effects[3,4]. Since active phytoconstituents from traditional medicinal plants are believed to possess lower side effects[5], these could be used to discover new drug molecules. The WHO has also recommended investigating hypoglycaemic agents from folk medicinal plants as antidiabetic drugs[6].
Species of genus Tinospora are widely used as a folk medicine for various purposes[7]. Tinospora species have been reported to possess antidiabetic, antioxidant, immunostimulatory and antiosteoporosis activities. Further, Ayurveda has listed T. cordifolia in the management of diabetes. T. capillipes and T. sinensis are also officially listed in Chinese Pharmacopoeia[7,8]. Borapetoside C is one of the active phytoconstituents from T. crispa while cordifolioside A and magnoflorine are the major phytoconstituents from T. cordifolia and were reported to cause a reduction of oxidative stress and regularise carbohydrate metabolism[9-11] in experimental models of T2DM. However, the molecular mechanism responsible for the blood glucose lowering effect has not been understood.
Hence, an in silico study was performed to screen borapetoside C, cordifolioside A, and magnoflorine as antidiabetics against known targets such as 17β-hydroxysteroid dehydrogenase, retinol binding protein, C-jun N-terminal kinase, cholesteryl ester transfer protein, Lamin A/C, protein kinase B, adiponectin, insulin degrading enzyme, PPARG, human glucose transporter and adenylate cyclase, which have been implicated in the pathogenesis of T2DM and associated complications. The results were compared to those of repaglinide and sitagliptin, 2 clinically used antidiabetic drugs.
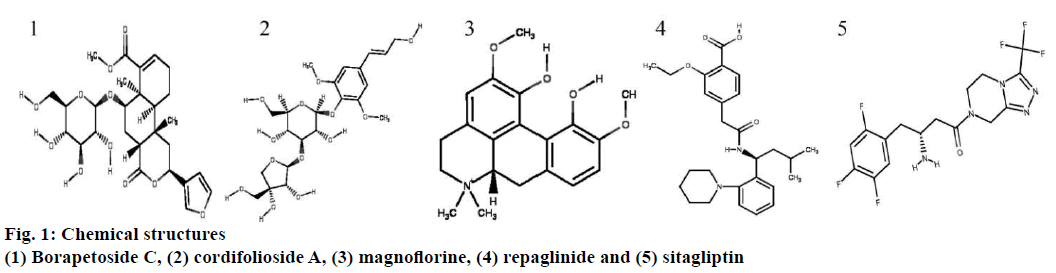
All 2D and 3D structures of borapetoside C, cordifolioside A, magnoflorine, repaglinide, and sitagliptin were retrieved from PubChem database (https://pubchem.ncbi.nlm.nih.gov/) in .sdf format and converted into .pdb and .mol using Discovery studios 2016. Canonical SMILES were also retrieved from PubChem database. Canonical SMILES explains the molecular structure as a graph with optional chiral indications. The energy of ligand molecules was minimized using mmff94 (https://open babel. readthedocs.io/en/latest/Forcefields/mmff94.html) force field and conjugate gradients as an optimization algorithm. Energy minimization is an important step in the preparation of a ligand to abolish clashes within the atoms of a ligand molecule and produce a reasonable staring pose. The energy minimized ligand molecules were then converted into AutoDock ligand in the format of .pdbqt. All the 2D structures of compounds are shown in fig. 1.
All the targets were retrieved from Protein Data Bank (https://www.rcsb.org). The retrieved 3D structures of proteins were combined with water and other hetero molecules. Discovery studio 2016 was used to remove water molecules and heteroatoms. Water molecules and other heteroatoms were removed to clear the binding pocket and make computations easier so that ligand can create satisfying interactions with the protein. The chain was selected based on their completeness of amino acid residue and presence of the active site. This modified protein was then saved in .pdb format. An online tool, Computed Atlas of Surface Topography of proteins[12] was used to predict a number of mouth opening, pocket MS area, pocket MS volume, mouth MS area and mouth MS circumference sum of each protein under the radius probe of 1.4 angstroms.
Drug-likeness character (Lipinski's rule of five) is associated with absorption, bioavailability, and biodistribution of the drug molecule from the human intestinal tract. This prediction is made before the molecule is synthesized in the laboratory or extracted from the natural source. Hence, molsoft (http://molsoft. com/), an online server was used to predict the druglikeness character of all the selected ligand molecules.
