- *Corresponding Author:
- Z. C. Li
Chemistry and Chemical Engineering School, Henan University of Technology, Zhengzhou 450001, Henan, P. R. China
E-mail: lizhicheng65@sina.com
| Date of Submission | 28 March 2015 |
| Date of Revision | 18 November 2014 |
| Date of Acceptance | 14 October 2013 |
| Indian J Pharm Sci 2015;77(2):196-201 |
Abstract
A series of new palmatine derivatives with alkyl or alkyl with N-heterocyclic structures were designed and synthesized at C-9-O according to the principle of association. These compounds were characterised by 1 H NMR, 13 C NMR, ESI-MS and elemental analysis, and tested for their antimicrobial activity in vitro to evaluate structure-activity relationships. The results indicated that 9-O-substituted palmatine derivatives exhibit varying degrees of antimicrobial activity. Antibacterial activities of compounds (3a-f) against Gram +ve bacteria increased 2- to 64-fold than that of palmatine. The compounds (3a-f) possessed relatively weaker inhibitory effects against Gram −ve bacteria and fungi than that against Gram +ve bacteria. Antimicrobial activities of compounds (5a-e) are lower than that of compounds (3a-f). Compound 3d showed the highest antimicrobial activity of all the compounds. The LD 50 values of compounds (3a-f) decreased as the alkyl side chain was elongated. Compound 3f showed least toxicity.
Keywords
9-O-substituted palmatine derivatives, antimicrobial activity, structure-activity relationships, synthesis, toxicity
Palmatine is one of the active constituents of Coptis chinensis Franch (Ranunculaceae), a traditional Chinese herbal medicine [1]. Palmatine, a member of quaternary protoberberine alkaloid (QPA) [2], exhibits a wide variety of pharmacological and biological activities such as antimicrobial [3], hypoglycemic [4], antiarrhythmic [5] and antioxidant [6] activities. And it has been used to treating various inflammatory diseases, such as surgical infection, bacillary dysentery, enteritis, gastritis, respiratory tract and urinary tract infection, pink eye and gynecological inflammation [7]. Efforts to improve the biological activity of palmatine have led to the development of derivatives by appropriate structural modification.
Substituted derivatives of QPA in the A, C or D ring exhibit changes in their pharmacological effects. Iwasa et al [8]. reported that dioxymethylene replacement at the C-2 and C-3 positions in the A ring, as well as 13-alkyl- or 8-alkyl-substitution [9,10] increased the antibacterial activity, but 13-hydroxysubstitued derivatives decreased the antibacterial activity [11]. Park et al. reported that benzyl introduced to C-13 of berberine and berberrubine increased the antifungal activities [12]. The antimicrobial activity of 3-alkoxyjatrorrhizine [13] or 8-alkylpalmatine [14] derivatives increased with the aliphatic chain elongation and then decreased gradually when the alkyl chain exceeds eight carbon atoms. Wang et al. [15] reported that 3-octyloxy-8-alkyljatrorrhizine derivatives displayed high antimicrobial activities, especially against Gram +ve bacteria.
In this study, we designed and synthesized a series of the new compounds mostly starting from the archetypal palmatine by introducing alkyl or alkyl with N-heterocyclic structures at C-9-O. Their antimicrobial activity and toxicity were tested in vitro to evaluate their structure-activity relationships.
Materials and Methods
Palmatine was procured from JieXiang Herb Material Co. Ltd., Sichuan, China. Alkyl bromides were prepared in the laboratory. Dibromoalkanes and substituent amines were obtained from Aladdin Reagents Co. Ltd., Shanghai, China. All other reagents and solvents used were of analytical grade purchased from Kermel Chemical Reagents Co. Ltd., Tianjin, China. The melting points were detected by microscope melting point apparatus and are uncorrected. 1H and 13C NMR spectra were recorded at 400 MHz on a Bruker-400 spectrometer using TMS as an internal standard and CD3OD or DMSO-d6 as solvent. MS (ESI) spectra were obtained using an Agilent LC-MS 6310 EV instrument. TLC analysis was used to confirm the purity of the compounds, which was performed on silica gel-GF254 thin layers. Elemental analyses were carried out with a Flash EA 1112 elemental analyser.
Five pathogenic microorganisms used in this investigation for the antibacterial assay were kindly provided by Biological Engineering School, Henan University of Technology. These were Gram +ve bacteria, Bacillus subtilis and Staphylococcus aureus, Gram –ve bacteria, Proteus mirabilis and Escherichia coli, and a fungus Candida albicans. The bacteria were grown in beef-extract peptone medium. Healthy Kunming mice of both genders (20±2 g, 7 weeks old) were purchased from the Laboratory Animal Centre of Zhengzhou University.
