- *Corresponding Author:
- H Padh
B. V. Patel Pharmaceutical Education and Research Development (PERD) Centre, Thaltej-Gandhinagar Highway, Thaltej, Ahmedabad-380 054, India
E-mail: perd@perdcentre.com
| Date of Submission | 4 May 2006 |
| Date of Revision | 30 January 2007 |
| Date of Acceptance | 10 March 2007 |
| Indian J Pharm Sci, 2007, 69 (2): 180-189 |
Abstract
Pharmacogenetics has revolutionized the way in which drug metabolism was looked upon in the pre-genomic era. Today with the advent of genomics it is possible to genotype an individual. The genetic differences, which determine the disposition of a given drug in an individual, ultimately give differences in drug response. Genotyping and phenotyping tests to predict dose requirement are now increasingly introduced during preclinical and clinical studies of new drugs. However, the hypothesis for the genotype and the phenotype correlation has to be established. Once this correlation will be established and technology to genotype advances enough to give predictive values over 90%, pharmacogenetics will enter the clinics. Pharmacogenetics will help to tailor the drug type and its dosage according to the individual's genotype. It will help to minimize adverse drug reactions as well as cases of non-responders to the drug therapy. In addition, this will provide better understanding of pharmacogenetic variations both within as well as among major populations/groups of the world.
Keywords
Pharmacogenetics, drugs, metabolism, cytochrome P450, genotype, phenotype
Our understandings of development of diseases have improved from gross anatomical or metabolic symptoms to much finer resolution at the level of cell and molecules. This is in post-genomic era, unlike in pre-genomic era, the diseases were described at a gross pathological level and the molecular mechanisms involved in the development of diseases remained less known leading to the concept of a uniform disease. Similarly patients were also considered homogenous in their response to drugs, leading to adaptation of universal therapeutic strategies. Today the scenario has changed. The 3.2 billion letter nucleotide base-pair sequence of the human genome is now available as a fundamental resource for scientific and medical discovery on many fronts [1,2]. Now the mechanism of a disease and the drug response are beinglooked upon from the molecular etiology of disease development and drug disposition. In post-genomic era, we are in a position to develop molecular pathological description revealing heterogeneity in diseasedevelopment as well as drug disposition.
Recently, concepts like genes and genomics are gaining popularity and their conceptual understanding is allowing us to obtain in-depth knowledge of the cause of disease, drug targeting and drug disposition. Genomics involves the systemic identification of all human genes and gene products, the study of human genetic variations, combined with changes in gene and protein expression over time. Genomics is revolutionizing the study of disease processes along with the development and rational use of the drugs.
Pharmacogenetics
The origin and development of pharmacogenetics is traced to the early hint by Garrod [3], Haldane [4], and later by Williams [5]. The concept was delineated by Motulsky [6] in 1957 and described as pharmacogenetics by Vogel in 1959 [7]. Resemblance of identical twins in drug metabolism as compared with non-identical twins established the importance of genetic inheritance in disposal of many drugs [8]. More recent developments have broadened pharmacogenetic approaches to include novel genomic scale techniques and introduction of the term pharmacogenomics in the 1990’s [9]. The term pharmacogenomics reflects the evolution of pharmacogenetics into the study of the spectrum of genes that determine drug response, including the assessment of the diversity of the human genome sequence and its clinical consequences [10].
Pharmacogenetics is the study of the linkage between an individual’s genotype with individual’s response to a foreign compound. Thus it deals with inherited variations responsible for variable drug effects. It offers the premise of explaining how the individual’s make-up of genes determines drug efficacy and toxicity.
Pharmacogenetics had its beginnings about 40 years ago when researchers realized that some adverse reactions were caused by genetically determined variations in enzyme activity [6,11]. For example, prolonged muscle relaxation after suxamethonium was explained by an inherited deficiency of a plasma cholinesterase, hemolysis and caused by antimalarials was recognized as being associated with inherited variants of glucose-6- phosphate dehydrogenase [12,13]. Similarly, inherited changes in a patient’s ability to acetylate isoniazid were found to be the cause of the peripheral neuropathy, which was due to this drug [14,15].
The relation between dose and blood concentration of pharmacologically active drug can be altered by the differences in metabolism among individuals. Genetic polymorphism of the enzymes involved in the drug metabolism can lead to poor or ultra-rapid metabolizer phenotypes. Thus, genetic variations of polymorphic drug- metabolizing enzymes for the prediction of an individual’s drug response are studied in pharmacogenetics. The knowledge of such variations can prevent adverse reactions or therapeutic failure when applied to dose regimen or drug selection [16,17].
Inefficacy and adverse drug Reactions (adrs)
It is a lesser-known fact that for a drug to be approved by the Food and Drug Administration (US), it has to show efficacy only in one third of the population. It is very much likely that this one third population may belong to one particular group showing good effect of the medication, but the same drug, even after being approvedby the FDA may have adverse or less effect or even maynot have effect at all in rest of the populations. Some very-well known examples of such cases are shown inTable 1 [18].
| Disease | Therapy | % of non-responders |
|---|---|---|
| Various cancers | Various | 70-100% |
| Asthma | Beta-2 agonists | 40-75% |
| Diabetes | Sulfonylureas | 25-50% |
| Depression | Tricyclics, | 20-40%, |
| Selective Serotonin Reuptake Inhibitors (SRRIs) | 25-50% | |
| Duodenal Ulcer | Proton pump inhibitors | 20-90% |
| Hypertension | Thiazides | 50–75% |
| Beta-blockers | 20–30% | |
| Angiotensin converting enzyme (ACE) inhibitors | 10–30% | |
| Angiotensin I antagonists | 10–30% | |
| Osteo/Rheumatoid Arthritis | Nonsteroidal Anti-Inflammatory Drugs (NSAID), | 20-50% |
| Cyclooxygenase-2 (COX-2) inhibitors |
Table 1: Examples Of Poor/Non-Responders Tovarious Therapies [18]
An ideal drug is one that effectively treats or prevents disease in all and has no adverse effect. However, it is very unlikely that a medication is effective and safe in all patients. Therefore, when a physician determines the dose of a drug, it is always a compromise between “not too high” and “not too low” to avoid toxicity and increase efficacy. Dealing with diversity in drug effects is a major problem in clinical medicine and in drug development [10].
