- Corresponding Author:
- P. Daisy
Department of Biotechnology and Bioinformatics, Bioinformatics centre (BIF), Holy Cross College (Autonomous), Tiruchirappalli-620 002
E-mail: daisylesslie@gmail.com
| Date of Submission | November 29, 2011 |
| Date of Revision | May 23, 2012 |
| Date of Acceptance | May 28, 2012 |
| Indian J Pharm Sci, 2012, 74 (3): 217-222 |
Abstract
Tuberculosis is a highly communicable and chronic respiratory disease caused by pathogenic bacterium Mycobacterium tuberculosis. The drug â?? resistant species of Mycobacterium tuberculosis are tough to cure due to its resistant activity toward potential drugs. Available inhibitors of tuberculosis include few antimicrobial fluoroquinolone agents like ciprofloxacin, ofloxacin, and moxifloxacin to treat resistant Mycobacterium strains. Literature study elucidates that macromolecular target namely, HtrA2 of Mycobacterium tuberculosis play a dual role of protease and chaperone. These two activities are dependent on temperature, with low temperatures promoting the chaperone function and high temperatures promoting serine protease activity. Under normal physiological conditions HtrA2 acts as a quality control factor and promotes cell survival. In the present investigation, we screened fluoroquinolone such as ciprofloxacin, moxifloxacin and ofloxacin and their analogues based on better Docking score, absorption, distribution, metabolism and excretion screening and Lipinski’s rule of 5, to find out their efficiency on resistant strain through in silico study. From the results observed, the analogues are suggested to be potent inhibitors of HtrA2 with sufficient scope for further exploration.
Keywords
Ciprofloxacin, HtrA2, moxifloxacin Mycobacterium tuberculosis, ofloxacin
The comeback of Mycobacterium tuberculosis with power of multiple drug resistance becomes a severe public health threat and so it has more comparable researches with a synergy between HIV and M. tuberculosis infection [1]. Forty years of research in tuberculosis (TB) drug discovery has failed in this decade due to rise of multiple drug-resistant strains [2]. The statistical growth of multi-drug resistant problems in TB treatment has led to an increase in demand to test new antitubercular drugs which overcomes drug resistance problem. Several approaches have been explored to conquer drug resistance, but limited numbers of successes are reported due to lack of understanding of how resistance emerges in bacteria upon drug treatment. Biological network analyses of the proteins involved are essential for gaining insights into the routes required for emergence of drug resistance. Studies on early bactericidal activity provide a fast and economic way to evaluate the clinical efficacy of potential agents for the treatment of tuberculosis [3]. Due to the destructions caused by M. tuberculosis, there is a need for the development and assessment of new antituberculosis agents that might contribute to trouble-free treatment in patients.
Fluoroquinolones (FQs) show the evidence of potent in vitro and in vivo antimycobacterial activity and there are significant efforts taken to include FQs new front-line and second-line agents [4,5]. The WHO recommends the researchers and doctors to use fluoroquinolone as second-line drugs for the treatment of multiple drug resistance-tuberculosis (MDR-TB). Based on the available literatures on bactericidal activity, FQs like ciprofloxacin (CIP), moxifloxacin (MFX) and ofloxacin (OFX) are approved for clinical studies and are now recommended as part of treatment for MDR-TB.
Ciprofloxacin has shown encouraging in vitro activity against M. tuberculosis [6]. It significantly enhances antibacterial potency. It has a broad-spectrum antimicrobial activity and is used in the treatment of tuberculosis. MFX has more activity than CIP and OFX resistant M. tuberculosis [7,8] and also has less potential to promote fluoroquinolone resistance [9-11]. It has also improved outcomes on patients with extensive drug resistance (XDR) tuberculosis. OFX has been identified as a new agent in the treatment of TB due to its significant bactericidal activity against M. tuberculosis. OFX has been well tolerated by patients (by absence of side effects) with low toxicity [12,13]. The excellent antimycobacterial activity of OFX has encouraged non-comparative clinical trials for the treatment of pulmonary tuberculosis [14].