Synthesis of palmatrubine
Palmatine (1, 1 g) was heated at 200-220° in a dry oven under vacuum (20-30 mmHg) for 20 min. [8] The crude product was chromatographed on a silica gel column, eluted with CH2Cl2/MeOH (8/1, v/v) to give a solid 2 with an yield of 69%; reddish brown solid; mp: 276-278°. 1H NMR (CD3OD, 400 MHz) δ: 9.43 (1H, s), 8.31 (1H, s), 7.69 (1H, d, J=8.48 Hz), 7.47 (1H, s), 7.24 (1H, d, J=8.48 Hz), 6.94 (1H, s), 4.72 (2H, t, J=6.06 Hz), 3.94 (3H, s), 3.91 (3H, s), 3.88 (3H, s), 3.19 (2H, t, J=6.08 Hz). MS (m/z, %): 338.2 (M+, 100). Elemental analysis: Found: C, 64.34%; H, 5.41%; N, 3.72%; calcd for C20H20ClNO4: C, 64.26%; H, 5.39%; N, 3.75%.
Synthesis of 9-O-alkylpalmatines
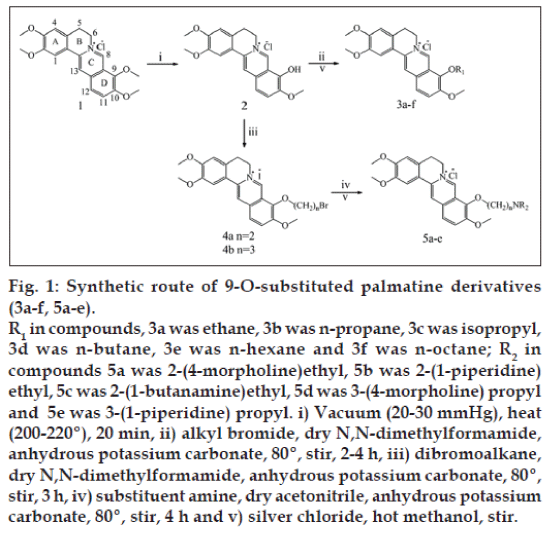
The alkyl bromide (1 ml) was added to a magnetically stirred solution of 2 (1 mmol) and anhydrous K2CO3 (2 mmol) in dry DMF (18 ml) [11]. The reaction mixture was heated at 80° for 2-4 h and the reaction was monitored by TLC. After cooling the solvent was evaporated under reduced pressure, the obtained crude product was chromatographed on a silica gel column, eluted with CH2Cl2/MeOH (30:1, v/v) to give 3a-f bromides. The bromides (3a-f) were treated with AgCl in hot MeOH to convert it to corresponding chlorides (fig. 1).
9-O-ethylpalmatine (3a) was obtained in an yield of 79%; light yellow solid; mp: 225-227°. 1H NMR (DMSO-d6, 400 MHz) δ: 9.81 (1H, s), 9.02 (1H, s), 8.22 (1H, d, J=9.16 Hz), 8.03 (1H, d, J=9.08 Hz), 7.71 (1H, s), 7.10 (1H, s), 4.98 (2H, d, J=5.96 Hz), 4.34-4.39 (2H, m), 4.06 (3H, s), 3.94 (3H, s), 3.87 (3H, s), 3.24 (2H, t, J=5.96 Hz), 1.47 (3H, t, J=7.0 Hz). 13C NMR (DMSO-d6, 100 MHz) δ: 151.96, 150.85, 149.22, 145.86, 143.04, 138.12, 133.62, 129.10, 127.11, 123.76, 122.31, 120.39, 119.43, 111.78, 109.26, 70.43, 57.51, 56.70, 56.36, 55.93, 26.47, 15.88. MS (m/z, %): 366.2 (M+, 100). Elemental analysis: Found: C, 65.79%; H, 6.05%; N, 3.51%; calcd for C22H24ClNO4: C, 65.75%; H, 6.02%; N, 3.49%.
