- *Corresponding Author:
- P. Khanal
Department of Pharmacology and Toxicology, KLE College of Pharmacy, KAHER, Belagavi-590010, India
E-mail: pukarkhanal58@gmail.com
| Date of Submission | 24 March 2018 |
| Date of Revision | 23 September 2018 |
| Date of Acceptance | 18 February 2019 |
| Indian J Pharm Sci 2019;81(2):326-332 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The present investigation dealt with determination of the binding affinities in silico of six limonoids from Neem, 7-deacetoxy-7-oxogedunin, 17-hydroxyazadiradione, nimolicinol, 6-acetynimbadiol, andirobin and gedunin to bind the protein kinase, pfpk5 from Plasmodium falciparum, compared to staurosporine, a well-known protein kinase inhibitor. Further, the molecules’ pharmacokinetics and toxicity was also evaluated. Among the six compounds, 7-deacetoxy-7-oxogedunin and nimolicinol showed the best binding affinity for pfpk5 via interactions of hydrogen and pi bond and were also predicted to be potent molecules. The current in silico study suggested that limonoids could be potential pfpk5 inhibitors, which could further be developed as antimalarial drugs. Going forward the in vitro and in vivo studies are needed to confirm the efficacy of limonoids as pfpk5 inhibitors.
Keywords
Azadirachta indica, docking, limonoids, malaria, pfpk5, Plasmodium falciparum
Malaria is a vector-borne disease transmitted by female Anopheles mosquito carrying a unicellular protozoan from the genus Plasmodium. There are a number of species of the genus Plasmodium but few of them are more responsible in transmitting disease in human population such as P. vivax, P. malariae, P. ovale, and P. falciparum [1]. World Health Organization estimates the presence of 500 million clinical cases of malaria leading to the death of approximately one million throughout the world [2]. The condition reported is even worse in the African continent. Malaria has been reported as the representative cause of death of children in underdeveloped countries [2]. Due to the rapid spread of the chloroquine-resistant P. falciparum followed by the emergence of many multidrug-resistance strains of Plasmodium, an urgent need for a new drug for treating drug-resistant malaria [1-3].
Azadirachta indica, commonly known as Neem belongs to the family Meliaceae and constitutes alkaloids, polyphenols, flavanoids, limonoids, steroids and carotenoids as active phytoconstituents. Neem has been reported to possess various medicinally important activities that include anticancer, neuroprotective, antiinflammatory, antifungal, antiviral, antibacterial and hepatoprotective as well as was also found to modulate numerous tumor suppressor genes and play a prime role in immunomodulation [4]. In vitro and in vivo studies suggested the potential applicability of limonoid as antimalarial agent [3,5,6]. Apart from this, limonoids have also been reported to possess potent antiproliferative and antioxidant activity [7].
Due to resistance of P. falciparum towards existing antimalarials, new targets and new potent molecules needed to be identified for the treatment of fever and other malarial symptoms. The cyclin-dependent kinases (cdks) are involved in the regulation of the cell cycle in many organisms. P. falciparum protein kinase 5 (pfpk5) is recognized as one of the novel targets in falciparum species [1]. Pfpk5 is one of the essential protein kinases isolated from P. falciparum, which is needed for the activation and maintenance of S-phase of the parasite [8].
As per reported literature, in silico investigation has not been performed to address the role of limonoids as antimalarial agent. Hence, the current study was aimed to investigate the absorption, distribution, metabolism, elimination and toxicity (ADMET) parameters and binding affinity of 7-deacetoxy-7- oxogedunin, 17-hydroxyazadiradione, nimolicinol, 6-acetynimbadiol, andirobin and gedunin to pfpk5. The binding affinity of all the compounds was compared to staurosporine [9], a known protein kinase inhibitor.