While poor/non-responder patients are deprived of the benefits of the therapy, a small fraction of responders maysometimes show adverse drug reactions (ADRs). ADR is one of the major public health problems. Reports indicate that ADRs are responsible for approximately 7% of hospitalization with fatalities occurring in 0.3% of the cases, making ADRs the fourth to sixth leading cause of death in the United States. As a whole, in US each year ADR- the United States. As a whole, in US each year ADR-associated hospitalizations cost from US$1.56 to $4 billion [10,19] Due to ADRs, many drugs have been withdrawn even after being approved by the FDA (Table 2) [20-27]
| Drug | Type of adverse drug reactions |
|---|---|
| Grepafloxacin | Cardiac-related fatalities |
| Cisapride | Cardiac arrhythmias |
| Terfenadine | Cardiac arrhythmias |
| Astemizole | Cardiac arrhythmias |
| Encainide | Proarrhythmias |
| Temafloxacin | Severe hemolytic-uremic syndrome |
| Zomepirac | Hepatotoxicity |
| Bromfenac | Hepatotoxicity |
| Ticrynafen | Hepatotoxicity |
| Benoxaprofen | Hepatotoxicity |
| Rofecoxib | Increased risk of cardiovascular events (including heart attack and stroke) |
Table 2: Drugs Withdrawn From The Market After Fda Approval [20-27]
Genome and Genetic Variability
The study of the genome supports the fundamental unity of human beings throughout the world. We all are 99.9% similar as far as the genome is concerned [1,28]. The remaining 0.1% of the genome leads to inter-individual differences in disease proneness and in drug response. Recently, the role of genetics has been evaluated in drug receptors, such as the β2-adrenoceptor, and drug transporters like the Multi-Drug Resistance gene (MDR1). The genetic variations of these and other drug-metabolizing enzymes or drug targets were highlighted when adverse drug reactions were observed in the individuals of the same families [29-32].
The most common form of DNA variation in the human genome is the Single Nucleotide Polymorphism (SNP) [33,34].SNP is a result of a mutation in which a single nucleotide is substituted by another nucleotide at a given position. SNPs occur once every 300-3,000 base-pairs if one compares the genomes of two unrelated individuals [35,36]. On an average it can be said that one SNP occurs in the sequence of 1000 base-pairs. Thus in the entire genome the estimate of the SNPs would be around 3.2 million in 3.2 billion nucleotides. SNPs are responsible for most of medically important SNPs/traits that are related to the of the predisposition individuals to certain diseases and differential drug response [37].
thePharmacogenetics focuses on SNPs for the simple and practical reason that they are both the most common and the most technically accessible class of genetic variants. For clinical correlation studies in relatively small populations, SNPs that occur at frequencies of greater than 10% are most likely to be useful, but rare SNPs with strong selection component and a more marked effect on phenotype are also equally important. Once a largenumber of these SNPs and their frequencies in different populations are known, they can be used to predict an individual’s genetic “fingerprint” with the probable drug . response. High-density maps of SNPs may allow using these SNPs as markers of drug responses even if the target remains unknown. This will provide a “drug response profile” associated with contributions from multiple genes [10,38,39]. Indeed, there is a rapidly growing effort to identify SNPs that will be useful for identifying patients who are at high risk to experience adverse drug reactions or to determine the best therapeutic approach for a given patient.[38,40] In future, SNPs will play a major role in establishing a genotype related to drug response.
With the advent of pharmacogenetics, now we are at a stage where we can develop a patient risk profile. The ability to predict inter-individual differences in drug efficacy or toxicity will thus be a realistic scenario for the future. Thus, genotyping procedures will play an important role in deciding therapeutic course. This is already a reality in few well-documented cases [10]
Genetic Variability in Pharmacokinetics (pk) and Pharmacodynamics (pd)
The in vivo response to drug treatment is a complex and highly dynamic process, which involves multiple factors. As many as 50 proteins, e.g. carrier proteins, transporters, metabolizing enzymes, receptors and their signal response to a drug. Many genes coding for these proteins contain polymorphisms that alter the activity or the level of expression of the encoded proteins. Thus, the response of a given drug in an individual reflects the interaction of multiple variable genetic factors that cause important variations in drug metabolism, distribution and action on its target [10].
Polymorphism in drug disposition - metabolism
Of all the different processes involved in the disposition of the drug in our body, metabolism is a major pathway for the elimination of drugs (Table 3) [31]. The extent of polymorphism in the genes encoding enzymes that metabolize drugs and other xenobiotics is more compared to other genes. Drug response is better correlated with polymorphism in these genes encoding enzymes. Hence it is important to study the genes responsible for drug metabolism. Several common genetic polymorphisms had been identified on a phenotypic basis prior to the characterization of relevant genes. The DNA samples from individuals of known phenotype after sequence analysis, led to the identification of the polymorphisms responsible for the phenotype, and the development of reliable genotyping assays based mainly on the polymerase chain reaction [10].