Various pharmacokinetic parameters of drug molecules and probable toxicity in different models were predicted by using an online server, admetSAR. Blood-brain barrier permeability, human intestinal absorptivity, caco-2 permeability, AMES toxicity, aqueous solubility, rat acute toxicity, and fish toxicity were predicted. The server was also used to predict the possible interaction of drug molecule with various isoenzymes. The prediction was based on a vector machine classification algorithm and in-house substructure pattern recognition method, which were built via regression methods[13]. Similarly, Escherichia coli toxicity of each molecule was predicted using abSYNTH[14]. The probable antidiabetic activity of these 3 molecules was also predicted via the PASS online. PASS online predicts about 4000 probable pharmacological activities with an average probability of 0.95 based on structure-activity relationships[15].
Docking was performed using AutoDock Vina. It uses sophisticated gradient optimization method to calculate grid maps and provides the cluster of results[16]. After the completion of docking, 10 docking poses of ligand were obtained. The pose with minimum binding energy was selected for visualizing the ligand-protein interaction.
Among the targeted protein molecules, insulindegrading enzyme scored the maximum number of mouth openings, pocket MS area, pocket MS volume, mouth MS area, and mouth MS circumference sum compared to other target molecules (Table 1).
| Protein | PDB ID | NMO | PMSA | PMSV | MMSA | MMSCS |
|---|---|---|---|---|---|---|
| 17ß-hydroxysteroid dehydrogenase | 1BHS | 2 | 1452.215 | 2214.501 | 156.5 | 80.50 |
| Retinol binding protein | 1GGL | 1 | 2096.563 | 2779.102 | 9.81 | 11.28 |
| C-Jun N-terminal kinase | 1JNK | 3 | 1630.603 | 3342.965 | 947.35 | 321.01 |
| Cholesteryl ester transfer protein | 2OBD | 3 | 3303.066 | 5518.994 | 122.36 | 77.92 |
| Lamin A/C | 3GEF | 5 | 1680.576 | 3682.228 | 764.62 | 309.74 |
| Protein kinase B | 3QKK | 2 | 1079.742 | 1770.330 | 87.37 | 59.78 |
| Adiponectin | 4DOU | 4 | 682.933 | 1110.525 | 144.90 | 99.10 |
| Insulin degrading enzyme | 4PF9 | 8 | 11882.211 | 32149.483 | 995.70 | 402.56 |
| PPARG | 4Y29 | 2 | 1492.833 | 1932.907 | 83.82 | 57.43 |
| Human glucose transporter | 5C65 | 2 | 1613.297 | 3310.845 | 668.11 | 194.83 |
| Adenylate cyclase | 5IV4 | 3 | 1108.655 | 1795.905 | 184.61 | 104.47 |
NMO: Number of mouth openings, PMSA: pocket MS area, PMSV: pocket MS volume, MMSA: mouth MS area and MMSCS: mouth MS circumference sum
Table 1: Number of binding pocket/openings, mouth volume area and circumference of each of the protein
All the selected molecules were able to interact with targeted protein molecules. The interaction between the ligand and protein is explicated by the binding energy and number of interactions; defined by the hydrogen bonds and pi interactions. Binding energy and number of hydrogen bond interaction of all compounds with each protein are shown in Table 2.