9-O-propylpalmatine (3b) was obtained in an yield of 82%; light yellow solid; mp: 226-228°. 1H NMR (DMSO-d6, 400 MHz) δ: 9.74 (1H, s), 9.03 (1H, s), 8.20 (1H, d, J=9.12 Hz), 8.02 (1H, d, J=9.08 Hz), 7.70 (1H, s), 7.08 (1H, s), 4.97 (2H, t, J=6.08 Hz), 4.26 (2H, t, J=6.78 Hz), 4.04 (3H, s), 3.92 (3H, s), 3.86 (3H, s), 3.23 (2H, t, J=6.06 Hz), 1.85-1.91 (2H, m), 1.05 (3H, t, J=7.38 Hz). 13C NMR (DMSO-d6, 100 MHz) δ: 152.12, 150.67, 149.34, 145.57, 143.46, 138.09, 133.81, 129.05, 127.31, 123.69, 122.11, 120.50, 119.43, 111.95, 109.63, 76.36, 57.65, 56.95, 56.44, 56.12, 26.54, 23.38, 10.71. MS (m/z, %): 380.3 (M+, 100). Elemental analysis: Found: C, 66.45%; H, 6.31%; N, 3.39%; calcd for C23H26ClNO4: C, 66.42%; H, 6.30%; N, 3.37%.
9-O-isopropylpalmatine (3c) was obtained in an yield of 81%; light yellow solid; mp: 228-230°. 1H NMR (DMSO-d6, 400 MHz) δ : 9.73 (1H, s) 9.07 (1H, s), 8.19 (1H, d, J=9.08 Hz), 8.03 (1H, d, J=9.04 Hz), 7.71 (1H, s), 7.08 (1H, s), 4.90-5.00 (3H, m), 4.03 (3H, s), 3.92 (3H, s), 3.85 (3H, s), 3.23 (2H, t, J=5.72 Hz), 1.36 (6H, t, J=3.04 Hz). 13C NMR (DMSO-d6, 100 MHz) δ: 152.13, 150.86, 149.35, 145.70, 142.10, 138.13, 133.81, 129.14, 127.12, 123.51, 123.10, 120.47, 119.47, 111.95, 109.60, 76.66, 57.56, 56.87, 56.43, 56.09, 26.55, 22.64, 16.58. MS (m/z, %): 380.2 (M+, 100). Elemental analysis: Found: C, 66.43%; H, 6.32%; N, 3.40%; calcd for C23H26ClNO4: C, 66.42%; H, 6.30%; N, 3.37%.
9-O-butylpalmatine (3d) was obtained in an yield of 84%; light yellow solid; mp: 225-227°. 1H NMR (DMSO-d6, 400 MHz) δ: 9.68 (1H, s), 8.96 (1H, s), 8.18 (1H, d, J=9.12 Hz), 8.01 (1H, d, J=9.08 Hz), 7.71 (1H, s), 7.08 (1H, s), 4.97 (2H, t, J=6.36 Hz), 4.34 (2H, t, J=6.74 Hz), 4.05 (3H, s), 3.94 (3H, s), 3.88 (3H, s), 3.25 (2H, t, J=6.32 Hz), 1.83-1.90 (2H, m), 1.48-1.57 (2H, m), 1.01 (3H, t, J=7.4 Hz). 13C NMR (DMSO-d6, 100 MHz) δ: 151.87, 150.67, 149.14, 145.60, 143.25, 137.99, 133.64, 128.94, 127.00, 123.73, 122.02, 120.48, 119.36, 111.71, 109.23, 74.42, 57.49, 56.77, 56.33, 55.99, 32.06, 26.46, 19.05, 14.26. MS (m/z, %): 394.4 (M+, 100). Elemental analysis: Found: C, 67.07%; H, 6.57%; N, 3.29%; calcd for C24H28ClNO4: C, 67.05%; H, 6.56%; N, 3.26%.
9-O-hexylpalmatine (3e) was obtained in an yield of 85%; light yellow solid; mp: 213-215°. 1H NMR (DMSO-d6, 400 MHz) δ: 9.73 (1H, s), 9.11 (1H, s), 8.18 (1H, d, J=9.12 Hz), 8.05 (1H, d, J=9.04 Hz), 7.70 (1H, s), 7.07 (1H, s), 4.98 (2H, t, J=5.64 Hz), 4.28 (2H, t, J=6.74 Hz), 4.03 (3H, s), 3.93 (3H, s), 3.85 (3H, s), 3.23 (2H, t, J=5.64 Hz), 1.83-1.90 (2H, m), 1.45-1.50 (2H, m), 1.29-1.38 (4H, m), 0.84-0.90 (3H, m). 13C NMR (DMSO-d6, 100 MHz) δ: 151.95, 150.72, 149.21, 145.71, 143.32, 138.08, 133.67, 129.04, 127.13, 123.71, 122.08, 120.47, 119.42, 111.77, 109.30, 74.75, 57.52, 56.77, 56.36, 56.00, 31.57, 29.97, 26.48, 25.44, 22.59, 14.43. MS (m/z, %): 422.3 (M+, 100). Elemental analysis: Found: C, 68.20%; H, 7.05%; N, 3.08%; calcd for C26H32ClNO4: C, 68.18%; H, 7.04%; N, 3.06%.
