- Corresponding Author:
- Supriya Mahajan
Mumbai Education Trust, Near Lilawati Hospital, Bandra, Mumbai-400 052, India
E-mail: supriya_ma2@yahoo.com
| Date of Submission | 04 February 2012 |
| Date of Revision | 12 January 2012 |
| Date of Acceptance | 29 September 2010 |
| Indian J Pharm Sci, 2012, 74 (1): 41-47 |
Abstract
A set of benzophenone derivatives was evaluated for the antimalarial activity against Plasmodium berghei in mice and the mean survival time of mice for all the compounds was determined. The QSAR analysis was carried out for the fourteen benzophenone derivatives using different physicochemical descriptors. The multiple linear regression analysis was used to correlate the physicochemical descriptors with the antimalarial activity of the benzophenone derivatives from the training set and the best QSAR model was developed, which was further used to predict the antimalarial activity of other compounds from the class of benzophenones. To confirm the predictivity of the best QSAR model, a new set (test set) of six compounds was designed, synthesized and evaluated for the antimalarial activity. A good correlation between the experimental and predicted antimalarial activities was obtained for the test set compounds in the validation procedure, indicating the high predictivity of the developed QSAR model. Five benzophenone derivatives, which showed good antimalarial activity, were further studied for their drug-likeliness characteristic and per cent oral absorption using software “QikProp”. It was observed that all the five benzophenone derivatives were found to be good drug candidates and showed good oral absorption.
Keywords
Antimalarial activity, benzophenone derivatives, physicochemical descriptors, QSAR, multiple linear regression analysis
Introduction
Mosquitoes are responsible for spreading some of the dreadful diseases like malaria (Anopheles), yellow fever, dengue (Aedes) and filaria (Culex). More than a million people die every year from mosquito borne diseases and million more are stricken [1]. Out of these four diseases, malaria is a devastating infectious disease affecting a significant portion of world population. Mostly, it is a cause of death and illness in children and adults in tropical countries [2]. Mortality, currently estimated at over a million people per year, has risen in recent years, probably due to increasing resistance to antimalarial medicines [3]. Thus, it is very essential to develop new antimalarials.
Design and development of new drugs is simplified and made more cost-effective because of the advances in the concepts of quantitative structure activity relationships (QSAR) studies. A methodology of QSAR studies is one of the approaches to the rational drug design. The introduction of Hansch model,in early 1960, enabled chemists to describe the structure activity relationships in quantitative terms and check those using statistical methods [4]. QSAR are statistically derived models that can be used to predict the biological activity of untested compounds from their molecular structures [5,6]. This concept helps to understand the role of physicochemical descriptors of compounds in determining their biological activity. Thus, the QSAR studies help in estimating the characteristics of the new and potent compounds, without synthesizing and evaluating their biological activity.
Ketones like exifone, a benzophenone derivative, and rufigallol, an anthraquinone derivative, have been reported to possess antimalarial activity in vitro [7-9]. Hence, it was decided to evaluate antimalarial activity of few more ketones from the benzophenone series and perform QSAR studies to identify the influence of the physicochemical descriptors on their antimalarial activity. Thus, the objective of the present work was to develop various QSAR models by simple and multiple linear regression methods and to use the best QSAR model for designing few more benzophenone derivatives and predicting their antimalarial activity.
Materials and Methods
Synthesis of compounds
All the twenty compounds, in the training and test sets, were synthesized in our laboratory by the general procedure reported by Mahajan et al [10].
Antimalarial activity of benzophenone derivatives: All the twenty compounds in the training and test sets were evaluated for their antimalarial activity using Rane’s test [11]. The institutional animal ethics committee of Haffkine Institute, Mumbai, India, approved the animal experiments.
The antimalarial activity was determined by using albino male mice against a malarial parasite Plasmodium berghei. The animals were infected with an intraperitoneal injection of 0.1 ml of citrated heart blood, containing a minimum of 1×106 parasitized red blood cells, drawn from the donor mice, infected one week earlier with Plasmodium berghei NK–65, procured from the Central Drug Research Institute, Lucknow, India. The test compounds were suspended in distilled water by the addition of few drops of tween-80 and a single dose of 160 mg/ kg was administered subcutaneously, 72 h after the infection. A group of six infected mice, treated with chloroquine, was used as a positive control. A group of six infected but untreated mice was used as a negative control. The mice were observed for forty days. The antimalarial activity of the synthesized compounds is expressed as the Mean survival time of mice (MST). Survival time of mice is the period from the day the mouse is inoculated to the day it is found dead. If the MST of the test compound is double the mean survival time of the negative control (MSTC), then the compound is considered to have good antimalarial activity.