| Enzyme | Drugs metabolized by the enzyme |
|---|---|
| N-Acetyltransferase 2 (NAT2) | Isoniazid, Sulfonamides |
| Cytochrome P450 2C9 (CYP2C9) | NSAIDs, Tolbutamide, Phenytoin, Warfarin |
| Cytochrome P450 2C19 (CYP2C19) | Omeprazole, Mephenytoin, Mephobarbital, Propranolol, Proguanil, Phenytoin |
| Cytochrome P450 2D6 (CYP2D6) | Antidepressants, Beta blockers, Debrisoquine, Dextromethorphan, Phenformin |
| Thiopurine methyltransferase (TPMT) | Mercaptopurine, Azathioprine, Thioguanine |
Table 3: Examples of Polymorphic Drug Metabolizing Enzymes [31]
Polymorphism in drug metabolizing enzymes (DMEs) have relatively clear phenotypes, hence genotyping them represents an area of importance in clinical medicine [41,42]. Liver is the main organ for drug metabolism. Liver metabolizing enzymes are mainly found in theendoplasmic reticulum of hepatocytes. As a result of genetic polymorphism these enzymes exhibit distinct phenotypes. Poor metabolism maybe the result of a codominant autosomal trait due to deletions, null or inactivating mutations in drug metabolizing enzymes giving rise to accumulation of the drug. Gene duplication, resulting in autosomal dominant trait, gives rise to ultra- rapid drug metabolism that result in reduced drug levels [43]. Liver metabolizing enzymes can be classfied into two groups: Phase I and phase II drug metabolizing enzymes.
Polymorphism in drug metabolizing enzymes -Phase I
The major modification that takes place by the Phase I enzymes is the change in the functional group, through hydrolysis, oxidation and reduction. Phase I metabolism is mostly carried out by the monooxgenase heme-thiolate protein superfamily, referred to as Cytochrome P450 (CYP). As CYP plays a very vital role in drug metabolism, researchers were prompted to undertake a detail search for polymorphism in the human CYP genes. detail search for polymorphism in the human CYP genes. metabolism of drugs and other xenobiotics has been studied [44]. They are likely to increase incidence of adverse effects or non-response to a wide variety of drugs that include antidepressants, amphetamines, many β-adrenoceptor antagonists [45]. Approximately 80 different forms of CYP have been characterized in human, each of which has distinct catalytic specificity and unique regulation [46]. Because of this diversity, the high frequency of CYP genetic variations may be explained by their redundant or dispensable nature. In human, the major CYP enzymes responsible for the metabolism of widely prescribed drugs are CYP3A4, CYP2D6, CYP2C19, CYP2C9, CYP1A1/2, CYP2B6, CYP2E1 and CYP2C8 [31]. Among these, CYP2D6, CYP2C9, CYP2C19 are highly polymorphic and responsible for most of the ADRs and inefficacy [10,46].
Many of these polymorphisms are functionally significant, often resulting in altered activity or complete absence of enzyme protein. Analysis of functional significance has been facilitated by the fact that well-established systems for expression of CYP are available together with robust assays for enzyme activity. Yet the complete understanding of all the aspects has not been achieved. Today it is possible to genotype an individual for a range of CYP polymorphisms and based on the metabolic capability of these enzymes, individuals are classified as extensive metabolizers (EMs) or poor metabolizers (PMs) for the given CYP. This information is of utility in predicting the metabolic capacity of an individual for a particular drugs metabolism by this CYP [47]
CYP2D6 comprises only of 1-2% of the total liver drug metabolizing enzymes, but is involved in the metabolism of more than 30% drusgs (Table 4) [48,49]. At least 16 different genetic variations including point mutations resulting in early stop codon or amino acid substitution, microsatellite nucleotide repeats, and gene amplifications or deletions are the causes of differences in the enzymatic activity of CYP2D6 resulting in either complete deficiency to ultra-rapid metabolism.
| CYP2D6 | Beta blockers | Antidepressants | Antipsychotics | Miscellaneous | Miscellaneous |
| Carvedilol | Amitriptyline | Haloperidol | Enacinide | Codeine | |
| S-metoprolol | Clomipramine | Perphenazine | Flecainide | Debrisoquine | |
| Propafenone | Desipramine | Risperidone | Perhexiline | Dextromethophan | |
| Timolol | Imipramine | Thioridazine | Propranolol | Phenformin | |
| Paroxetine | Sparteine | Tramadol | |||
| CYP2C9 | NSAIDs | Sulfonylureas | Angiotensin II blockers | Miscellaneous | Miscellaneous |
| Diclofenac | Glyburide | Losartan | Phenytoin | Amitriptyline | |
| Ibuprofen | Glibenclamide | Irbesartan | Rosiglitazone | Celecoxib | |
| Lornoxicam | Glipizide | Tamoxifen | Fluoxetine | ||
| Meloxicam | Glimepiride | Torsemide | Fluvastatin | ||
| S-naproxen | Tolbutamide | S-warfarin | Glyburide | ||
| Piroxicam | Nateglinide | ||||
| Suprofen | |||||
| CYP3A,4,5,7 | Macrolide | HMG CoA Reductase Steroid 6 | Miscellaneous | Miscellaneous | |
| antibiotics | Inhibitors | Beta-OH | |||
| Clarithromycin | Atorvastatin | Estradiol | Alfentanyl | Lidocaine | |
| Erythromycin | Cerivastatin | Hydrocortisone | Aripiprazole | Nateglinide | |
| Telithromycin | Lovastatin | Progesterone | from | Odanestron | |
| Simvastatin | Testosterone | Buspirone | Propranolol | ||
| Cafergot | Quinine | ||||
| Caffeine | Salmeterol | ||||
| Cilostazol | Sildenafil | ||||
| Cocaine | Tamoxifen | ||||
| Dapsone | Taxol | ||||
| Dextromethorphan | Terfenadine | ||||
| CYP2C19 | Proton pump Inhibitors | Anti-epileptics | Miscellaneous | Miscellaneous | Miscellaneous |
| Lansoprazole | Diazepam | Amitriptyline | Indomethacin | Primidone | |
| Omeprazole | Phenytoin | Carisoprodol | R-mephobarbital | Progesterone | |
| Pantoprazole | S-mephenytoin | Citalopram | Moclobemide | Proguanil | |
| Rabeprazole | Phenobarbitone | Hexobarbital | Nelfinavir | Propranolol | |
| Nilutamide | Teniposide | ||||
| R-warfarin |
Table 4: Examples Of The Drugs Metabolized By Cyp2d6, Cyp3a4 And Cyp2c1949
The CYP2D6 poor-hydroxylator phenotype, the so-called sparteine-debrisoquine allele, is caused by homozygous inheritance of inactivating alleles. The frequency of this trait varies extensively among ethnic groups, ranging from 6% in Black Africans to 51% in Asians (Table 5) [13-50].Consequently, clinical responses to standard doses of Consequently, clinical responses to standard doses of chlorpromazine, codeine and antiarrhythmics, vary from increased risk of ADRs at recommended doses in poor increased risk of ADRs at recommended doses in poor metabolizers [10,51].