The success of M. tuberculosis as a pathogen is caused, in part, by its ability to survive in macrophages and establish long-term, persistent infection in the host during periods of control by the cell-mediated immunity. Members of the high temperature requirement A (HtrA) family are envelope-associated serine proteases that perform crucial functions, involving protein quality control in the periplasmic space, acting as both molecular chaperones and proteases [15,16]. Proteases are amongst the numerous factors that may be important for bacterial survival in vivo. HtrA2 is a serine protease and chaperone. At low temperatures it acts as a chaperone, whereas at high temperatures it behaves as a protease. Intriguingly, the protease activity appears to be important in protecting bacteria against heat shock. It is a part of a two operon consisting of MprA-MprB, which is a two component system required by M. tuberculosis for growth in in vivo during the persistent stage of infection [17]. Low oxygen, existence in the granulomatous tissue contributes to the persistence of infection by HtrA2, and its virulence is caused mainly by their role in stress resistance and survival.
HtrA2 protein is one of the aetiologies for mycobacterium infections. Targeting this protein will be helpful in treating the infection caused by M. tuberculosis. Determining the binding structures in the active site of HtrA2 and exploring the interactions for the original drugs and their analogues are essential, to improve the design of second-generation inhibitors. In the present study the fluoroquinolone drugs CIP, MFX, OFX and also their analogues were targeted for HtrA2 through in silico approach to find out its inhibitory activity against HtrA2.
Materials and Methods
Ligand preparation
The structure of CIP, MFX and OFX and their analogues were retrieved from PUB_CHEM database by performing substructure screening. The ligands were prepared using LigPrep 2.3 [18] module of the Schrödinger suite [19] using Merck molecular force field. MMFF was used for 2D to 3D conversion of ligand molecules and optimized potential for liquid simulation (OPLS) force field was used for conformation analysis through Confgen [20,21]. LigPrep generated multiple conformations using confgen, and ionisation states were generated for all the compounds by using Epik 2. A single ligand was searched for multiple conformations based on torsional angles [22].
Protein preparation
The structure of HtrA2 protein (PDB ID: 2Z9I) was downloaded from the Protein Data Bank and imported and prepared by a multistep process through the protein preparation wizard of Maestro (9.0). It was especially used to obtain the optimised and minimised energy conformation of the protein. Firstly, the bond order in the protein was assigned, Hydrogen atoms were added and the water molecules which did not participate in interactions were removed. Following the above steps of preparation, the protein was subjected for energy minimisation using Schrödinger implementation of OPLS-2005 force field with implicit solvation.
Active site prediction
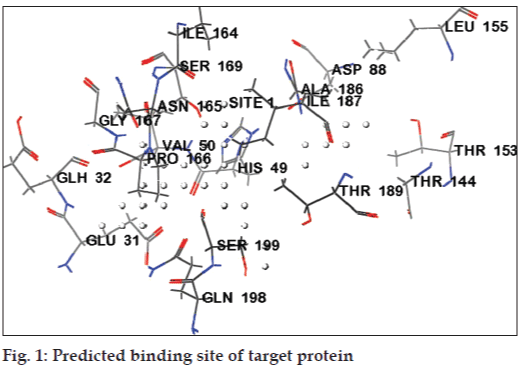
Active site is a pocket pouch present on the protein structure that has the tendency to accept the ligand molecules within it. The Sitemap applies theoretical methods and predicts the most accurate binding site [23]. The OPLS-AA force field generates site points, possible for ligand interaction within the protein. The sitemap gives an idea about positions favourable for a donor, acceptor and hydrophobic group to be present in the receptor. The maps were very useful for analysing the interactions of ligands with the receptor.
Molecular docking studies
Docking studies on LigPrep treated ligands were carried out to predict the binding pocket of 2Z9I using the docking program, Glide [24]. Glide used a series of hierarchical filters to search for possible locations for the ligand in the active site region of the receptor [25]. For the grid-based ligand docking, the receptor grid generation file was used. For protein structure, a grid box of 30×30×30 Å3 with a default inner box (10×10×10 Å3) was centered on the corresponding ligand. Default parameters were used, and no constraints were included. The processes of virtual screening were carried in three phases, docking simulations of varying precisions and computational intensities. All 300 prepared ligands were docked using the high-throughput virtual screening (HTVS) docking protocol which was computationally the least intense process intended for rapid screening of the ligands. Ligands were selected for further successive steps on the basis of the docking score; the cut-off was set as -5.0. 97 ligands selected had a docking score below -5.0 and were docked in the same receptor using the standard precision (SP) docking protocol in which the docking scores cut-off was set to -7.0 and 26 ligands were selected for further steps. Screened ligands (26 compounds) were docked using the extra precision (XP) docking protocol a more powerful and discriminative procedure, that too longer to run than SP docking. After removing the duplicates, top 5 ligands from XP docking were considered for visual inspection.