Materials and Methods
Preparation of ligand for bioactive molecules:
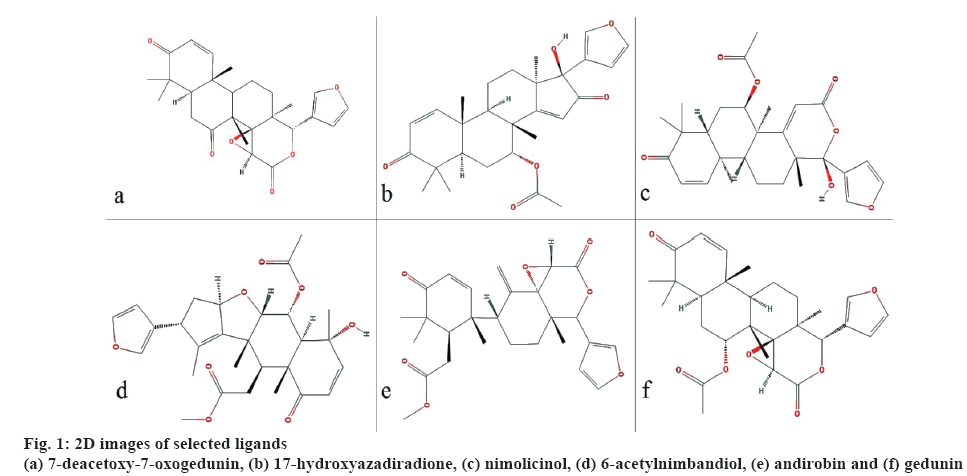
All the 3D structures of the bioactive molecules were retrieved from PubChem in structural data format (SDF) and converted to protein data bank (PDB) format using Discovery studio 4.0. Open Babel graphical user interface (GUI) was used to convert SDF format into MOL, which was used for the prediction of Escherichia coli toxicity. Canonical simplified molecular line-entry system (SMILES) were retrieved from PubChem to predict pharmacokinetic and drug-likeness character. The retrieved biomolecules were then minimized using Open Babel using mmff94 force field and conjugate gradients as an optimization algorithm which is available in PyRx 0.8. The 2D images of the selected ligands are shown in figure 1.
Preparation of macromolecule:
Target pfpk5 (PDB ID: 1OB3) was retrieved from the PDB (https://www.rcsb.org) website and viewed under Discovery studio 4.0. The retrieved protein was solved by X-ray diffraction technique at 1.99 Å resolution, with R-values i.e. R work 0.19 and R free 0.23. The retrieved molecules were complex with water molecules and hetero-atoms. Hence, Discovery studio 4.0 was used to remove hetero atoms and water molecules to avoid docking interference and saved in the PDB format.
Active site prediction:
Presence of the active site in protein plays an important role in protein and ligand interaction. Hence online CASTp server i.e. Compound Atlas of Surface Topography of proteins (http://cast.engr.uic.edu) was used to predict the active site in the pfpk5 protein (PDB ID: 1OB3). The tool detects all the possible pockets in the protein structure. The server also provides the information regarding the SA, Richards' surface (solvent accessible surface) and MS, Connolly's surface (molecular surface). The first pocket was then chosen as the most favourable active site for the docking studies.
ADMET and drug-likeness predictions of ligand:
The pharmacokinetic properties such as absorption, distribution, metabolism, and excretion and toxicity studies of bioactive molecule play an important role in the drug development steps. Hence, all the possible pharmacokinetic parameters (ADMET) and toxicity of selected biomolecules were predicted using admetSAR in different models. Similarly, an online server ABSYNTH was used to predict E. coli toxicity. Further, druglikeness character was predicted using MolSoft.
Ligand protein docking:
Docking was performed by using PyRx 0.8. After the completion of docking, autodock preferences were obtained for both ligand and target in PDBQT format. Open babel GUI was used to convert preferred protein target (PDBQT) into PDB. Protein and ligand were viewed under Chimera 1.11.2 and the complex was saved in PDB format. The pose of minimum binding energy was chosen as the best interaction.
Results and Discussion
Estimating the binding affinity of a small molecule in the active site of the receptor helps to understand ligand protein interaction. The interaction between protein and ligand occurs via a hydrogen bond and pi-interaction. Among the six selected bioactives, two compounds showed the highest binding affinity i.e. –5.5 kcal/mol by 7-deacetoxy-7-oxogedunin and nimolicinol. Similarly, 6-acetylnimbandiol showed the least binding affinity i.e. –4.6 kcal/mol (Table 1).