| Allele | Mutation | Consequence | Allele frequency (%) | ||
|---|---|---|---|---|---|
| Caucasians | Asians | Black Africans | |||
| CYP2D6*2xN | Gene duplication/Multi-duplication | Increased enzyme activity | 1-5 | 0-2 | 2 |
| CYP2D6*3 | 2634A deletion | Frame shift | 2 | 0 | 0 |
| CYP2D6*4 | Defective splicing | Inactive enzyme | 12-21 | 1 | 2 |
| CYP2D6*5 | Gene deletion | No enzyme | 2-7 | 6 | 4 |
| CYP2D6*10 | P34S, S486T | Unstable enzyme | 1-2 | 51 | 6 |
| CYP2D6*17 | T107I, R296C, S486T | Altered affinity for substrates | 0 | 0 | 20-30 |
Table 5: Frequencies of Cyp2d6 Alleles In Different Populations [13,50]
CYP2C9 metabolizes a number of commonly prescribed drugs such as S-warfarin, phenytoin, tolbutamide and a range of nonsteroidal anti-inflammatory drugs (Table 4) [49,52].CYP2C9 which metabolizes the active enantiomer, S- warfarin in the warfarin therapy is associated with hemorrhagic complications. This analysis was based on Caucasian population, studied retrospectively within a system of protocol-driven anticoagulation management. The two most common variant alleles are CYP2C9*2 (R144C) and CYP2C9*3 (I359L) and, among northern Europeans, over 30% of the population has one or two of these alleles, with the overall allele frequency of CYP2C9*2 approximately 0.10 compared with 0.08 for CYP2C9*353-55. The distribution of CYP2C9 alleles varies with ethnicity, but the overall frequency of variant alleles (CYP2C9*2 and CYP2C9*3) seems to be ~30% in the general population [56-59].
CYP3A4 is involved in the metabolism of >50% of all the drugs prescribed today (see Table 4) [49,51]. The inter-individual differences in the pharmacokinetics of these drugs is thought to be related to variation in CYP3A4 enzyme activity [60-62]. The variation may be caused by age and disease-related differences, by drugs inducing or repressing transcription/translation, or by genetic polymorphisms. Although the CYP3A4 gene was initially thought not to be very polymorphic, recent reports have described three genetic variants of this gene: CYP3A4*1B, CYP3A4*2, and CYP3A4*363. The allelic frequency for the CYP3A4*1B allele, which contains an A290G substitution in the promoter region of CYP3A4 was found to be more than 54% in African Americans whereas in American and European Caucasians it was reported to be 4-5%. This mutation was not found in Chinese and Japanese Americans [64-67].
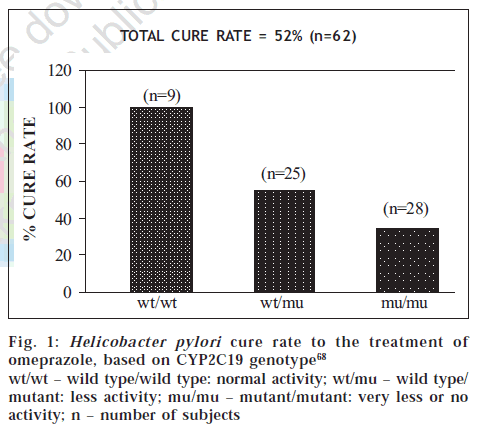
CYP2C19 metabolizes (S)-mephenytoin and other substrates (Table 4) [49]. CYP2C19 contains several inactivating mutations, whose frequencies vary among ethnic populations [13]. Variant alleles from CYP2C19*2 to CYP2C19*8 that are responsible for the PM phenotype have been identified. Variations have been observed both at the phenotypic as well as genotypic levels and it represents wide inter-ethnic differences. For example, polymorphism of CYP2C19 cause differences in cure rate of ulcers caused by Helicobacter pylori (fig. 1) with omeprazole [68]. Five-fold differences have been observed in PMs compared to EMs in the metabolism of the Proton Pump Inhibitors (PPIs) like omeprazole, lansoprazole and pantoprazole. Such polymorphism results in resistance to treatment at a standard dose regimen in nearly 20% of European Caucasians, and in an even higher percentage of Asians [69] .
Figure 1: Helicobacter pylori cure rate to the treatment of omeprazole, based on CYP2C19 genotype [68]
wt/wt – wild type/wild type: normal activity; wt/mu – wild type/ mutant: less activity; mu/mu – mutant/mutant: very less or no activity; n – number of subjects
Polymorphism in drug metabolizing enzymes -Phase II
Phase II conjugation of drugs can occur alone or after phase I metabolism. In order to enhance drug excretion in urine or bile, phase II enzymes link large polar moieties to drug molecules thus rendering drugs water soluble ready for excretion. Several of these proteins have an important role in deciding drug response, such as uridine diphosphate glucuronosyltransferase 1A1 (UGT1A1), N-acetyl transferase 2 (NAT2) and thiopurine S-methyltransferase (TMPT).