Absorption, distribution, metabolism and excretion screening
The QikProp program [26] was used to predict the ADME properties of the analogues. All the analogues were neutralised before being used by QikProp. The neutralizing step was essential, as QikProp was unable to neutralise a structure and, therefore no property will be generated in normal mode. It predicted physicochemical significant descriptors and pharmacokinetically relevant properties. QikProp provided ranges for comparing particular molecule properties with those of 95% of known drugs. It also evaluated the acceptability of analogues based on Lipinski’s rule of 5 [27,28] which was essential to ensure drug-like pharmacokinetic profile while using rational drug design.
Results and Discussion
The druggability site for the target protein (PDB ID: 2Z9I) was predicted using SiteMap program in Schrödinger. The in silico predicted active sites for target protein (PDB ID: 2Z91) were GLU31, GLH32, VAL50, HIS49, ASP88, THR153, THR189, LEU155, THR144, IIE187, SER199, GLN198, ASN165, IIE164, PRO166, and GLY167 (Table 1). The active site residues were represented in fig. 1. Docked sites of the ligand interacting with receptor were, found to be located in the amino acid residues ASP88, GLU31, GLH32, GLY167, GLN198, THR189, SER199, ILE189, HIS49, ASN48, SER69 and SER143. The major docking interactions between the fluroquinolones analogues and the target protein were predicted to be in the regions of GLU31, GLH32, GLY167, GLN198, THR189, and SER199. The docking result demonstrates that drugs and analogues bind into the same binding site of the receptor, which is predicted by SiteMap algorithm.
| In silico predicted active site residues | Docked residues |
|---|---|
| GLU31, GLH32, VAL50, HIS49, | ASP88, GLH32, GLU31, GLY167, |
| ASP88, THR153, LEU155, THR144, | GLN198, THR189SER199, |
| IIE187, SER199, GLN198, ASN165, | IIE189, HIS49, ASN48, SER169, |
| IIE164, PRO166, GLY167 | GLH198, SER143 |
Table 1: Predicted active site residues and docked residues.
To gain insights into the structural basis for its activity, FQs analogues were docked into the active site of the target protein. A correlation was calculated by Glide score and other docking parameters.
For theoretical prediction of analogues, mainly four parameters were taken for consideration, these included G-score, Glide energy, hydrogen bonds and Emodel. On the basis of these parameters the binding affinity of ligands toward the receptor are discussed here. The docking results are revealed in Table 1. From a total of 300 compounds, best five compounds were selected based on docking results. Higher negative value of G-score indicated enhanced binding affinity of the ligand with the receptor. In this in silico effort, we short-listed the compounds keeping the G-score value of more than -10 as underlying criteria. The preferred ligands PUB_CHEM ID 16043034, 465157, 16095387, 16042854 and 25139637 had a G-score value of -12.156895, -11.410010, -10.602376, -10.526178, and -10.009522, respectively. The G-score values of the above-mentioned three FQs (CIP, MFX and OFX) were comparatively lesser than their analogues with the scoring of -4.802, -3.609 and -3.588.