| Ligand molecules | PubChem ID | Binding energy (kcal/mol) |
|---|---|---|
| 7-deacetoxy-7-oxogedunin | 71300386 | –5.5 |
| 17-hydroxyazadiradione | 52951892 | –4.9 |
| Nimolicinol | 184937 | –5.5 |
| 6-acetylnimbandiol | 52952216 | –4.6 |
| Andirobin | 266444 | –4.9 |
| Gedunin | 12004512 | –5 |
| Staurosporine* | 44259 | –4.66 |
*A known protein kinase inhibitor; docked against the pfpk5 under the same settings of the grid box. All the ligands were docked against pfpk5 protein (PDB ID: 1OB3)
Table 1: Ligand Protein Binding Energy
Among the selected bioactive molecules, 17-hydroxyazadiradone scored highest druglikeness character i.e. 0.63. E. coli toxicity prediction helps to predict the concentration of drug molecule required to inhibit the growth of the prokaryotic cell. It also helps to predict the affinity of the molecule to act on the single cell stage. In our study, 7-deacetoxy-7-oxogedunine scored the highest value for E. coli toxicity i.e. 0.4 g/l and the lowest value was scored by 6-acetylnimbandiol i.e. 0.21 g/l (Table 2).
| Compounds name | Molecular formula | E. coli IC50 (gm/l) | Lipinski rule of five | Drug-likeness score | |||
|---|---|---|---|---|---|---|---|
| Molecular weight no | No. of HBA | No. of HBD | XLog P | ||||
| Acceptable Values for Lipinski Rule of Five | - | - | <500 | <10 | <5 | <5 | - |
| 7-deacetoxy-7-oxogedunin | C26H30O6 | 0.4 | 438.20 | 6 | 0 | 2.82 | -0.76 |
| 17-hydroxyazadiradione | C28H34O6 | 0.29 | 466.24 | 6 | 1 | 3.63 | 0.63 |
| Nimolicinol | C28H34O7 | 0.37 | 482.23 | 7 | 1 | 3.56 | 0.45 |
| 6-acetynimbadiol | C28H34O8 | 0.21 | 498.23 | 8 | 1 | 2.96 | -0.01 |
| Andirobin | C27H32O7 | 0.38 | 468.21 | 7 | 0 | 3.70 | -1.00 |
| Gedunin | C28H34O7 | 0.37 | 482.23 | 7 | 0 | 3.41 | -0.42 |
IC50 is inhibitory concentration 50, HBA is hydrogen bond acceptor, and HBD is hydrogen bond donor, BBB is blood brain barrier, HIA is human intestinal absorption, C-2P is Caco-2 permeability, AMEST is AMES Toxicity, AOT is acute oral toxicity, RAT is rat acute toxicity (LD50, mol/kg)
Table 2: Drug-Likeness and Toxicity Prediction on E. Coli
Using admetSAR, pharmacokinetic parameters were predicted in three different models i.e. blood-brain barrier, human intestinal absorption and Caco-2 permeability. The study showed that all limonoids tested exhibited positive result to cross bloodbrain barrier with the highest probability of 0.86 for 7-deacetoxy-7-oxogedunin and lowest probability of 0.67 for andirobin. Similarly, all the selected limonoids exhibited positive result for human intestinal absorption with the highest probability of 1 for 7-deacetoxy-7- oxogedunin and gedunin and lowest probability of 0.98 for andirobin. On looking to the Caco-2 permeability, 7-deacetoxy-7-oxogedunin showed a positive result with a score of 0.5 whereas the rest of the molecules tested scored negative result. Similarly, all the selected molecules showed a negative result for Ames toxicity and carcinogenicity. Similarly, nimolicinol showed the highest probability for acute oral toxicity i.e. 0.61 with a lethal dose (LD50) of 4.2 mol/kg. Similarly, among the selected molecules, gedunin scored highest Tetrahymena pyriformis toxicity (pIGC50) at 1.27μg/l (Table 3).