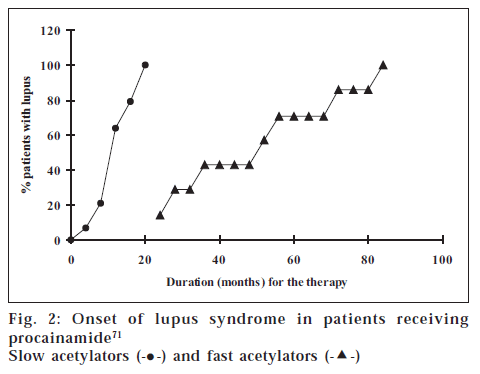
UGT1A1 detoxifies various lipophilic chemicals and endogenous substances including bilirubin. This gene contains polymorphism in a promoter region,which alters the level of expression of the encoded enzyme, resulting in a wide variation of drug metabolism [70]. The homozygous mutation in inactivated NAT2 results in a slow-inactivator phenotype. This phenotype is responsible for increased dose-dependent toxicity due to the accumulation of drugs such as in hydralazine-induced lupus, isoniazid-induced neuropathies, and sulfonamide-induced hypersensitivity reactions in some ethnic groups PATIENTS WITH INFLAMMATORY BOWEL DISEASE72 Side effects acetylators (fig. 2, Table 6) [31,71,72]. Polymorphic variation of TMPT can reduce biotransformation of thiopurine drugs, such as azathioprine and 6-mercaptopurine, and can lead to potentially fatal hematopoietic toxicity in some patients [73].
| Side effects | % Frequency of side effects | |
|---|---|---|
| Slow acetylators | Fast acetylators | |
| Cyanosis | 9 | 1 |
| Hemolysis | 5 | 0 |
| Transient reticulocytosis | 6 | 0 |
Table 6: Adverse Effects To Sulfasalazine Inpatients With Inflammatory Bowel Disease [72]
Figure 2: Onset of lupus syndrome in patients receiving procainamide [71]
Slow acetylators (-•-) and fast acetylators (- -)
-)
Correlation between Phenotype and genotype
The cited examples of both the phase I as well as phase II enzymes lead us to conclusion that with the help of pharmacogenetics, we can predict and classify individuals for their metabolic capabilities. However, there are a few important questions yet to be answered. As discussed earlier in the case of NAT2, adverse effects such as hydralazine-induced lupus, isoniazid-induced neuropathies, and sulfonamide-induced hypersensitivity reactions were observed in some ethnic groups perhaps due to the accumulation of drugs. The question is whether the adverse effects are really due to higher concentration of the drugs in slow acetylators (fig. 2, Table 6) [31,71,72].
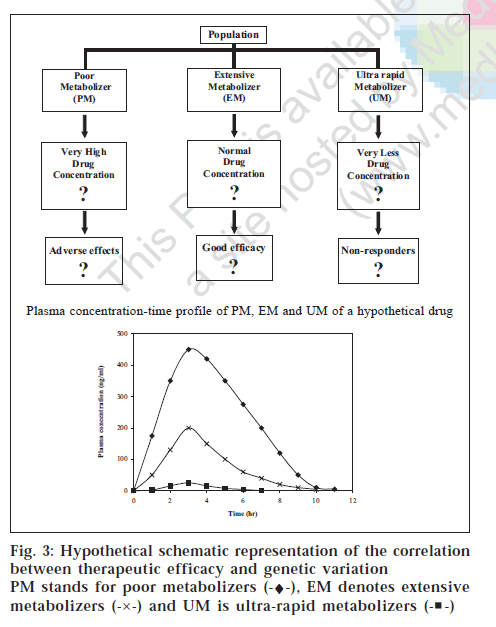
We have earlier seen that extensive metabolizers show poor efficacy, as in case of CYP2C19 genotype and cure rate of ulcers (fig. 1) [68]. Consequently, the question is whether the poor/non-responder phenotype is due to lower concentration of drug in extensive metabolizer. The hypothesis (fig. 3) based on phenotype-genotype data is yet to be carefully scrutinized before its implication can be translated into clinical practice.
Pharmacogenetics In Clinics Reality Test
For application of pharmacogenetics in clinic we need good correlation of genotyping test with therapeutic outcome. In some cases, as discussed earlier, we have good correlation between two and for others we need to await data from clinics which assure us that positive and negative predictive values of such tests are close to 90%. For establishing phenotype-genotype correlation, one requires a vast data set, preferably large prospective studies. Thus before realizing the utility of pharmacogenetics concepts, one needs to test the hypothesis for different drugs as well as for different polymorphisms. The lack of large prospective studies to evaluate the impact of genetic variation on efficacy of drug therapy is one reason for the slow acceptance of these principles.
The example of the CYP2D6 polymorphism again provides incontrovertible clinical data for these concepts. The majority of patients (~90%) require 75-150 mg/day of nortriptyline to reach a “therapeutic” plasma steady-state concentration of 200-600 nmol/l (50-150 μg/l), but poor metabolizer individuals need only 10-20 mg/day, to reach the same levels. Ultra-rapid metabolizers on the other extreme may require 300-500 mg or even >500 mg per day to reach the same plasma concentration [74]. Without knowing about the genotype or phenotype of the patient, poor metabolizers will be overdosed and be at high risk of drug toxicity, whereas ultra rapid metabolizers will be under-dosed and will show inefficacy. Clinical observations have repeatedly confirmed these predictions. Hence it can be concluded that it depends much on the drug in question. Another example is where the efficacy depends on the active metabolite and not the drug (e.g., morphine from codeine). Poor metabolizers will have no drug effect and ultra-rapid metabolizer may have exaggerated drug responses [75]
One would predict that in cases were therapeutic window is narrow, toxicity window is large and overlapping with therapeutic window, time of treatment is critical, and the cost of the treatment is high, pharmacogenetics would have wider application. It is already a reality in few cases/therapeutics and with time will find its rightful place in many more.