| PDB ID | Binding energy and number of hydrogen bond of protein molecules with each ligand | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Borapetoside C | Cordifolioside A | Magnoflorine | Repaglinide | Sitagliptin | ||||||
| BE (kcal/mol) | NHB | BE (kcal/mol) | NHB | BE (kcal/mol) | NHB | BE (kcal/mol) | NHB | BE (kcal/mol) | NHB | |
| 1BHS | -6.9 | 2 | -9.1 | 1 | -7.6 | 0 | -8 | 1 | -8.7 | 7 |
| 1GGL | -5.1 | 4 | -5.1 | 0 | -4.9 | 0 | -4.1 | 2 | -4.6 | 3 |
| 1JNK | -5.9 | 3 | -5.7 | 1 | -5.3 | 1 | -5.3 | 3 | -6.1 | 2 |
| 2OBD | -5 | 1 | -5 | 1 | -5.4 | 0 | -4.7 | 1 | -5.3 | 3 |
| 3GEF | -6.4 | 6 | -6.6 | 3 | -5.6 | 2 | -5.4 | 1 | -6.4 | 6 |
| 3QKK | -6.9 | 2 | -6.9 | 0 | -5.9 | 2 | -6.1 | 2 | -7.1 | 4 |
| 4DOU | -6.6 | 3 | -6.7 | 0 | -6.3 | 1 | -6.5 | 2 | -7.1 | 1 |
| 4PF9 | -7.4 | 4 | -7.5 | 1 | -6.7 | 2 | -6.5 | 1 | -7.2 | 3 |
| 4Y29 | -6.1 | 3 | -6.4 | 0 | -6.2 | 4 | -5.3 | 3 | -6.4 | 1 |
| 5C65 | -6.7 | 4 | -6.5 | 3 | -5.5 | 3 | -5.2 | 1 | -6.4 | 3 |
| 5IV4 | -5.1 | 1 | -5.4 | 2 | -5.1 | 1 | -5 | 1 | -5.8 | 4 |
BE: binding energy, NHB: number of hydrogen bond
Table 2: Binding energy and number of hydrogen bonds of ligand protein interaction
Borapetoside C scored the highest binding affinity with two proteins i.e. retinol binding protein (–5.1 kcal/mol) and human glucose transporter (–6.7 kcal/mol), cordifolioside A scored highest binding affinity with 17β-hydroxysteroid dehydrogenase (–9.1 kcal/mol), retinol binding protein (–5.1 kcal/mol), Lamin A/C (–6.6 kcal/mol), insulin degrading enzyme (–7.5 kcal/mol) and PPARG (–6.4 kcal/mol). Further, magnoflorine scored highest binding affinity with only one protein molecule cholesteryl ester transfer protein (–5.4 kcal/mol). Sitagliptin scored the highest binding affinity with five protein molecules, C-jun N-terminal kinase (–6.1 kcal/mol), protein kinase B (–7.1 kcal/mol) and adiponectin (–7.1 kcal/mol), PPARG (–6.4 kcal/ mol) and adenylate cyclase (–5.8 kcal/mol). Binding affinity of individual compounds with each target and a respective number of hydrogen bond interactions are summarized in Table 2. Among the phytoconstituents tested, magnoflorine scored highest drug-likeness value of 0.80 without any violation of the rule of five. However, borapetoside C violated two rules, the molecular weight and the number of hydrogen bond acceptors, while cordifolioside A violated three rules, the molecular weight, the number of hydrogen bond acceptors and the number of hydrogen bond donors (Table 3). Most drug molecules under development fail in clinical trials due to poor pharmacokinetics and high toxicity[17]. Hence, each individual molecule was predicted for pharmacokinetic, i.e. absorption, distribution, metabolism, excretion and toxicity (ADMET) using regression models. Magnoflorine was predicted to cross blood brain barrier but not the human intestinal tract. All compounds were predicted to be non-mutagenic and non-carcinogenic.