Computational studies
Maestro, the molecular modeling software from Schrödinger Inc., USA, was used to obtain different physicochemical descriptors for the synthesized compounds and also to develop quantitative structure activity relationships models. Maestro provides a graphical user interface for all Schrödinger computational programs like LigPrep,QikProp, Strike, etc. The 3D molecular structures of twenty benzophenones derivatives were initially built in Maestro. The structures of these compounds were then refined using the program LigPrep, which helps to determine different conformers, ionization states, tautomer states and potential energy of molecules.
Determination of physicochemical descriptors
The structure of a molecule is expressed quantitatively in terms of its physicochemical descriptors, which are lipophilic, electronic and steric in nature. The physicochemical descriptors govern the biological activity of the compounds. Physicochemical descriptors like molecular weight, molar volume, dipole moment, electron affinity and ionization potential were obtained using the program QikProp and are summarized in Table 1. For the QSAR studies by multiple linear regression (MLR) analysis method, the descriptors were selected based on the results of the inter-correlation matrix between the descriptors. For the true correlation between the physicochemical descriptors and the antimalarial activity, the descriptors selected for MLR analysis in QSAR, should not be inter-correlated (r2<0.6). The inter-correlation matrix for various descriptors is presented in Table 2.
| Compd. No. | Descriptor obtained by usingsoftware “LigPrep” | Descriptors obtained by using software “QikProp” | ||||||
|---|---|---|---|---|---|---|---|---|
| PE | MW | DM | MV | IP | EA | LogPo/w | ||
| 1 | 71.98 | 212.00 | 5.31 | 760.0 | 9.3 | 0.53 | 2.64 | |
| 2 | 71.19 | 232.66 | 4.13 | 744.2 | 9.4 | 0.77 | 3.14 | |
| 3 | 72.72 | 230.69 | 4.00 | 781.4 | 9.5 | 0.74 | 3.72 | |
| 4 | 71.96 | 210.27 | 4.42 | 797.3 | 9.6 | 0.54 | 3.53 | |
| 5 | 73.98 | 182.22 | 4.00 | 677.0 | 9.8 | 0.60 | 3.27 | |
| 6 | 88.95 | 212.24 | 3.30 | 751.1 | 9.3 | 0.53 | 2.90 | |
| 7 | 73.57 | 251.11 | 1.95 | 765.6 | 9.6 | 0.90 | 3.94 | |
| 8 | 86.78 | 272.21 | 6.74 | 822.3 | 10.3 | 2.20 | 1.38 | |
| 9 | 85.62 | 212.25 | 5.96 | 732.9 | 7.7 | 0.29 | 1.10 | |
| 10 | 105.76 | 258.31 | 4.13 | 912.9 | 9.1 | 0.77 | 4.48 | |
| 11 | 115.20 | 272.34 | 4.33 | 972.8 | 9.1 | 0.75 | 4.88 | |
| 12 | 104.74 | 292.76 | 3.87 | 957.1 | 9.2 | 0.88 | 5.06 | |
| 13 | 115.20 | 303.31 | 7.00 | 985.4 | 9.4 | 1.67 | 3.80 | |
| 14 | 78.29 | 214.22 | 3.61 | 722.7 | 9.2 | 0.66 | 1.64 | |
PE is the potential energy, MW is the molecular weight, DM is the dipole moment, MV is the molar volume, IP is the ionization potential, EA is the electron affinity, and Log Po/w is the partition coefficient in octanol/water.