At present, phenotyping test methods are established which allow one to classify patients as PMs or EMs, but these require the administration of a specific marker drug or test drug, collection of urine, blood, or saliva for analysis of drug and metabolite concentrations. Moreover, these tests are time consuming, expensive and subject to drug-drug interactions or other influences. Genotyping tests have the advantage of having to be done only once in a lifetime and provides unequivocal genetic information.However, at times phenotype may deviate from genotype for a simple reason that phenotype is manifestation of an enzyme or protein which itself can be subjected to activation, inhibition, induction or suppression. Phenotype is also influenced by other xenobiotics in the system. Prediction is that with better understanding of correlation between phenotype and genotype, the trend will develop for genotyping as test of predictive of phenotype.
It is also perhaps naïve to think that a complex physio/ pharmacologic response of body to a drug can be linked to one gene or a single locus. Multiple loci and other parameters may be responsible for the toxicity or the inefficacy. However, it has been seen that drug metabolism and drug metabolizing enzymes are critical and perhaps sole determinant of drug response, at least in few well-established cases. In such cases the predictive values of the tests is very high in acceptable range (Table 7) [73]. The low predictive values in some cases can be attributed to the fact that metabolism is a multi-step process involving many factors which also have to be understood and incorporated in consideration to achieve better predictive values [76].
| Polymorphic gene/enzyme | PPV* | NPV† | |
|---|---|---|---|
| Antidepressants | (CYP2D6) | 63 | 80 |
| Clozapine | Serotonin | 51 | 73 |
| Isoniazid | N-acetyl transferase | 24 | 94 |
| Leukotriene | Arachidonate 5- | ||
| inhibitor | lipoxygenase (ALOX5) | 91 | 48 |
| Oral contraceptive | Factor V Leiden | 27 | 92 |
| Tacrine | Apolipoprotein E4 | 83 | 60 |
| Warfarin | (CYP2C9) | 16 | 97 |
*PPV-Positive predictive value, †NPV-Negative predictive value
Table 7: Predictive Values For Some of The Genomic Tests [73]
Pharmacogenetics and genomics in general have ethical, legal, financial and behavioral aspects that have not been analyzed. In addition, genotyping technology platforms are relatively new and their ruggedness and reliability will improve with time. Therefore, the promise of improved health care through personalized drug treatment remains a realistic scenario in many areas of medicine [10].
Conclusion
Pharmacogenetics has revolutionized the way in which drug metabolism was looked upon in the pre-genomic era. Today with the advent of genomics it is now possible to genotype an individual for his metabolic capacity for a given drug.
Their influences on the pharmacokinetics or a specific receptor function can now be predicted. Molecular studies in pharmacogenetics started with the initial cloning and characterization of the drug-metabolizing enzyme CYP2D6 [29,77] and now have been extended to numerous human genes, including about 20 drug-metabolizing enzymes, drug receptors and several drug transport systems [31]. Genotyping and phenotyping tests to predict dose requirements are now being increasingly introduced into preclinical studies of drugs and into the clinical routine, e.g., in the choice and initial dose determination of antidepressants [78]. Another important aspect of pharmacogenetics is the realization that all pharmacogenetic variations occur with different frequencies among sub-populations of different ethnic or racial origin. For instance, striking ethnic differences exist in the frequency of slow acetylators of isoniazid due to mutations of NAT2, in poor metabolizers of warfarin due to mutations of CYP2C9, omeprazole due to polymorphism of CYP2C19, and of ultra-rapid metabolizers due to duplication of CYP2D6 gene [29,31,79,80]. Some of the mutations of these genes have a very rare occurrence in other ethnic populations.
Pharmacogenetics will help to tailor the drug dosage according to the individual genotype. It will help in minimizing the adverse drug reactions and will reduce the cases of inefficacies. People will be saved from the unnecessary exposure of drugs and their relative side effects. In future, individual will carry a genetic profile and the physician will demand for such profiles before prescribing the medication [81].
References
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science 2001;291:1304-51.
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al . Initial sequencing and analysis of the human genome. Nature 2001;409:860-921.
- Garrod AE. Inborn factors in disease: An assay. Oxford University Press: 1931.
- Haldane JB. Disease and evolution. La RicercaScientifica 1949;19:68-75.
- Williams RJ. Biochemical Individuality. John Wiley and Sons: New York; 1956.
- Motulsky AG. Drug reactions, enzymes, and biochemical genetics. JAMA 1957;165:835-7.
- Vogel F. Moderneprobleme der humangenetik.Ergebnisse der innerenmedizin und kinderheilkunde 1959;12:65-126.
- Vesell ES. Twin studies in pharmacogenetics. Hum Genet 1978;1:19-30.
- Motulsky A. Pharmacogenetics and ecogenetics to pharmacogenomics. Genetica e BiotechnologienellaMedicina. Available from: http://www.histmed.it/ab-motulsky.htm.
- Licinio J, Wong M. Pharmacogenomics: The search for individualized therapies. Wiley-VCH Verlag GmbH, Weinheim, Germany; 2002.
- Kalow W. Pharmacogenetics. Heredity and the response to drugs. W.B. Saunders Company: Philadelphia, London; 1962.
- Lehmann H, Ryan E. The familial incidence of low pseudocholinesterase level. Lancet 1956;271:124.
- Kalow W, Staron N. On distribution and inheritance of atypical forms of human serum cholinesterase, as indicated by dibucaine numbers. Can J Med Sci 1957;35:1305-20.
- Blum M, Demierre A, Grant DM, Heim M, Meyer UA. Molecular mechanism of slow acetylation in man.ProcNatlAcadSci 1991;88:5237-41.
- Hughes HB, Biehl JP, Jones AP, Schmidt LH. Metabolism of isoniazid in man as related to the occurrence of peripheral neuritis. Am Rev Tuberc 1954;70:266-73.
- Weinstein JN. Pharmacogenomics-Teaching old drugs new tricks. N Engl J Med 2000;343:1408-9.
- Linder MW, Prough RA, Valdes R. Pharmacogenetics: A laboratory tool for optimizing therapeutic efficiency. ClinChem 1997;43:254-66.