| Compounds name | Models used for pharmacokinetic and toxicity studies | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BBB | HIA | C-2P | AMEST | Carcinogen | AOT | RAT(LD50, mol/kg) | THPT(pIGC50, μg/l) | |||||||
| R | P | R | P | R | P | R | P | R | P | R | P | |||
| 7-deacetoxy-7-oxogedunin | + | 0.86 | + | 1.00 | + | 0.5 | - | 0.85 | - | 0.92 | III | 0.41 | 2.98 | 1.13 |
| 17-hydroxyazadiradione | + | 0.85 | + | 1.00 | - | 0.62 | - | 0.81 | - | 0.94 | I | 0.45 | 3.67 | 1.25 |
| Nimolicinol | + | 0.8 | + | 0.99 | - | 0.67 | - | 0.78 | - | 0.95 | I | 0.61 | 4.20 | 1.22 |
| 6-acetynimbadiol | + | 0.74 | + | 0.98 | - | 0.52 | - | 0.88 | - | 0.91 | III | 0.57 | 3.47 | 1.14 |
| Andirobin | + | 0.67 | + | 0.98 | - | 0.54 | - | 0.76 | - | 0.93 | I | 0.34 | 3.55 | 1.24 |
| Gedunin | + | 0.85 | + | 1.00 | - | 0.56 | - | 0.85 | - | 0.92 | III | 0.37 | 3.07 | 1.27 |
THPT is tetrahymena pyriformis toxicity (pIG50, μg/l) and R is results, P is probability, + is positive and – is negative.
Table 3: Pharmacokinetic Parameters and Toxicity Prediction of Selected Molecules
All the selected molecules were found to be substrates and inhibitors of P-glycoprotein (p-gp). The compounds were found to be non-substrate for isoenzymes CYP2C9 and CYP2D6. However, all the molecules were a substrate for CYP3A4. All the bioactive molecules were noninhibitors for CYP2C9, CYP2D6, and CYP2C19 isoenzymes. The selected biomolecules for docking studies were also non-inhibitors for the isoenzyme CYP1A2 except 17-hydroxyazadiradione. Similarly, CYP3A4 isoenzyme was inhibited by all the molecules tested except 17-hydroxyazadiradione. All our bioactives were also found to be non-inhibitor for renal organic cation transporter (Table 4).
| Substrate and isoenzymes | Compounds name | ||||||
|---|---|---|---|---|---|---|---|
| 7-deacetoxy-7-oxogedunin | 17-hydroxyazadiradione | Nimolicinol | 6-acetynimbadiol | Andirobin | Gedunin | ||
| P-gp Substrate | R | S | S | S | S | S | S |
| P | 0.65 | 0.74 | 0.74 | 0.66 | 0.76 | 0.69 | |
| P-gp Inhibitor I | R | I | I | I | I | I | I |
| P | 0.84 | 0.88 | 0.83 | 0.90 | 0.95 | 0.93 | |
| P-gp Inhibitor II | R | I | I | I | I | I | I |
| P | 0.88 | 0.96 | 0.93 | 0.96 | 0.98 | 0.96 | |
| CYP2C9 Substrate | R | NS | NS | NS | NS | NS | NS |
| P | 0.76 | 0.79 | 0.82 | 0.81 | 0.85 | 0.77 | |
| CYP2D6 Substrate | R | NS | NS | NS | NS | NS | NS |
| P | 0.80 | 0.90 | 0.9 | 0.90 | 0.86 | 0.83 | |
| CYP3A4 Substrate | R | S | S | S | S | S | S |
| P | 0.71 | 0.76 | 0.73 | 0.65 | 0.75 | 0.72 | |
| CYP1A2Inhibitor | R | NI | I | NI | NI | NI | NI |
| P | 0.77 | 0.55 | 0.66 | 0.89 | 0.74 | 0.81 | |
| CYP2C9 Inhibitor | R | NI | NI | NI | NI | NI | NI |
| P | 0.85 | 0.55 | 0.65 | 0.77 | 0.75 | 0.83 | |