| Ligand molecules | PubChem CID | Lipinski rule of five | DLS | ECT | Log S (mg/l) | |||
|---|---|---|---|---|---|---|---|---|
| M. W. | NHBA | NHBD | Log P | |||||
| <500 | <10 | <5 | <5 | |||||
| Borapetoside C | 101697033 | 536.23 | 11 | 4 | 0.63 | 0.15 | 0.19 | 235.03 |
| Cordifolioside A | 45359937 | 504.18 | 13 | 7 | -2.60 | 0.57 | 0.17 | 166434.48 |
| Magnoflorine | 73337 | 342.17 | 4 | 2 | 2.77 | 0.80 | 0.45 | 52245.2 |
| Repaglinide | 65981 | 452.27 | 4 | 2 | 5.80 | 0.92 | 0.43 | 1.34 |
| Sitagliptin | 4369359 | 407.12 | 4 | 2 | 1.43 | 0.49 | 0.46 | 6.77 |
MW: molecular weight, NHBA: number hydrogen bond acceptors, NHBD: number hydrogen bond donors, DLS; drug-likeness score and ECT: E. coli toxicity IC50 (g/l)
Table 3: Druglikeness of compounds
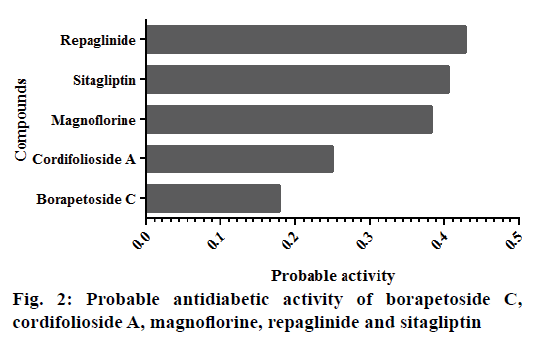
Cordifolioside A scored highest binding affinity to the majority of the selected protein molecules, but this parameter alone could not be considered for the drug development process. Hence, Lipinski's rule of five, a qualitative parameter, was used to understand the druglikeness character of each molecule. Magnoflorine scored highest drug-likeness value compared to other compounds tested. Further, pharmacological activity prediction of all the phytoconstituents suggested that magnoflorine to be a potent antidiabetic molecule as shown in fig. 2. Personalized medicine concept is based on mutagenicity involving the genes that encode CYP2D6 and CYP2C19[18]. Current study showed that the selected phytoconstituents were not a substrate for CYP2D6 nor inhibitors of CYP2D6 and CYP2C19. However, predicted result showed that CYP2C19 mutagenicity test is necessary for sitagliptin.
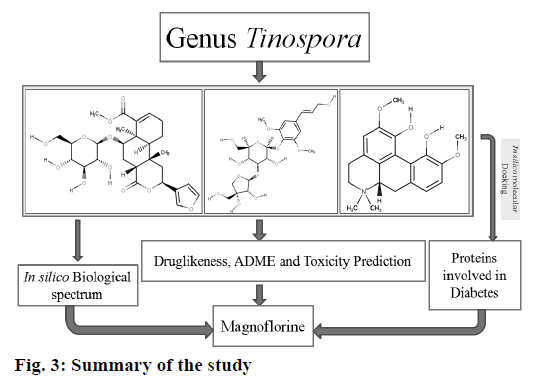
The target protein molecules play a crucial role in the pathogenesis of diabetes. Alteration of these proteins or inhibition of their synthesis might be help in the management of diabetes. Hence, the current study demonstrated that magnoflorine to be a potential molecule that could alter majority of the proteins that have been implicated in the pathogenesis of diabetes. The summary of the current study is shown in fig. 3.
Borapetoside C, cordifolioside A and magnoflorine are the three molecules reported to possess antidiabetic activity from various modes. Borapetoside C is reported to lower the blood glucose by enhancing insulin sensitivity and GLUT2 expression followed by Akt phosphorylation[10]. Similarly, cordifolioside A is reported to improve insulin signaling and hepatic metabolism during insulin resistance. Further, it is reported to lower the blood glucose and cholesterol level[19]. Magnoflorine is reported to have a direct effect on carbohydrate metabolism, increase the insulin sensitivity and reverse diabetes-associated alterations in lipid profile and oxidative stress[20]. Repaglinide is reported to stimulate the release of insulin from beta cells of the pancreas by increasing the intracellular calcium level[21]. Sitagliptin is a selective DPP-4 inhibitor to minimize the inactivation of various incretin hormones, which are involved in the regulation of glucose homeostasis[22]. Further, the role of selected target protein molecules in the pathogenesis of diabetes mellitus is summarized in Table 4.