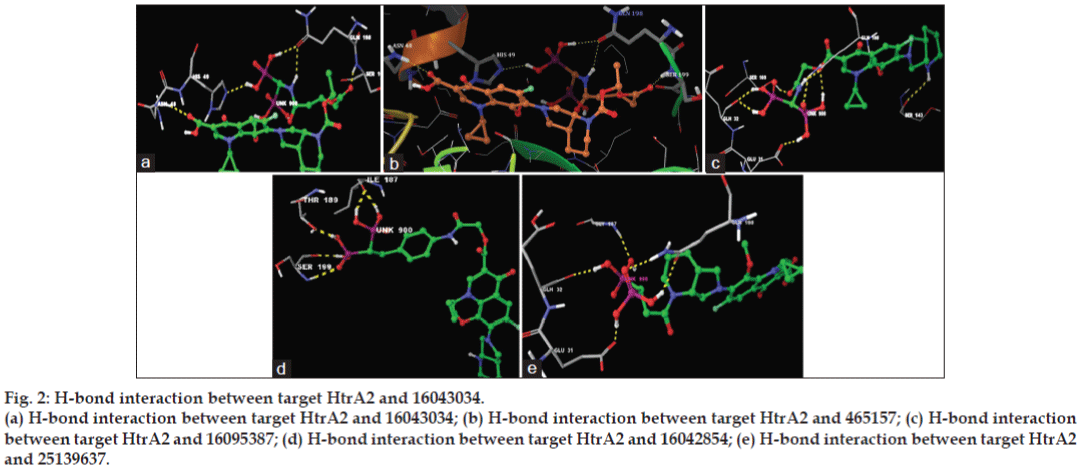
The minimum glide energy required for the formation of complex between ligand and the receptor indicates excellent binding affinity. Very low energy indicates that the ligand is buried in the cavity of the receptor. The glide binding energy of the fluroquinolone analogues as ligands was found to be -74.759915, -49.698217, -74.042655, -67.789424, and -66.310752. FQs such as CIP, MFX and OFX possessed low glide energy in the range between -30 and -32. In our present investigation, the five fluoroquinolone analogues ligands had high-quality binding affinity with the receptor HtrA2. hydrogen bond interaction can act either as antagonist or agonist for a ligand with receptor. On analysing the docking results, all the five compounds had more than four H-bond interactions with the receptor indicating that if the H-bond interaction was more, the binding affinity of the ligand was higher. hydrogen bond interaction in target protein with the ligand (PUB_CHEM ID: 16043034) had four hydrogen bond interactions. Ligand (PUB_CHEM ID: 465157) and the target protein had eight hydrogen bond interactions, Ligand (PUB CHEM_ID: 16095387) with the target protein had six H-bond interactions, Ligand (PUB_ CHEM ID: 16042854) and the protein had four hydrogen bond interactions, and finally Ligand (PUB_CHEM ID: 25139637) with the target protein had five hydrogen bond interactions. All the five ligands had more number of hydrogen bond interactions with the receptor (fig. 2). The distance of the H-bonds were less than three which indicated the presence of favourable interactions between ligand and receptor. The last parameter Emodel had a more significant weighting of the force field components (electrostatic and van der Waals energies), which made it well-suited for comparing conformers. Therefore, Glide uses Emodel to pick the ‘best’ pose of a ligand (pose selection), and then ranks these best poses against one another with Glide_Score. The low Emodel value indicates the best binding affinity between protein and ligand. The Glide Emodel values of −100.390545, −60.559325, −103.356869, −81.831223, −94.630635, respectively, for 5 best compounds (fluroquinolone analogues) were very low and are represented in Table 2 suggesting an energetically favourable interaction with the active site when compared with the Emodel score of CIP (-38.634), MFX (-38.506), and OFX (-38.917) shown in Table 3.
| Ligand no. | Ligand atom | Protein residue | H-bond length | Docking score | Glide energy | Emodel |
|---|---|---|---|---|---|---|
| 16043034 (MFX) | O | ASN 48 (H) | 2.007 | −12.156895 | −74.759915 | −100.390545 |
| H | HIS 49 (N) | 2.108 | ||||
| H | GLN198 (O) | 1.837 | ||||
| O | SER 199 (H) | 1.2041 | ||||
| 465157 (OFX) | H | THR 189 (O) | 2.207 | −11.410010 | −49.698217 | −60.559325 |
| N | SER199 (O) | 2.296 | ||||
| N | GLN 198 (H) | 2.342 | ||||
| H | GLN198 (O) | 2.409 | ||||
| H | GLU31 (O) | 1.862 | ||||
| H | GLH32 (O) | 1.967 | ||||
| H | ASP88 (O) | 2.066 | ||||
| O | GLY167 (H) | 2.086 | ||||
| 16095387 (MFX) | H | GLH 32 (O) | 1.658 | −10.602376 | −74.042655 | −103.356869 |
| O | SER 169 (H) | 2.046 | ||||
| H | GLU 31 (O) | 1.733 | ||||
| O | GLN198 (H) | 3.261 | ||||
| H | GLN198 (O) | 1.814 | ||||
| H | SER 143 (O) | 2.432 | ||||
| 16042854 (MFX) | H | ILE 187 (O) | 1.624 | −10.526178 | −67.789424 | −81.831223 |
| H | THR 189 (O) | 1.989 | ||||
| O | SER 199 (H) | 2.305 | ||||
| H | SER 199 (O) | 1.746 | ||||
| 25139637 (MFX) | H | GLU 31 (O) | 1.503 | −10.009522 | −66.310752 | −94.630635 |
| H | GLH 32 (O) | 1.611 | ||||
| O | GLY167 (H) | 1.869 | ||||
| O | GLN198 (H) | 2.201 | ||||
| H | GLN 198 (O) | 2.124 |
MFX=Moxifloxacin, OFX=Ofloxacin
Table 2: Docking results of best five analogues.