Table 1: Physicochemical Descriptors Obtained For The Fourteen Benzophenone Derivatives
| PE | MW | DM | MV | Log Po/w | IP | EA | |
|---|---|---|---|---|---|---|---|
| PE | 1 | ||||||
| MW | 0.8 | 1 | |||||
| DM | 0.3 | 0.2 | 1 | ||||
| MV | 0.9 | 0.9 | 0.2 | 1 | |||
| Log Po/w | 0.5 | 0.5 | -0.4 | 0.67 | 1 | ||
| IP | -0.0 | 0.1 | 0.1 | -0.1 | 0.04 | 1 | |
| EA | 0.3 | 0.6 | 0.4 | 0.35 | -0.11 | 0.62 | 1 |
Table 2: Inter-Correlation Matrix For Different Physicochemical Descriptors
Development of different quantitative structure activity relationships models
The QSAR studies were carried out to correlate physicochemical descriptors of fourteen synthesized benzophenone derivatives from the training set with their antimalarial activity, expressed as Log MST. The physicochemical descriptors were taken as the independent variables and the antimalarial activity was taken as the dependent variable.
Various QSAR models were developed by correlating either one (simple linear regression analysis) or more than one (multiple linear regression analysis) physicochemical descriptors at a time, with antimalarial activity of these fourteen compounds. The correlation was studied using the program Strike from Schrödinger Inc., USA. The best QSAR model (described by Equation 1, under Results and Discussion) was chosen based on the statistically significant values of correlation coefficient (r), square of the correlation coefficient (r2), Fischer’s value of significance (F), leave one out cross validated (q2) value and standard error (s).
Validation of the best QSAR model
In QSAR modeling, it is very important to validate the relevance of the resulting best QSAR model. The internal and external validation was performed to assess the predictivity of the best QSAR model. The internal validation was performed by using the training set compounds. The antimalarial activity of 14 compounds in the training set was predicted using the best QSAR model (Eqn. 1). The experimental antimalarial activity of these 14 compounds was then correlated with their predicted antimalarial activity using the software, Strike.
The most important validation is the external validation, which consists of making predictions for an independent set (test set) of compounds, not used in the training set, from the best QSAR model. For external validation of the best QSAR model (Eqn. 1), six new compounds were designed based on their predicted antimalarial activities. These compounds were further synthesized in our laboratory and tested for their antimalarial activity. The experimental antimalarial activity of these compounds was then correlated with their predicted antimalarial activity using the software, Strike.
Determination of Lipinski’s value and per cent human oral absorption
The Lipinski’s rule of 5 (Lipinski’s value) and the per cent human oral absorption for the five benzophenone derivatives (“Active” compounds from the training and test sets), which were found to posses good antimalarial activity, were predicted using the software, QikProp, Lipinski’s rule of 5 helps in evaluating the drug-likeliness characteristic of a compound, and the per cent human oral absorption (% OA) describes the extent of oral absorption of a compound.
Results and Discussion
The antimalarial activity of all the fourteen compounds (training set) is summarized in Table 3. From the results of the antimalarial studies, it was observed that compounds 1 and 13 were “Active”, as the mean survival time of mice treated with these compounds was more than double the mean survival time of mice in the untreated control group of mice. These compounds were found to possess good antimalarial activity. Compounds 2, 3, 4 and 12 were found to be marginally “Inactive” since the mean survival time of mice treated with these compounds was little lesser than double the mean survival time of the untreated mice. All the other compounds were found to be “Inactive” as the mean survival time of the mice treated with these compounds was less than double the mean survival time of the untreated mice. Mice treated with chloroquine (positive control) were found to be alive throughout the observation period of 40 days.

| Compd. No. | Name of compound | MST (Days) | MSTC (Days) | Remarks | Log MST |
|---|---|---|---|---|---|
| 1 | Benzophenone | 17.0 | 8.0 | Active | 1.2330 |
| 2 | 4-Methylbenzophenone | 15.2 | 8.0 | Inactive | 1.1818 |
| 3 | 4-Methoxybenzophenone | 14.2 | 8.0 | Inactive | 1.1522 |
| 4 | 4-Chlorobenzophenone | 15.4 | 8.0 | Inactive | 1.1875 |
| 5 | 4-Nitrobenzophenone | 13.2 | 8.0 | Inactive | 1.1205 |
| 6 | 4,4′-Dichlorobenzophenone | 13.6 | 8.8 | Inactive | 1.1335 |
| 7 | 4-Methoxy-4′-methylbenzophenone | 13.0 | 8.8 | Inactive | 1.1139 |
| 8 | 4-Phenylbenzophenone | 7.8 | 6.5 | Inactive | 0.8920 |
| 9 | 4-Phenyl-4′-methylbenzophenone | 9.6 | 6.5 | Inactive | 0.9822 |
| 10 | 4-Phenyl-4′-chlorobenzophenone | 5.8 | 6.5 | Inactive | 0.7634 |
| 11 | 4-Phenyl-4′-nitrobenzophenone | 10.0 | 6.5 | Inactive | 1.0000 |
| 12 | 4-Hydroxybenzophenone | 19.0 | 10.0 | Inactive | 1.2787 |
| 13 | 4-Hydroxy-4′-methylbenzophenone | 27.2 | 10.0 | Active | 1.4345 |
| 14 | 3,5-Dihydroxybenzophenone | 16.4 | 10.0 | Inactive | 1.2148 |
| Standard | Chloroquine | All mice were alive for 40 days | |||
MST is the mean survival time of mice for the benzophenone derivatives at the dose of 160 mg/kg body weight of a mouse and MSTC is the mean survival time for the untreated control group of mice.