| Protein name | Role in Diabetes |
|---|---|
| 17ß-Hydroxysteroid dehydrogenase | It is involved in steroidogenesis and metabolism of lipids and its elevation leads to insulin resistant and obesity. The level of enzyme gets elevated in women of post-menopausal stage[23] |
| Cellular retinol binding protein | It is involved in the lipid homeostasis under diet induced obesity. The protein is upregulated in adipose tissue in insulin resistant state[24] |
| C-Jun N-Terminal Kinase | It stimulates serine phosphorylation to inhibit insulin stimulated tyrosine phosphotylation[25] |
| Cholesteryl ester transfer protein | It involved in remodelling of triglycerides and lipoproteins[26] |
| Lamin A/C | It is associated for loss of subcutaneous adipose tissue, insulin resistance, dyslipidaemia and T2DM[27] |
| Protein kinase B | It activates phosphodiesterase enzyme and involved in the regulation of insulin, glucose and Camp[28] |
| Adiponectin | It plays an important role in the development of atherosclerosis and insulin resistance[29] |
| Insulin degrading enzyme | It is involved in the metabolism of insulin[30] |
| PPARG | It is involved in the modulation of adipose tissue, fat and carbohydrate. The level of PPARG increases with insulin resistance[31] |
| Human glucose transporter | It is involved in phosphorylation of glucose by increasing glycolytic flux. Expression of this protein is decreased in Obesity and T2DM[32] |
| Adenylate cyclase | The protein is involved in enhancing the insulin secretion and increases cAMP level in ß-cells[33] |
Table 4: Details of the role of target proteins in diabetes.
In conclusion, the current study projected magnoflorine as a potential molecule to interact with majority of proteins that are implicated in the pathogenesis DM. The current study also reported that these predictions were comparable to those of repaglinide and sitagliptin. The findings of current study is based only on computational predictions, which needs to be further proven by well designed wet-lab protocols.
Conflicts of interest:
The authors declare that there is no conflict of interest.
References
- Hameed I, Masoodi SR, Mir SA, Nabi M, Ghazanfar K, Ganai BA. Type 2 diabetes mellitus: From a metabolic disorder to an inflammatory condition. World J Diabetes 2015;6(4):598-612.
- Ogurtsova K, Fernandes JDR, Huang Y, Linnenkamp U, Guariguata L, Cho NH, et al. IDF Diabetes Atlas: Global estimates for the prevalence of diabetes for 2015 and 2040. Diabetes Res Clin Pract 2017;128:40-50.
- Upadhyay J, Polyzos SA, Perakakis N, Thakkar B, Paschou SA, Katsiki N, et al. Pharmacotherapy of type 2 diabetes: An update. Metabolism 2018;78:13-42.
- Krentz AJ, Bailey CJ. Oral antidiabetic agents: Current role in type 2 diabetes mellitus. Drugs 2005;65(3):385-411.
- Moreira DDL, Teixeira SS, Monteiro MHD, De-oliveira ACAX, Paumgartten FJR. Traditional use and safety of herbal medicines. Rev Bras Farmacogn 2014;24(2):248-57.
- Diabetes mellitus. Report of a WHO expert committee. World Health Organization. 1965. p. 1-44. Available from: https://apps.who.int/iris/bitstream/handle/10665/38442/WHO_TRS_310.pdf;jsessionid=2C4D0847578CDB3E30D98B6B54F12241?sequence=1.
- Chi S, She G, Han D, Wang W, Liu Z, Liu B. Genus Tinospora: Ethnopharmacology, Phytochemistry, and Pharmacology. Evid Based Complement Alternat Med 2016;2016:1-32.
- Nadig PD, Revankar RR, Dethe SM, Narayanswamy SB, Aliyar MA. Effect of Tinospora cordifolia on experimental diabetic neuropathy. Indian J Pharmacol 2012;44(5):580-83.
- Sangeetha MK, Balaji Raghavendran HR, Gayathri V, Vasanthi HR. Tinospora cordifolia attenuates oxidative stress and distorted carbohydrate metabolism in experimentally induced type 2 diabetes in rats. J Nat Med 2011;65(3-4):544-50.
- Ruan CT, Lam SH, Chi TC, Lee SS, Su MJ. Borapetoside C from Tinospora crispa improves insulin sensitivity in diabetic mice. Phytomedicine 2012;19(8-9):719-24.
- Patel MB, Mishra S. Hypoglycemic activity of alkaloidal fraction of Tinospora cordifolia. Phytomedicine 2011;18(12):1045-52.
- Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: Computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res 2006;34:W116-8.
- Cheng F, Li W, Zhou Y, Shen J, Wu Z, Liu G, et al. admetSAR: A Comprehensive Source and Free Tool for Assessment of Chemical ADMET Properties. J Chem Inf Model 2010;52(11):3099-105.
- Planson AG, Carbonell P, Paillard E, Pollet N, Faulon JL. Compound Toxicity Screening and Structure-Activity Relationship Modeling in Escherichia coli. Biotechnol Bioeng 2012;109(3):846-50.