9-O-octylpalmatine (3f) was obtained in an yield of 86%; light yellow solid; mp: 210-212°. 1H NMR (DMSO-d6, 400 MHz) δ: 9.75 (1H, s), 9.09 (1H, s), 8.21 (1H, d, J=9.12 Hz), 8.05 (1H, d, J=9.08 Hz), 7.72 (1H, s), 7.10 (1H, s), 4.99 (2H, t, J=5.72 Hz), 4.30 (2H, t, J=6.72 Hz), 4.05 (3H, s), 3.94 (3H, s), 3.87 (3H, s), 3.25 (2H, t, J=5.78 Hz), 1.84-1.91 (2H, m), 1.44-1.50 (2H, m), 1.28-1.35 (8H, m), 0.87(3H, d, J=6.8 Hz). 13C NMR (DMSO-d6, 100 MHz) δ: 151.97, 150.75, 149.22, 145.77, 143.32, 138.13, 133.65, 129.10, 127.18, 123.69, 122.10, 120.43, 119.43, 111.79, 109.27, 74.73, 57.52, 56.71, 56.36, 55.99, 31.77, 29.99, 29.31, 29.20, 26.48, 25.78, 22.60, 14.47. MS (m/z, %): 450.3 (M+, 100). Elemental analysis: Found: C, 69.21%; H, 7.48%; N, 2.89%; calcd for C28H36ClNO4: C, 69.19%; H, 7.47%; N, 2.88%.
Synthesis of 9-O-bromoalkylpalmatines (4a-b)
A solution of 2 (5 mmol), anhydrous K2CO3 (10 mmol) and dibromoalkane (5 ml) in dry DMF (40 ml) was heated at 80° for 3 h and then Et2O was added. The resulting solid was filtered. The crude product was chromatographed on a silica gel column, eluted with CH2Cl2/MeOH (30/1, v/v) to give 4a-b.
9-O-2-bromoethylpalmatine (4a) was obtained in an yield of 61%; light yellow solid; mp: 218-220°. 1H NMR (DMSO-d6, 400 MHz) δ: 9.84 (1H, s), 9.03 (1H, s), 8.23 (1H, d, J=9.04 Hz), 8.06 (1H, d, J=9.12 Hz), 7.71 (1H, s), 7.10 (1H, s), 4.96 (2H, t, J=6.12 Hz), 4.63 (2H, t, J=5.68 Hz), 4.07 (3H, s), 3.98 (2H, t, J=5.7 Hz), 3.94 (3H, s), 3.87 (3H, s), 3.25 (2H, t, J=6 Hz).
9-O-3-bromopropylpalmatine (4b) was obtained in an yield of 67%; light yellow solid; mp: 223 -225°. 1H NMR (DMSO-d6, 400 MHz) δ: 9.80 (1H, s), 9.03 (1H, s), 8.23 (1H, d, J=9.2 Hz), 8.04 (1H, d, J=9.12 Hz), 7.71 (1H, s), 7.10 (1H, s), 4.97 (2H, t, J=6.2 Hz), 4.43 (2H, t, J=6.06 Hz), 4.07 (3H, s), 3.94 (3H, s), 3.87 (3H, s), 3.85 (2H, t, J=6.58 Hz), 3.25 (2H, t, J=6.18 Hz), 2.39-2.45 (2H, m).
Synthesis of 9-O-aminoalkylpalmatines
The substituent amine (1 mmol) was added to a magnetically stirred solution of 4a or 4b (1 mmol) and anhydrous K2CO3 (2 mmol) in dry acetonitrile (15 ml). The reaction mixture was heated at 80° for 4 h and the reaction was monitored by TLC. The resulting solid was filtered at room temperature. The crude product was chromatographed on an Al2O3 column, eluted with CHCl3/MeOH (9:1, v/v) to give the compounds 5a-e bromides. The bromides (5a-e) were treated with AgCl in hot MeOH to convert it to corresponding chlorides.