Table 3: Observed (Experimental) Antimalarial Activity Of The Benzophenone Derivatives In Mice
The observed antimalarial activity of fourteen compounds was correlated with different physicochemical descriptors listed in Table 1, to develop QSAR models. Physicochemical descriptors are expressed as numerical values that encode different structural features of the molecules. The activity data of the standard drug, chloroquine, was not included in the development of QSAR model as it belonged to a different structural series.
The selection of the number of physicochemical descriptors was done based on the ratio of the number of physicochemical descriptors used for the correlation to the number of compounds, which is usually 1: 5. In this perspective, the number of physicochemical descriptors considered initially, should not be higher than one-fifth of the compounds in the training set [5]. Thus, for the development of the best QSAR model for the training set consisting of 14 compounds, the antimalarial activity can be correlated at a time with 3 descriptors. Physiochemical descriptors having high inter-correlation (r2>0.6) were eliminated from the multiple linear regression (MLR) analysis study, as they lead to the false correlation. It can be seen from the correlation matrix that the physicochemical descriptors (potential energy (PE), dipole moment (DM) and electron affinity (EA)), which were used in the development of the best QSAR model in the present work, are independent (r2<0.6) of each other. The values of inter-correlation coefficients for the correlation of PE with DM, PE with EA and DM with EA are shown in Table 2 as bold figures.
For the training set consisting of 14 compounds, the best QSAR model was obtained when potential energy, dipole moment and electron affinity were correlated simultaneously with the antimalarial activity. The best QSAR model is expressed by Eqn. 1.
Log MST = 2.356 – 0.00133 PE + 0.00241 DM– 0.3476 EA. Eqn. 1
(n=14, r=0.9455, r2=0.8941, q2=0.734, s=0.0672, F=18.8)
In Eqn. 1, PE is the potential energy, DM is the dipole moment, EA is the electron affinity, n is the number of compounds, r is the Pearson’s correlation coefficient, r2 is the square of Pearson’s correlation coefficient, q2 is the leave one out cross validated r2 value, s is the standard error and F is the Fischer’s value of significance.
In Eqn. 1, r2 is greater than 0.8, s is small and F-value is higher as compared to the tabulated F-value. Thus, Eqn. 1 was found to be statistically significant. If the value of q2 is greater than 0.6, it indicates the suitability of a model for predicting the antimalarial activity of other compounds from the same series. In equation 1, q2 = 0.734, which was higher compared to the value of q2 obtained for correlation of antimalarial activity with other physicochemical descriptors. Hence, Eqn. 1 was considered to be the best QSAR model.
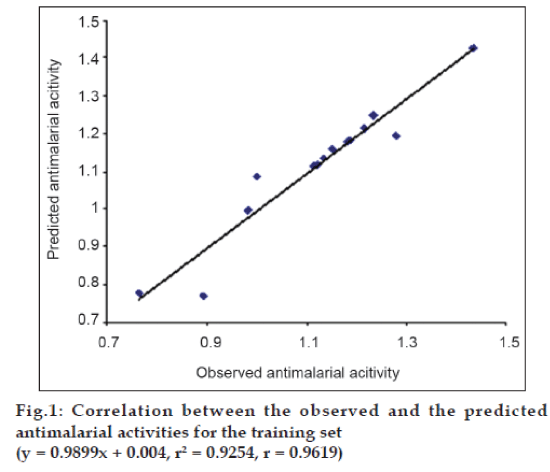
The internal validation of the above mentioned best QSAR model was first performed by predicting Log MST values for all the 14 compounds in the training set, using the software, Strike. The predicted values for the training set were then correlated with the observed (experimentally obtained) Log MST values using the same software. The observed and predicted Log MST values for the training set are summarized in Table 4. Good correlation between the observed and the predicted antimalarial activities was obtained, as the correlation coefficient (r) was 0.9619 and the square of correlation coefficient (r2) was 0.9254. This correlation is represented graphically in fig. 1.