- Silber BM. Pharmacogenomics, biomarkers, and the promise of personalized medicine. In :Kalow W, Meyer U, editors. Pharmacogenomics. Marcel Dekker Publishers: New York; 2000.
- Lazarou J, Pomeranz BH, Corey PN. Incidence of adverse drug reactions in hospitalized patients: A meta-analysis of prospective studies. JAMA 1998;279:1200-5.
- US. Food and drug administration. Available from: http://www.fda.gov/medwatch/safety/1998/duract3.htm.
- US. Food and drug administration. Available from: http://www.fda.gov/medwatch/safety/1998/propul.htm.
- Rubinstein E, Camm J. Cardiotoxicity of fluoroquinolones. J AntimicrobChemother 2002;49:593-6.
- COX-2 selective and non-selective non-steroidal anti-inflammatory drugs (NSAIDs). Available from: http://www.fda.gov/cder/drug/infopage/cox2/default.htm.
- Monahan BP, Ferguson CL, Cleave ES, Lloyd BK, Troy J, Cantilena LR. Torsade de pointes occurring in association with terfenadineuse. JAMA 1990;264:2788-90. Back to cited text no. 24
- Rubinstein E. History of quinolones and their side effects. Chemotherapy 2001;47:3-8
- Griffin MR. The COX-2 report: The good, the bad, and the unknown. JCOM 2000;7:62-6.
- Antiarrhythmics. Available from: http://www.aic.cuhk.edu.hk/web8/antiarrhythmics.htm
- Sachidanandam R, Weissman D, Schmidst SC, Kakol JM, Stein LD, Marth G, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 2001;409:928-33.
- Meyer UA, Zanger UM.Molecular mechanisms of genetic polymorphisms of drug metabolism.Annu Rev PharmacolToxicol 1997;37:269-96.
- Nebert DW. Polymorphisms in drug-metabolizing enzymes: What is their clinical relevance and why do they exists? Am J Hum Genet 1997;60:265-71.
- Evans WE, Relling MV. Pharmacogenomics: Translating functional genomics into rational therapeutics. Science 1999;286:487-91.
- Meyer UA. Pharmacogenetics and adverse drug reactions. Lancet 2000;356:1667-71.
- Kuhlmann J. Alternative strategies in drug development: Clinical pharmacological aspects. Int J ClinPharmacolTher 1999;37:575-83.
- Craig AM, Malek M. Market structure and conduct in the pharmaceutical industry. PharmacolTher 1995;66:301-37.
- Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, et al . A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 2001;409:928-33.
- Kruglyak L, Nickerson DA. Variation is spice of life. Nat Genet 2001;27:234-6.
- Stoneking M. Single nucleotide polymorphisms: From the evolutionary past. Nature 2001;409:821-2.
- Roses AD. Pharmacogenetics and the practice of medicine. Nature 2000;405:857-65.
- The SNP Consortium Ltd. Single nucleotide polymorphism for biomedical research. Available from: http://snp.cshl.org.
- Pfost DR, Boyce-Jacino MT, Grant DM. A SNPshot: Pharmacogenetics and the future of drug therapy. Trends Biotechnol 2000;18:334-8.
- Ingelman-Sundberg M. Pharmacokinetics: An opportunity for a safer and more efficient pharmacotherapy. J Intern Med 2001;250:186-200.
- Russell A, Wilke David MR, Jason HM. Combinatorial pharmacogenetics. Nat Rev Drug Discov 2005;4:911-8.
- Gonzalez FJ, Skoda RC, Kimura S, Umeno M, Zangar UM, Nebert DW, et al. Characterization of the common genetic defect in humans deficient in debrisoquine metabolism. Nature 1988;331:442-6.
- Human Cytochrome P450 (CYP) Allele Nomenclature Committee. Available from: http://www.imm.ki.se/CYPalleles.
- Daly AK. Molecular basis of polymorphic drug metabolism. J Mol Med 1995;73:539-53.
- Nelson DR, Koymans L, Kamataki T, Stegeman JJ, Feyereisen R, Waxman DJ, et al. P450 superfamily: Update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics 1996;6:1-42.
- Daly AK. Pharmacogenetics of the major polymorphic metabolizing enzymes.FundamClinPharmacol 2003;17:27-41.
- Shimada T, Yamazaki H, Mimura M, Inui Y, Guengerich FP. Interindividual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals: Studies with liver microsomes of 30 Japanese and 30 Caucasians. J Pharm ExpTher 1994;270:414-23.
- Drug Interactions. Available from: http://medicine.iupui.edu/flockhart.
- Ingelman-Sundberg M. Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): Clinical consequences, evolutionary aspects and functional diversity. Pharmacogenomics J 2005;5:6-13.
- Rendic S, Di Carlo FJ. Human cytochrome P450 enzymes: A status report summarizing their reactions, substrates, inducers, and inhibitors. Drug Metab Rev 1997;29:413-580.
- Lee CR, Goldstein JA, Pieper JA. Cytochrome P450 2C9 genetic polymorphisms: A comprehensive review of the in vitro and human data. Pharmacogenetics 2002;12:251-63. Back to cited text no. 52
- Aithal GP, Day CP, Kesteven PJ, Daly AK. Association of polymorphisms in the cytochrome P450 CYP2C9 with warfarin dose requirement and risk of bleeding complications. Lancet 1999;353:717-9.
- Scordo MG, Aklillu E, Yasar U, Dahl ML, Spina E, Ingelman-Sundberg M. Genetic polymorphism of cytochrome P450 2C9 in a Caucasian and a black African population. Br J ClinPharmacol 2001;52:447-50.
- Taube J, Halsall D, Baglin T. Influence of cytochrome P-450 CYP2C9 polymorphisms on warfarin sensitivity and risk of over-anticoagulation in patients on long-term treatment. Blood 2000;96:1816-9.