9-O-2-(4-morpholine)ethylpalmatine (5a) was obtained in an yield of 25%; light yellow solid; mp: 225-227°. 1H NMR (DMSO-d6, 400 MHz) δ: 9.88 (1H, s), 9.06 (1H, s), 8.21 (1H, d, J=9.16 Hz), 8.04 (1H, d, J=9.12 Hz), 7.73 (1H, s), 7.11 (1H, s), 4.98 (2H, t, J=6.04 Hz), 4.44 (2H, t, J=5.32 Hz), 4.06 (3H, s), 3.94 (3H, s), 3.87 (3H, s), 3.42 (4H, s), 3.25 (2H, t, J=6.82 Hz), 2.76 (2H, t, J=5.3 Hz), 2.39 (4H, s). 13C NMR (DMSO-d6, 100 MHz) δ: 152.21, 150.71, 149.43, 146.10, 143.45, 138.11, 133.66, 129.08, 127.23, 123.74, 122.54, 120.41, 119.52, 112.04, 109.68, 70.63, 70.32, 66.73, 58.18, 57.62, 56.87, 56.47, 56.19, 53.67, 26.62. MS (m/z, %): 450.4 (M+, 100). Elemental analysis: Found: C, 66.75%; H, 6.65%; N, 7.30%; calcd for C27H32ClNO5: C, 66.73%; H, 6.64%; N, 2.88%.
9-O-2-(1-piperidine)ethylpalmatine (5b) was obtained in an yield of 28%; light yellow solid; mp: 250-253°. 1H NMR (DMSO-d6, 400 MHz) δ: 10.24 (1H, s), 9.08 (1H, s), 8.25 (1H, d, J=9.12 Hz), 8.08 (1H, d, J=9.04 Hz), 7.73 (1H, s), 7.11 (1H, s), 5.04 (2H, t, J=9.96 Hz), 4.66 (2H, t, J=6.68 Hz), 4.09 (3H, s), 3.94 (3H, s), 3.87 (3H, s), 3.65-3.68 (4H, m), 3.26 (2H, t, J=5.64 Hz), 3.09-3.17 (2H, m), 1.73-1.90 (6H, m). 13C NMR (DMSO-d6, 100 MHz) δ: 152.02, 150.53, 149.26, 146.14, 138.18, 133.53, 129.08, 127.03, 124.18, 120.34, 119.39, 111.83, 109.25, 57.62, 56.68, 56.38, 55.85, 53.54, 29.49, 26.47, 22.57, 14.44. MS (m/z, %): 448.3 (M+, 100). Elemental analysis: Found: C, 69.50%; H, 7.11%; N, 2.90%; calcd for C28H34ClNO4: C, 69.48%; H, 7.08%; N, 2.89%.
9-O-2-(1-butanamine)ethylpalmatine (5c) was obtained in an yield of 29%; light yellow solid; mp: 221-223°. 1H NMR (DMSO-d6, 400 MHz) δ: 10.25 (1H, s), 9.09 (1H, s), 8.25 (1H, d, J=9.16 Hz), 8.08 (1H, d, J=9.16 Hz), 7.73 (1H, s), 7.11 (1H, s), 5.05 (2H, t, J=6.1 Hz), 4.60 (2H, t, J=4.62 Hz), 4.08 (3H, s), 3.94 (3H, s), 3.87 (3H, s), 3.51 (2H, t, J=4.4 Hz), 3.26 (2H, t, J=5.88 Hz), 3.09 (2H, t, J=7.9 Hz), 1.69-1.76 (2H, m), 1.36-1.42 (2H, m), 0.95 (3H, t, J=7.36 Hz). 13C NMR (DMSO-d6, 100 MHz) δ: 152.08, 150.35, 149.28, 146.05, 142.20, 138.33, 133.63, 129.13, 127.18, 124.35, 121.60, 120.33, 119.37, 111.86, 109.30, 69.78, 57.70, 56.70, 56.39, 55.72, 47.48, 47.20, 27.94, 26.44, 19.88, 14.03. MS (m/z, %): 437.4 (M, 100). Elemental analysis: Found: C, 66.05%; H, 7.05%; N,5.93%; calcd for C26H33ClN2O4: C, 66.02%; H, 7.03%; N, 5.92%.