| CYP2D6 Inhibitor | R | NI | NI | NI | NI | NI | NI |
| P | 0.93 | 0.91 | 0.93 | 0.94 | 0.91 | 0.93 | |
| CYP2C19Inhibitor | R | NI | NI | NI | NI | NI | NI |
| P | 0.8 | 0.63 | 0.78 | 0.86 | 0.7 | 0.76 | |
| CYP3A4 Inhibitor | R | I | NI | I | I | I | I |
| P | 0.8 | 0.51 | 0.5 | 0.58 | 0.88 | 0.8 | |
| ROCT | R | NI | NI | NI | NI | NI | NI |
| P | 0.85 | 0.8 | 0.8 | 0.84 | 0.77 | 0.84 | |
ROCT is renal organic cation transporter, R is results, P is probability, S is substrate, NS is non-substrate, I is inhibitor and NI is non-inhibitor
Table 4: Interaction of the Drug Molecules with Various Enzymes
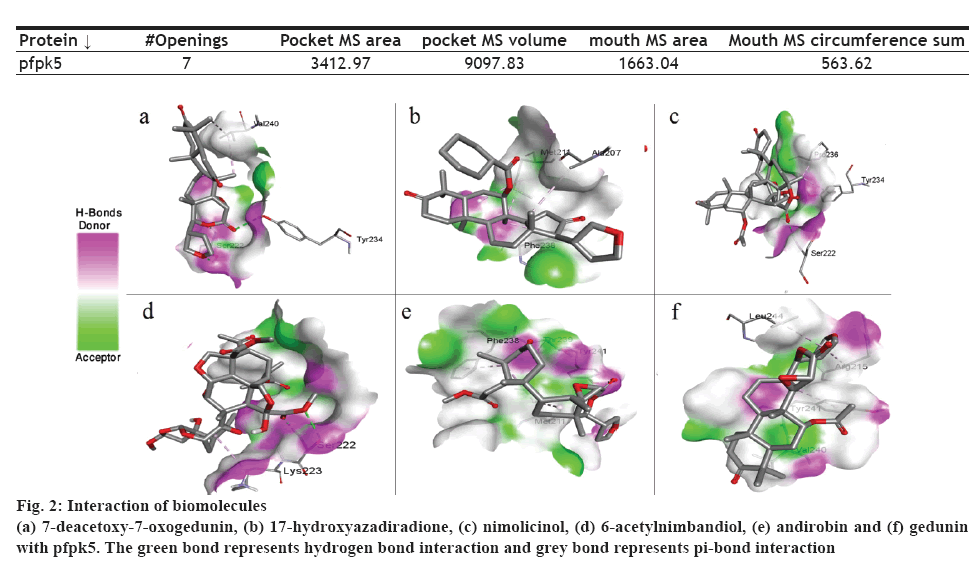
The presence of a binding site on a protein plays an important role in determining binding affinity of the drug molecule. Protein pfpk5 had a total 7 mouth openings, mouth MS area was 1663.04, pocket MS area was 3412.97 and pocket MS volume was 9097.83 (Table 5). The selected target pfpk5 well known as Plasmodium falciparum protein kinase 5 has been reported to activate or maintain the S-phase of the parasite [6]. The predicted parameters for the proteins showed the chosen protein (PDB ID: 1OB3) was suitable for docking study due to the completeness of the domain and low-resolution value (Table 5).
| Protein ↓ | #Openings | Pocket MS area | pocket MS volume | mouth MS area | Mouth MS circumference sum |
|---|---|---|---|---|---|
| pfpk5 | 7 | 3412.97 | 9097.83 | 1663.04 | 563.62 |
Table 5: Binding Cavity Analyses
In this context, PyRx 0.8 was used for the docking study. All the selected molecules were able to bind to pfpk5. This binding affinity represented that all the selected bioactive molecules were able to fit into the binding cavity/pocket of the targeted protein. The bioactive compounds were able to interact with the protein molecule via a hydrogen and pi-bond (figure 2). On assessing the binding affinity (Table 1), 7-deacetoxy-7-oxogedunin and nimolicinol showed better affinity i.e. –5.5 kcal/mol for both compounds to pfpk5 target. Staurosporine, a well-known protein kinase inhibitor was also docked with the target protein under the same setting and the binding energies were found to be comparable.
Lipinski rule of five describes the drug-likeness character to reflect if a compound can be a suitable drug candidate with respect to factors like bioavailability and biodistribution. According to Lipinski rule of five, only two violations are accepted to show the drug-likeness character of the molecule [10]. None of the selected biomolecules violated any rule of five. However, only two biomolecules i.e. 17-hydroxyazadiradione and nimolicinol scored positive drug-likeness score. The highest drug-likeness score obtained was 0.63 by 17-hydroxyazadiradione (Table 2).