| Compd. No. | Observed | Predicted antimalarial |
|---|---|---|
| antimalarial activity | activity (Log MST) | |
| (Log MST) | ||
| 1 | 1.233 | 1.2478 |
| 2 | 1.1818 | 1.1774 |
| 3 | 1.1522 | 1.1594 |
| 4 | 1.1875 | 1.1820 |
| 5 | 1.1205 | 1.1165 |
| 6 | 1.1335 | 1.1325 |
| 7 | 1.1139 | 1.1132 |
| 8 | 0.892 | 0.7704 |
| 9 | 0.9822 | 0.9960 |
| 10 | 0.7634 | 0.7750 |
| 11 | 1.000 | 1.0845 |
| 12 | 1.2787 | 1.1940 |
| 13 | 1.4345 | 1.4253 |
| 14 | 1.2148 | 1.2123 |
Table 4: Comparison Between The Observed And The Predicted Antimalarial Activities Using The Best Qsar Model For The Trainingset Of Benzophenone Derivatives
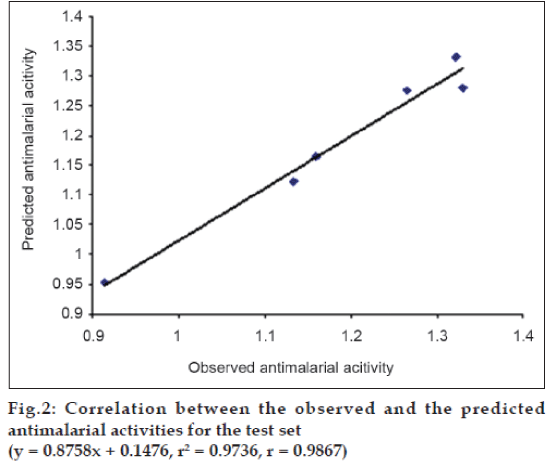
The best QSAR model, expressed by equation 1 was then used to predict the antimalarial activity of the test set compounds. This step was undertaken to perform external validation of the best QSAR model. The test set includes the set of compounds whose structures are similar to the training set compounds and whose biological activity is not evaluated. There should be more than five compounds in the test set. The compounds showing high predicted antimalarial activity were synthesized in our laboratory. The Log MST values for these six compounds were also determined experimentally and it was observed that out of six compounds three compounds showed good antimalarial activity. The observed and the predicted Log MST values for the test set compounds were correlated using the program, Strike. These values are listed in Table 5 and the correlation between them is represented graphically in fig. 2. The higher correlation coefficient value (r= 0.9867 and r2 = 0.9736) for this correlation indicated good predictivity of the best QSAR model (Eqn. 1). Hence, it can be used successfully to predict the antimalarial activity of the other molecules from this class.