9-O-3-(4-morpholine)propylpalmatine (5d) was obtained in an yield of 30%; light yellow solid; mp: 236-237°. 1H NMR (DMSO-d6, 400 MHz) δ: 9.83 (1H, s), 9.08 (1H, s), 8.22 (1H, d, J=9.08 Hz), 8.05 (1H, d, J=9.08 Hz), 7.73 (1H, s), 7.10 (1H, s), 5.00 (2H, s), 4.38 (2H, t, J=5.86 Hz), 4.06 (3H, s), 3.94 (3H, s), 3.87 (3H, s), 3.64-3.71 (4H, m), 3.25 (2H, t, J=5.54 Hz), 2.50-2.67 (6H, m), 2.12-2.17 (2H, m). 13C NMR (DMSO-d6, 100 MHz) δ: 151.98, 150.60, 149.23, 145.76, 143.10, 138.17, 133.66, 129.10, 127.16, 123.86, 121.99, 120.44, 119.43, 111.82, 109.32, 72.62, 70.27, 65.46, 57.59, 56.76, 56.38, 55.95, 54.64, 52.90, 52.47, 29.47, 26.46. MS (m/z, %): 464.4 (M+, 100). Elemental analysis: Found: C, 67.29%; H, 6.86%; N, 2.83%; calcd for C28H34ClNO5: C, 67.26%; H, 6.85%; N, 2.80%.
9-O-3-(1-piperidine)propylpalmatine (5e) was obtained in an yield of 29%; light yellow solid; mp: 236-237°. 1H NMR (DMSO-d6, 400 MHz) δ: 9.89 (1H, s), 9.07 (1H, s), 8.24 (1H, d, J=9.08 Hz), 8.06 (1H, d, J=9.16 Hz), 7.73 (1H, s), 7.11 (1H, s), 5.04 (2H, t, J=5.68 Hz), 4.39 (2H, t, J=5.66 Hz), 4.07 (3H, s), 3.94 (3H, s), 3.87 (3H, s), 3.50-3.55 (2H, m), 3.26 (2H, t, J=5.74 Hz), 2.95-2.97 (2H, m), 2.32 (2H, d, J=1.72 Hz), 1.75-1.83 (6H, m), 1.41-1.45 (2H, m). 13C NMR (DMSO-d6, 100 MHz) δ: 151.96, 150.45, 149.21, 145.74, 142.86, 138.17, 133.62, 129.09, 127.06, 123.98, 121.82, 120.40, 119.38, 111.80, 109.25, 71.97, 57.61, 56.75, 56.37, 55.91, 53.78, 52.62, 29.47, 26.42, 24.95, 22.96, 21.87. MS (m/z, %): 462.4 (M+, 100). Elemental analysis: Found: C, 69.95%; H, 7.30%; N, 2.83%; calcd for C29H36ClNO4: C, 69.93%; H, 7.29%; N, 2.81%.
Antimicrobial activity
The antimicrobial activity of synthesized compounds (3a-f, 5a-e) were determined by using paper disc method and 2-fold serial dilution test. [16] The compounds (1, 3a-f, 5a-e) were dissolved in MeOH- 1% DMF at a concentration of 10 mg/ml; 50 μg of the solution was applied to a paper disc (8 mm), and blank paper disc was prepared in MeOH-1% DMF only as a negative control; palmatine (1) was used as standard; the paper discs were dried 12 h at 60° and placed on an agar plate seeded with the test microorganism; after incubation for 24 h at 37°, the diameter of the inhibitory zone around the paper disc was measured. 2-fold serial dilution test: The compounds (1, 3a-f) were dissolved in H2O containing 1% DMF and 0.8% Tween 80 at the initial dosage 2000 μg/ml; the solution was prepared concentrations from 1000 to 0.97 μg/ml by a 2-fold serial dilution; blanks were prepared in H2O containing 1% DMF and 0.8%. Tween 80 only as a negative control; palmatine (1) was used as standard; the several 96-well plates, in which each well contained an appropriate growth medium with a different concentration of the respective test compounds, were incubated with the test microorganism at 37° for 24 h; the microbial growth was examined by measuring the absorbance at 655 nm with a spectrophotometer. Minimum inhibitory concentration (MIC) was defined as the lowest concentration of the test compounds, which did not induce visible growth in comparison with a blank experiment after incubation time. All experiments were run in triplicate.
Toxicity study
The median lethal dose (LD50) was determined to evaluate the toxicity of compounds according to Karber [17]. Healthy Kunming mice of both genders were randomly divided into 37 groups with 10 mice each. Animal care and toxicity test procedure were carried out according to a reported method [18]. The dosages of compounds given were designed according to Table 1. The normal control group was fed with the same volume of physiological saline. Mice were housed in stainless cages in a room with controlled temperature (25°) and humidity (40-60%) and 12 h light/dark cycle. The protocol complied with the guidelines of Zhengzhou City Laboratory Animal Administration Committee of China for the care and use of laboratory animals. Animals were then kept under observation for 7 days to record toxicity and total mortality.
| Compound | Dose (mg/kg) | ||||
|---|---|---|---|---|---|
| 1, 3a, 3b, 3e, 3f | 1180 | 847 | 603 | 430 | 310 |
| 3c, 3d | 790 | 560 | 400 | 285 | 205 |
Table 1: Dosages of compounds
Results and Discussion
The synthetic strategies used for the preparation of 9-O-substituted palmatine derivatives were illustrated in fig. 1. The compounds were purified by column chromatography using suitable solvent system. The 1H NMR and 13C NMR spectrum showed that some new proton and carbon atom signals of CH2/CH3 appeared. MS tested compounds formula. Elemental analysis tested elements ratio of the compounds. The results displayed that a series of new Palmatine derivatives were synthesized at C-9-O.