+| Ligand name | Ligand atom | Protein residue | H-bond length | Docking score | Glide energy | Emodel |
|---|---|---|---|---|---|---|
| Ciprofloxacin | H | HIS-49 | 2.911 | −4.802 | −30.851 | −38.634 |
| H | SER-185 | 2.144 | ||||
| O | SER199 | 1.868 | ||||
| Moxifloxacin | O | SER199 | 1.928 | −3.609 | −32.822 | −38.506 |
| O | THR-189 | 2.019 | ||||
| H | ASP-88 | 1.960 | ||||
| Ofloxacin | H | ASP-88 | 2.157 | −3.588 | −32.194 | −38.917 |
| O | THR-189 | 2.045 | ||||
| O | SER-199 | 2.013 |
Table 3: Docking results of ciprofloxacin, moxifloxacin and ofloxacin
Figure 2: H-bond interaction between target HtrA2 and 16043034.
(a) H-bond interaction between target HtrA2 and 16043034; (b) H-bond interaction between target HtrA2 and 465157; (c) H-bond interaction
between target HtrA2 and 16095387; (d) H-bond interaction between target HtrA2 and 16042854; (e) H-bond interaction between target HtrA2
and 25139637.
A number of physical descriptors as well as pharmaceutically relevant properties of 26 fluroquinolone analogues using QikProp are analysed, amongst which significant descriptors are reported and are important for predicting the drug-like properties of molecules. The properties includes H-bond Donor (0.0-6.0), H-Acceptor (2.0-20.0), Predicted water/gas partition coefficient. (QPlogpw) (4.0-45.0), Predicted octanol/water partition coefficient (QPlogPo/w) (–2.0 to 6.5), Predicted aqueous solubility (QPlogS) (–6.5 to 0.5).
Amongst the tested compounds (PUB CHEM ID: 16043034, 465157, 16095387, 16042854, and 25139637) (Table 4), were found to be the most potent where as other compounds showed an average activity against M. tuberculosis. In comparison with the ADME properties of the original fluroquinolones (Table 5), their drug analogues showed better activity.
| Compound number | H-bond donor | H-bond acceptor | QPlogPw (4.0-45) | QPlogPo/w (−2.0 to 6.5) | QPlogS (−6.5 to 0.5) |
|---|---|---|---|---|---|
| 16043034 | 0.25 | 17.5 | 20.558 | 3.067 | −4.029 |
| 465157 | 17 | 26.9 | 52.864 | −8.764 | −0.177 |
| 16095387 | 1.25 | 15.5 | 21.922 | −0.158 | −3.738 |
| 16042854 | 2 | 16.25 | 21.628 | 1.605 | −5.428 |
| 25139637 | 0 | 15.25 | 19.93 | 2.541 | −2.667 |
Table 4: Predicted absorbtion, disturbation, metabolism, excretion properties of fluoroquinolones analogs using qikprop.
| Compound name | H-bond donor | H-bond acceptor | Qplogpw (4.0–45) | QPlogPo/w (−2.0 to 6.5) | QPlogS (−6.5 to 0.5) |
|---|---|---|---|---|---|
| Ciprofloxacin | 1 | 6 | 9.951 | 0.280 | −3.792 |
| Moxifloxacin | 1 | 6.75 | 10.506 | 0.957 | −4.640 |
| Ofloxacin | 0 | 7.25 | 9.558 | 0.380 | −3.262 |
Table 5: Predicted absorption, distribution, metabolism and excretion properties of ciprofloxacin, moxifloxacin and ofloxacin.
The protein ligand interaction plays a significant role in structural based drug designing. In this approach, H-bonding, Glide energy score, docking results and ADME properties are kept as a support for our work through which, we predicted that fluoroquinolone analogues of MFX and OFX possess better antibacterial activity than the CIP by having good binding affinity with target protein and it could be used as potential drugs for second-generation drug development than the already existing inhibitors of M. tuberculosis. This study further requires experimental validation to establish these said analogues as more potent drugs for the treatment.
Acknowledgement
The financial support is extended by the Biotechnology Information System (BTIS), Department of Biotechnology (DBT), Ministry of Science and Technology, Government of India, India is acknowledged.