- Higashi MK, Veenstra DL, Kondo LM, Wittkowsky AK, Srinouanprachanh SL, Farin FM, et al . Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA 2002;287:1690-8.
- Hillman MA, Wilke RA, Caldwell MD, Berg RL, Glurich I, Burmester JK. Relative impact of covariates in prescribing warfarin according to CYP2C9 genotype.Pharmacogenetics 2004;14:539-47.
- Gage BF, Eby C, Milligan PE, Banet GA, Duncan JR, McLeod HL. Use of pharmacogenetics and clinical factors to predict the maintenance dose of warfarin.ThrombHaemost 2004;91:87-94.
- Daly AK. Pharmacokinetics of the major polymorphic metabolizing enzymes.FundamClinPharmacol 2003;17:27-41.
- De Wildt SN, Kearns GL, Leeder JS, Van Den Anker JN. Cytochrome P450 3A: Ontogeny and drug disposition. ClinPharmacokinet 1999;37:485-505.
- Thummel KE, Shen DD, Podoll TD, Kunze KL, Trager WF, Hartwell PS, et al. Use of midazolam as a human cytochrome P450 3A probe. I. In vitro-in vivo correlations in liver transplant patients. J PharmacolExpTher 1994;271:549-56.
- Lindholm A, Henricsson S, Lind M, Dahlqvist R. Intraindividual variability in the relative systemic availability of cyclosporin after oral dosing. Eur J ClinPharmacol 1988;34:461-4.
- Rebbeck TR, Jaffe JM, Walker AH, Wein AJ, Malkowicz SB. Modification of clinical presentation of prostate tumors by a novel genetic variant in CYP3A4. J Natl Cancer Inst 1998;90:1225-9.
- Sata F, Sapone A, Elizondo G, Stocker P, Miller VP, Zheng W, et al. CYP3A4 allelic variants with amino acid substitutions in exons 7 and 12: Evidence for an allelic variant with altered catalytic activity. ClinPharmacolTher 2000;67:48-56.
- Ball SE, Scatina J, Kao J, Ferron GM, Fruncillo R, Mayer P, et al. Population distribution and effects on drug metabolism of a genetic variant in the 5' promoter region of CYP3A4. ClinPharmacolTher 1999;66:288-94.
- Westlind A, L φfberg L, Tindberg N, Andersson TB, Ingelman-Sundberg M. Interindividual differences in hepatic expression of CYP3A4: Relationship to genetic polymorphism in the 5'-upstream regulatory region. BiochemBiophys Res Commun 1999;259:201-5.
- Van Schaik RH, De Wildt SN, Van Iperen NM, Uitterlinden AG, Van Den Anker JN, Lindemans J. CYP3A4-V polymorphism detection by PCR-restriction fragment length polymorphism analysis and its allelic frequency among 199 Dutch Caucasians. ClinChem 2000;46:1834-6.
- Furuta T, Ohashi K, Kamata T, Takashima M, Kosuge K, Kawasaki T, et al . Effect of genetic differences in omeprazole metabolism on cure rates for Helicobacter pylori infection and peptic ulcer. Ann Int Med 1998;129:1027-30.
- Andersson T, Regardh CG, Dahl-Puustinen ML, Bertilsson L. Slow omeprazole metabolizers are also poor S-mephenytoinhydroxylators. The Drug Monit 1990;12:415-6.
- Mackenzie PI, Owens IS, Burchell B, Bock KW, Bairoch A, Belanger A, et al. The UDP glycosyltransferase gene super-family: Recommended nomenclature update based on evolutionary divergence. Pharmacogenetics 1997;7:255-69.
- Woosley RL, Drayer DE, Reidenberg MM, Nies AS, Carr K, Oates JA.Effect of acetylator phenotype on the rate at which procainamide induces antinuclear antibodies and the lupus syndrome. N Engl J Med 1978;298:1157-9.
- Das KM, Eastwood MA, McManus JP, Sircus W. Adverse reactions during salicylazosulfapyridine therapy and the relation with drug metabolism and acetylator phenotype. N Engl J Med 1973;289:491-5.
- Relling MV, Hancock ML, Rivera GK, Sandlund JT, Ribeiro RC, Krynetski EY, et al. Mercaptopurine therapy intolerance and heterozygosity at the thiopurine S-methyltransferase gene locus. J Natl Cancer Inst 1999;91:2001-8.
- Bertilsson L, Dahl ML, Tybring G. Pharmacogenetics of antidepressants: clinical aspects. ActaPsychiatrScand 1997;96:14-21.
- Fagerlund TH, Braaten O. No pain relief from codeine…? An introduction to pharmacogenomics.ActaAnaesthesiolScand 2001;45:140-9.
- Neil AT. Will genetics revolutionize public health? Genetics and public policy studies, Institute of genetic medicine. Available from: http://www.uni-bielefeld.de/ZIF/KG/2003PHG/Holtzman_Freitag.pdf
- Gonzalez FJ, Skoda RC, Kimura S, Umeno M, Zanger UM, Nebert DW, et al. Characterization of the common genetic defect in humans deficient in debrisoquine metabolism. Nature 1988;331:442-6.
- Kirchheiner J, Brosen K, Dahl ML, Gram LF, Kasper S, Roots I, et al. CYP2D6 and CYP2C19 genotype-based dose recommendations for antidepressants: A first step towards subpopulation-specific dosages. ActaPsychiatrScand 2001;104:173-92.
- Ingelman-Sundberg M, Oscarson M, McLellan RA. Polymorphic human cytochrome P450 enzymes: An opportunity for individualized drug treatment. Trends PharmacolSci 1999;20:342-9.
- Shubha Rani, Padh H. Inter-individual variation in pharmacokinetics of proton pump inhibitors in healthy Indian males. Indian J Pharm Sci 2006;68:754-59.
- Padh H. Human genome project and pharmacogenomics: leading towards individualized medication. Indian Drugs 2001;38:160-63.
 -)
-)