- Filimonov DA, Lagunin AA, Gloriozova TA, Rudik AV, Druzhilovskii DS, Pogodin PV, et al. Prediction of the biological activity spectra of organic compounds using the pass online web resource. Chem Heterocycl Compd 2014;50(3):444-57.
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem 2010;31(2):455-61.
- Kennedy T. Managing the drug discovery/development interface. Drug Discov Today 1997;2(10):436-44.
- Annemans L, Redekop K, Payne K. Current methodological issues in the economic assessment of personalized medicine. Value Heal 2013;16(6S):S20-6.
- Stohs SJ, Ray S. Anti-diabetic and Anti-hyperlipidemic Effects and Safety of Salacia reticulata and Related Species. Phyther Res 2015;29(7):986-95.
- Ajiboye T, Uwazie JN, Haliru FZ, Ibitoye OB, Sunmonu T, Yakubu M, et al. Mechanisms of action of columbamine, jatrorrhizine and magnoflorine as antidiabetic agents. Recent Progress Med Plant 2017;45:221-32.
- Ambavane V, Patil R, Ainapure SS. Repaglinide: a short acting insulin secretagogue for postprandial hyperglycaemia. J Postgrad Med 2002;48:246.
- Gallwitz B. Review of sitagliptin phosphate: a novel treatment for type 2 diabetes. Vasc Health Risk Manag 2007;32):203-10.
- Cho MK. Decreased Expression of Type 5 17ß-Hydroxysteroid Dehydrogenase (AKR1C3) Protein Identified in Human Diabetic Skin Tissue. Ann Dermatol 2013;25(4):423-7.
- Zizola CF, Schwartz GJ,Vogel S. Cellular retinol-binding protein type III is a PPAR gamma target gene and plays a role in lipid metabolism. Am J Physiol Endocrinol Metab 2008;295(6):E1358-68.
- Lee YH, Giraud J, Davis RJ, White MF. C-Jun N-terminal kinase (JNK) mediates feedback inhibition of the insulin signaling cascade. J Biol Chem 2003;278(5):2896-902.
- Kimak E, Bylina J, Solski J, Halabis M, Baranowicz-Gaszczyk I, Ksiazek A. Association between lipids, lipoproteins composition of HDL Particles and triglyceride-rich lipoproteins, and LCAT and CETP activity in post-renal transplant patients. Cell Biochem Biophys 2013;67(2):695-702.
- Mesa JL, Loos RJF, Franks PW, Ong KK, Luan J, Rahilly SO, et al. Lamin A/C polymorphisms, type 2 diabetes, and the metabolic syndrome: case-control and quantitative trait studies. Diabetes 2007;56(3):884-9.
- Kallan NC, Spencer KL,Blake JF,Xu R, Heizer J, Bencsik JR et al. Discovery and SAR of spirochromane Akt inhibitors. Bioorg Med Chem Lett 2011;21(8):2410-14.
- Lihn AS, Pedersen SB, Richelsen B. Adiponectin: Action, regulation and association to insulin sensitivity. Obes Rev 2005;6(1):13-21.
- Ito S, Ohtsuki S, Murata S, Katsukura Y, Suzuki H, Funaki M et al. Involvement of insulin-degrading enzyme in insulin-and atrial natriuretic peptide-sensitive internalization of amyloid-ß peptide in mouse brain capillary endothelial cells. J Alzheimers Dis 2014;38(1):185-200.
- Zheng W, Qiu L,Wang R,Feng X, Han Y,Zhu Y et al. Selective targeting of PPAR? by the natural product chelerythrine with a unique binding mode and improved antidiabetic potency. Sci Rep 2015;5:12222.
- Simpson IA,Dwyer D,Malide D, Moley KH, Travis A,Vannucci SJ. The facilitative glucose transporter GLUT3: 20 years of distinction. Am J Physiol Endocrinol Metab 2008;295(2):E242-53.
- Lee YS, Jun HS. Glucagon-Like Peptide-1 Receptor Agonist and Glucagon Increase Glucose-Stimulated Insulin Secretion in Beta Cells via Distinct Adenylyl Cyclases. Int J Med Sci 2018;15(6):603-9.