Assessment of toxicity in a single cell stage helps to understand the toxic character of drug molecule at the cellular level. Hence E. coli model was utilized to predict the toxicity of biomolecules to act over a single cell stage. E. coli toxicity prediction web server (ABSYNTH) is based on QSAR models and was built by utilizing a clustering algorithm. This helps in finerefinement of heterologous expression leading to the metabolic pathway design [11]. From the result, it was observed that 6-acetylnimbandiol could be the choice of the molecule as an antimicrobial agent due to the least concentration required for the inhibition of the E. coli (Table 2).
Assessment of the pharmacokinetic and pharmacodynamic parameters play important role in drug development steps [12]. AdmetSAR predicts the pharmacokinetic and toxicity parameters of the drug molecule. The database also includes qualitative and quantitative regressive models to estimate the ecological and mammalian ADMET properties for the work of fiction molecule [13]. Since all the active biomolecules scored positive result to cross bloodbrain barrier, the possible pharmacological effects in the central nervous system need to be further studied. Similarly, all active biomolecules scored positive result for human intestinal absorptivity. Hence all the compounds could get absorbed through the gastrointestinal tract. Similarly, all the selected compounds scored negative result for Ames toxicity and carcinogenic character. Hence all the compounds are found to be suitable for human consumption. Among the six compounds, 7-deacetoxy-7- oxogedunin, 6-acetylnimbadiol, and gedunin were under the class III for acute oral toxicity. Similarly, 17-hydroxyazadiradione, nimolicinol and andirobin were under class I. On looking at the toxicity, the highest dose of LD50 for rat acute toxicity was scored by nimolicinol i.e. 4.1965 mol/kg. Similarly, gedunin scored the highest concentration (pIGC50) i.e. 1.2730 μg/l for Tetrahymena pyriformis toxicity (Table 3).
In the present study, all active biomolecules were found to be substrate and inhibitor for p-gp. p-gp, also known as ATP-binding cassette transporter, has a role to exclude toxins and xenobiotics out of cells. In vitro and in vivo studies demonstrate that absorption and disposition of the drug molecules are modulated by p-gp. Drug-drug interaction and pharmacological efficacy of the drug are due to extent of affinity to act as a substrate and inhibit towards p-gp [14]. Though utilization of the p-gp inhibitors has been made for the enhancement of drug absorption for poor intestinal absorbable drugs [15], it may cause the increase in bioavailability of the drugs leading the risk of adverse effect if the individual is under poly-pharmacy [16]. Apart from showing the substrate affinity towards p-gp, the inhibitory action of the drug molecule to inhibit various isoenzymes is also associated with drug interaction [17].
Today, due to the concept of personalized medicine in the pharmacotherapy of many disease conditions[18], it is very essential to assess the affinity of the drug molecules for various isoenzymes which are actively involved in drug metabolism steps. The concept of personalized medicine is based on the test of mutations in genes that encode CYP2D6 and CYP2C19 [19]. All the selected compounds were found to be substrate and inhibitor for p-gp. Hence prediction was made for selected biomolecules could be well metabolized via the enzymes but due to p-gp inhibition, bioactive molecules may get involved in the adverse drug reactions if the patient is under polypharmacy. Since bioactive molecules are p-gp inhibitors, they could be co-administered with the drugs that are having lower bioavailability. Among the selected bioactive, 17-hydroxyazadiradione scored positive result to inhibit CYP1A2. Similarly, all molecules were found to be inhibitors for CYP3A4 except 17-hydroxyazadiradione. Since the molecules are not having an affinity towards CYP2D6 and CYP2C19, the personalized prescription may not be needed (Table 4).
In conclusion, the results of the current study demonstrated that two active molecules i.e. 7-deacetoxy-7-oxogedunin and nimolicinol with pfpk5 exhibited highest binding affinity interaction via hydrogen bonding and pi-interaction. Prediction of the druglikeness character of the molecule has also helped to identify the 17-hydroxyazadiradione giving the good water solubility (LogS) value, which would help in the minimizing the utilization of the solvents for drug formulation. We found none of the compounds selected are a carcinogen and CYP450 isoenzyme non-inhibitors suggesting having lower drug toxicity effects. We expect that limonoids from Azadirachta indica are to be further investigated via wet lab experiments, which would help in the development of pfpk5 inhibitors as an antimalarial agent.