The data for antimicrobial activity of 9-O-substituted derivatives was shown in Tables 2 and 3. Diameter of inhibitory zone formed by the compounds was presented Table 2. 9-O-substituted palmatine derivatives displayed a good antimicrobial activity against different bacteria strains at the dose level of 10 mg/ml. Antimicrobial activities of the compounds (3a-f) were much better than that of palmatine (1). The compounds (3a-f) possessed relatively weaker inhibitory effect against Gram −ve bacteria and fungi. These results might suggest that the mode of action in Gram +ve bacterium is different from that in Gram −ve bacterium and fungi. The compound 3c was much stronger than 3b, suggesting that increased the spatial structure was good for antimicrobial activities at the same number of carbon atoms of aliphatic chain. Antimicrobial activities of the compounds (5a-e) is low, suggesting that substituents for hydrophilic groups at C-9-O are not conducive to increasing antimicrobial activity, which is consistent with what has been reported [19]. The minimum inhibitory concentrations (MIC) of 9-O-substituted palmatine derivatives are listed in Table 3. The results on microbes obtained by 2-fold serial dilution test are in accord with those obtained by paper disc method. Antimicrobial activities of the compounds (3a-f) against Gram +ve organisms increased 2- to 64-fold than that of palmatine (1), and which increased as the alkyl sidechain was elongated and then decreased gradually when the aliphatic chain exceeded four carbon atoms. Antibacterial activities of the compounds (3a-f) against Gram −ve bacteria and fungi increased 1- to 16-fold than that of palmatine (1), and which gradually increased as the aliphatic chain increased and tend to be stable. These results might further suggest that the mode of action in Gram +ve bacterium is different from that in Gram −ve bacterium and fungi.
| Compound (50 μg/disc) |
Gram +ve bacteria | Gram −ve bacteria | Fungi C. albicans | ||
|---|---|---|---|---|---|
| S. aureus | B. subtilis | E. coli | P. mirabilis | ||
| Control | – | – | – | – | – |
| 1 | 16.5 | 15.5 | 10.0a | 9.0a | b |
| 3a | 18.0 | 16.5 | 10.5a | 9.0a | 8.5a |
| 3b | 19.0 | 17.5 | 11.0a | 9.5a | 9.5a |
| 3c | 19.5 | 18.0 | 12.0a | 11.0a | 10.0a |
| 3d | 21.0 | 20.5 | 15.0a | 13.5a | 11.5a |
| 3e | 17.5 | 17.0 | 15.5a | 14.0a | 12.0 |
| 3f | 17.0 | 16.5 | 16.0a | 14.0a | 12.5a |
| 5a | 11.0 | 10.5 | b | b | b |
| 5b | 13.5 | 11.5 | 9.0a | 8.5a | b |
| 5c | 14.5 | 13.5 | 9.5a | 9.0a | 9.0a |
| 5d | 12.5 | 11.0 | 8.5a | b | b |
| 5e | 14.0 | 12.0 | 9.0a | 8.5a | b |
Table 2: Diameters of inhibitory zones of 9‑o‑substituted palmatine derivatives
| Compound | Gram +ve bacteria | Gram −ve bacteria | Fungi C. albicans | ||
|---|---|---|---|---|---|
| S. aureus | B. subtilis | E. coli | P. mirabilis | ||
| Control | – | – | – | – | – |
| 1 | 250 | 500 | 1000 | 1000 | 1000 |
| 3a | 125 | 125 | 1000 | 1000 | 500 |
| 3b | 62.5 | 31.25 | 500 | 500 | 250 |
| 3c | 15.6 | 15.6 | 250 | 125 | 125 |
| 3d | 3.91 | 7.8 | 125 | 125 | 125 |
| 3e | 31.25 | 15.6 | 62.5 | 62.5 | 62.5 |
| 3f | 62.5 | 62.5 | 62.5 | 62.5 | 62.5 |
Table 3: Minimum inhibitory concentration of 9‑o‑substituted palmatine derivatives.