| Compd. No. | Name of compound | Remarks | Observed antimalarial activity(Log MST) | Predicted antimalarial activity(Log MST) |
|---|---|---|---|---|
| 15 | 4,4′-Dimethylbenzophenone | Active | 1.2648 | 1.2750 |
| 16 | 4-Methyl-4′-chlorobenzophenone | Active | 1.3222 | 1.3319 |
| 17 | 4-Methoxy-4′-chlorobenzophenone | Inactive | 0.9130 | 0.9514 |
| 18 | 3,5-Dinitrobenzophenone | Inactive | 1.1335 | 1.1223 |
| 19 | 3,5-Diaminobenzophenone | Inactive | 1.1583 | 1.1635 |
| 20 | 4-Hydroxy-4′-chlorobenzophenone | Active | 1.3304 | 1.2789 |
Table 5: Comparison Between The Observed And The Predicted Antimalarial Activities Using The Best Qsar Model For The Test Set Of Benzophenone Derivatives
From the best QSAR model, it is obvious that the potential energy (PE), dipole moment (DM) and electron affinity (EA) in combination, govern the antimalarial activity of the benzophenone derivatives. Overall potential energy of the molecule is important for its reactivity or the stability. The negative value of the coefficient of the potential energy indicates that molecules with lower potential energy would lead to higher antimalarial activity. Dipole moment indicates the polar nature and orientation behavior of a molecule in an electrostatic field. In Eqn. 1, the coefficient of dipole moment is positive. Thus, it shows a positive correlation with the antimalarial activity, i.e. antimalarial activity increases with the increase in the dipole moment of a compound. This also suggests that the higher the polarity of the benzophenone derivatives, the higher is their antimalarial activity. Electron affinity of a molecule gives information about the ability of an atom to accept electrons. There is an inverse relation between the electron affinity and the antimalarial activity of benzophenones, since the coefficient of electron affinity is negative in Eqn. 1. This indicates that the electron donating or electron releasing substituents are desirable for getting higher antimalarial activity from the benzophenone series. Out of twenty synthesized benzophenone derivatives (from both the training set and test set), five compounds showed good antimalarial activity. The compound 1 (benzophenone), compound 13 (4-hydroxy-4′-methylbenzophenone), compound 15 (4, 4′-dimethylbenzophenone), compound 16 (4-methyl-4′-chlorobenzophenone) and compound 20 (4-hydroxy-4′-chlorobenzophenone), which were polar molecules, bearing electron releasing functional groups and having comparatively lower potential energies, were found to posses good antimalarial activity.
Further, when Lipinski’s value and per cent oral absorption were predicted for the five “Active” benzophenone derivatives from both the training set and test set, it was observed that all the five compounds showed Lipinski’s value as zero and hence, they were predicted to exhibit drug-like characteristics. Also, all these compounds showed 100% oral absorption. Hence, these five compounds were found to be suitable drug candidates.
The QSAR studies helped in predicting good antimalarials, since out of fourteen benzophenones from the training set only two were active, whereas in the test set three out of six compounds were found to be active. Thus, the rate of success to obtain newer antimalarials was increased from 14.28 to 50 per cent.
Acknowledgements
Authors deeply acknowledge All India Council of Technical Education (AICTE), New Delhi, India, for providing financial assistance for this project.
References
- Lee B, Singh A, Chiang P, Kemp S, Goldman E, Weinho I, et al.Antimalarial activities of novel synthetic cysteine protease inhibitors.Antimicrob Agents Chemother 2003;47:3810-4.
- World Health Organization, WHO guidelines for the treatment ofmalaria. Geneva: WHO; 2006.
- Geyer JA, Prigge ST, Waters NC. Targeting malaria with specific CDKinhibitors. BiochimBiophysActa 2005;1754:160-70.
- Wolf M, Kubiyini H, editors. Burger’s Medicinal chemistry and drugdiscovery. New York: A Wiley Interscience; 1995.
- Roy PP, Paul S, Mitra I, Roy K. Two novel parameters for validationof predictive QSAR models. Molecules 2009;14:1660-701.
- Konovalov DA, Llewellyn LE, Heyden YV, Coomans DJ. Robustcross-validation of linear regression QSAR models. ChemInf Model2008;48:2081-94.
- Winter RW, Cornell KA, Johnson LL, Isabelle LM, HinnrichsDJ, Riscoe MK. Hydroxy-anthraquinones as antimalarial agents.Bioorg Med ChemLett 1995;5:1927-32.
- Winter RW, Cornell KA, Johnson LL, Ignatushchenko M, Hinrichs DJ,Riscoe MK. Potentiation of the antimalarial agent rufigallol. AntimicrobAgents Chemother 1996;40:1408-11.
- Mahajan SS, Kamath VR, Ghatpande SS. Synergistic antimalarialactivity of ketones with rufigallol and vitamin C. Parasitol2005;131:459-66.
- Mahajan SS, Ghatpande SS. Synthesis of arylketones using EnvirocatEPZG catalyst. Ind J Chem 2005;44B:188-92.
- Osdene TS, Russell PB, Rane L. 2,4,7–Triamino-6-ortho-substitutedaryl pteridines. A new series of potent antimalarial agents. J Med Chem1967;10:431-4.