References
- Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 1998;393:537-44.
- Khasnobis S, Escuyer VE, Chatterjee D. Emerging therapeutic targets in tuberculosis: Post-genomic era. Expert OpinTher Targets 2002;6:21-40.
- Gumbo T, Louie A, Deziel MR, Drusano GL. Pharmacodynamicevidence that ciprofloxacin failure against tuberculosis is not due to poor microbial kill but to rapid emergence of resistance. Antimicrob Agents Chemother 2005;49:3178-81.
- Bryskier A, Lowther J. Fluoroquinolones and tuberculosis. Expert OpinInvestig Drugs 2002;11:233-58.
- Ginsburg AS, Grosset JH, Bishai WR. Fluoroquinolones, tuberculosis, and resistance. Lancet Infect Dis 2003;3:432-42.
- Collins CH, Uttley AH. In vitro susceptibility of mycobacteria to ciprofloxacin. J AntimicrobChemother 1985;16:575-80.
- Kam KM, Yip CW, Cheung TL, Tang HS, Leung OC, Chan MY. Stepwise decrease in moxifloxacin susceptibility amongst clinical isolates of multidrug-resistant Mycobacterium tuberculosis: Correlation with ofloxacin susceptibility. Microb Drug Resist 2006;12:7-11.
- Von Groll A, Martin A, Jureen P, Hoffner S, Vandamme P, Portaels F, et al. Fluoroquinolone resistance in Mycobacterium tuberculosis and mutations in gyrA and gyrB. Antimicrob Agents Chemother 2009;53:4498-500.
- Dong Y, Xu C, Zhao X, Domagala J, Drlica K. Fluoroquinolone action against mycobacteria: Effects of C-8 substituents on growth, survival, and resistance. Antimicrob Agents Chemother 1998;42:2978-84.
- Hooper DC. Minimizing potential resistance: The molecular view – A comment on Courvalin and Trieu-Cuot. Clin Infect Dis 2001;33:S157-60.
- Rodríguez JC, Cebrián L, López M, Ruiz M, Jiménez I, Royo G. Mutant prevention concentration: Comparison of fluoroquinolones and linezolid with Mycobacterium tuberculosis. J AntimicrobChemother 2004;53:441-4.
- Berning SE. The role of fluoroquinolones in tuberculosis today. Drugs 2001;61:9-18.
- Gillespie SH, Kennedy N. Fluoroquinolones: A new treatment for tuberculosis? Int J Tuberc Lung Dis 1998;2:265-71.
- Tsukamura M. In vitroantimycobacterial activity of a new antibacterial substance DL-8280 – Differentiation between some species of mycobacteria and related organisms by the DL-8280 susceptibility test. MicrobiolImmunol 1983;27:1129-32.
- Pallen MJ, Wren BW. The HtrA family of serine proteases. MolMicrobiol 1997;26:209-21.
- Page MJ, Di Cera E. Serine peptidases: Classification, structure and function. Cell Mol Life Sci 2008;65:1220-36.
- Zahrt TC, Deretic V. Mycobacterium tuberculosis signal transduction system required for persistent infections. ProcNatlAcadSci USA 2001;98:12706-11.
- LigPrep, version 2.3. Schrödinger, LLC: New York; 2009.
- Maestro, version 8.5. Schrödinger LLC: New York; 2008.
- ConfGen, version 2.2. Schrödinger, LLC: New York; 2011.
- Watts KS, Dalal P, Murphy RB, Sherman W, Friesner RA, Shelley JC. ConfGen: A Conformational Search Method for Efficient Generation of Bioactive Conformers. J ChemInf Model 2010;50:534-46.
- Hayes MJ, Stein M, Weiser J. Accurate Calculations of Ligand Binding Free Energies: Chiral Separation with Enantioselective Receptors. J PhysChem 2004;108:3572-80.
- Sitemap, version 2.4. Schrödinger, LLC: New York; 2008.
- Glide, version 5.0. Schrödinger, LLC: New York; 2008.
- Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, et al. Glide: a new approach for rapid, accurate docking and scoring.1. Method and assessment of docking accuracy. J Med Chem 2004;47:1739-49.
- Duffy EM, Jorgensen WL. Prediction of Properties from Simulations: Free Energies of Solvation in Hexadecane, Octanol, and Water. J Am ChemSoc 2000;122:2878-88.
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ.Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 1997;23:3-25.
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 2001;46:3-26