Acknowledgments
The authors thank the KLE College of Pharmacy, KAHER Belagavi for providing necessary facilities for the successful completion of the research work.
Conflict of interest
The authors declare that they have no conflict of interest.
Financial support and sponsorship
Nil.
References
- Prabhu P, Patravale V. Novel targets for malaria therapy. Curr Drug Targets 2011;12(14):2129-43.
- Negi A, Bhandari N, Shyamlal BRK, Chaudhary S. Inverse docking based screening and identification of protein targets for Cassiarin alkaloids against Plasmodium falciparum. Saudi Pharm J 2018;26(4):546-67.
- Bickii J, Njifutie N, Ayafor Foyere J, Basco LK, Ringwald P. In vitro antimalarial activity of limonoids from Khaya grandifoliola C.D.C. (Meliaceae). J Ethnopharmacol 2000;69(1):27-33.
- Alzohairy MA. Therapeutics role of Azadirachta indica (Neem) and their active constituents in diseases prevention and treatment. Evid Based Complement Altern Med 2016;2016:7382506.
- Pereira TB, Rocha LF, Amorim RCN, Melo MRS, Souza RCZ De, Eberlin MN, et al. In vitro and in vivo anti-malarial activity of limonoids isolated from the residual seed biomass from Carapa guianensis (Andiroba) oil production. Malar J 2014;13:317.
- Khalid SA, Farouk A, Geary TG, Jensen JB. Potential antimalarial candidates from African plants: An in vitro approach using Plasmodium falciparum. J Ethnopharmacol 1986;15(2):201-9.
- Priyadarsini RV, Murugan RS, Sripriya P, Karunagaran D, Nagini S. The neem limonoids azadirachtin and nimbolide induce cell cycle arrest and mitochondria-mediated apoptosis in human cervical cancer (HeLa) cells. Free Radic Res 2010;44(6):624-34.
- Tamaoki T, Nomoto H, Takahashi I, Kato Y, Morimoto M, Tomita F. Staurosporine, a potent inhibitor of phospholipid/Ca++dependent protein kinase. Biochem Biophys Res Commun. 1986;135(2):397-402.
- Graeser R, Wernli B, Franklin RM, Kappes B. Plasmodium falciparum protein kinase 5 and the malarial nuclear division cycles. Mol Biochem Parasitol 1996;82(1):37-49.
- Lipinski CA. Lead- and drug-like compounds: The rule-of-five revolution. Drug Discov Today Technol 2004;1(4):337-41.
- Planson AG, Carbonell P, Paillard E, Pollet N, Faulon JL. Compound toxicity screening and structure-activity relationship modeling in Escherichia coli. Biotechnol Bioeng 2012;109(3):846-50.
- Tuntland T, Ethell B, Kosaka T, Blasco F, Zang R, Jain M, et al. Implementation of pharmacokinetic and pharmacodynamic strategies in early research phases of drug discovery and development at novartis institute of biomedical research. Front Pharmacol 2014;5:1-16.
- Cheng F, Li W, Zhou Y, Shen J, Wu Z, Liu G, et al. admetSAR: A comprehensive source and free tool for assessment of chemical ADMET properties. J Chem Inf Model 2012;52(11):3099-105.
- Lin JH, Yamazaki M. Role of P-glycoprotein in pharmacokinetics: Clinical implications. Clin Pharmacokinet 2003;42(1):59-98.
- Amin ML. P-glycoprotein inhibition for optimal drug delivery. Drug Target Insights 2013;2013(7):27-34.
- Finch A, Pillans P. P-glycoprotein and its role in drug-drug interactions. Aust Prescr 2014;37(4):137-39.
- Ogu CC, Maxa JL. Drug interactions due to cytochrome P450. Proc (Bayl Univ Med Cent) 2000;13(4):421-23.
- Redekop WK, Mladsi D. The Faces of Personalized Medicine: A Framework for Understanding Its Meaning and Scope. Value Health 2013;16(6):S4-9.
- Annemans L, Redekop K, Payne K. Current methodological issues in the economic assessment of personalized medicine. Value Health 2013;16(6):S20-6.