The median lethal dose (LD50) data of 9-O-substituted derivatives towards mice are shown in Table 4. The toxicity of compounds (3a-f) decreased as the alkyl sidechain was elongated. The toxicity of compounds (3d-f) was lower than that of palmatine. Compounds 3f displayed the lowest toxicity.
| Compound | 1 | 3a | 3b | 3c | 3d | 3e | 3f |
|---|---|---|---|---|---|---|---|
| LD50 | 670.1 | 431.5 | 647.3 | 642.3 | 743.2 | 782.7 | 945.7 |
LD50 is the median lethal dose in mg/kg
Table 4: Ld50 of compounds 3a‑f
Acknowledgements
This work was financially supported by the Key Scientific and Technological Project of Henan Province (Grant No. 102102210027, 092102210007), the Scientific and Technological Project of Zhengzhou City (Grant No. 20120725).
References
- Yuan LJ, Tu DW, Ye XL, Wu JP. Hypoglycemic and hypocholesterolemic effects of Coptis chinensis Franch inflorescence. Plant Foods Hum Nutr 2006;61:139-44.
- Grycová L, Dostál J, Marek R. Quaternary protoberberine alkaloids. Phytochem 2007;68:150-75.
- Yu Y, Yi ZB, Liang YZ. Main antimicrobial components of Tinospora capillipes, an their mode of action against Staphylococcus aureus. FEBS Lett 2007;581:4179-83.
- Fu Y, Hu BR, Tang Q, Fu Q, Xiang JZ. Hypoglycemic activity of jatrorrhizine. Acta Med Univ Sci Tech Huazhong 2005;25:491-3.
- Han H, Fang DC. The blocking and partial agonistic actions of jatrorrhizine on alpha-adrenoceptors. Acta Pharmacol Sin 1989;10:385-9.
- Račkováa L, Májekováa M, Koštálováb D, Štefeka M. Antiradical and antioxidant activities of alkaloids isolated from Mahonia aquifolium structural aspects. Bioorg Med Chem 2004;12:4709-15.
- Yang Y, Ye XL, Li XG. Antimicrobial effect of four alkaloids from Coptidis Rhizome. Lishizhen. Med Mater Med Res 2007;18:3013-4.
- Iwasa K, Nishiyama Y, Ichimaru M, Moriyasu M, Kim HS, Wataya Y, et al. Structure-activity relationships of quaternary protoberberine alkaloids having an antimalarial activity. Eur J Med Chem 1999;34:1077-83.
- Iwasa K, Kamigauchi M, Sugiura M, Nanba H. Antimicrobial Activity of Some 13-alkyl substituted protoberberinium Salts. Planta Med 1997;63:196-8.
- Iwasa K, Lee DU, Kang SI, Wiegrebe W. Antimicrobial activity of 8-alkyl and 8-phenyl substituted berberines and their 12-bromo derivatives. J Nat Prod 1998;61:1150-3.
- Iwasa K, Kamigauchi M, Ueki M, Taniguchi M. Antibacterial activity and structure-activity relationships of berberine analogs. Eur J Med Chem 1996;31:469-78.
- Park PD, Lee JH, Kim SH, Kang TH, Moon JS, Kim SU. Synthesis of 13-(substituted benzyl) berberine and berberrubine derivatives as antifungal agents. Bioorg Med Chem Lett 2006;16:3913-6.
- Wang LJ, Ye XL, Li XG, Sun QL, Yu G, Cao XG, et al. Synthesis and antimicrobial activity of 3-alkoxyjatrorrhizine derivatives. Planta Med 2008;74:290-2.
- Ma TW, Ye XL, Li XG, Zhang BS, Jiang XF, Chen Z. Synthesis and antimicrobial activity of 8-alkylpalmatine derivatives. Lett Drug Des Discov 2011;8:464-8.
- Wang LJ, Ye XL, Chen Z, Li XG, Sun QL, Zhang BS, et al. Synthesis and antimicrobial activity of 3-octyloxy-8-alkyljatrorrhizine derivatives. J Asian Natur Prod Res 2009;11:365-70.
- Ma XR. Drug microbial test handbook. Beijing: People Health Publishing House; 2001.
- Zhou LG. Drug toxicology. Beijing: People’s Medical Publishing House; 2003. 18.
- GB 5193.3-2003. Acute Toxicity Test. Beijing: Standards Press of China; 2005.
- Yang Y. Study on the synthesis of 8-alkyl-berberine homologues and their pharmacological activities. Chongqing: Southwest University; 2008.