- *Corresponding Author:
- K. Kannabiran
Department of Biomedical Sciences, School Bio-Sciences and Technology, VIT University, Vellore-632 014, India
E-mail: kkb@vit.ac.in
| Date of Submission | 27 June 2017 |
| Date of Revision | 03 April 2018 |
| Date of Acceptance | 09 October 2018 |
| Indian J Pharm Sci 2018;80(6):1093-1099 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The present study was aimed to screen and identify antibacterial bioactive nitrogenous secondary metabolites produced by the marine Streptomyces sp., VITLGK012. The antibacterial potential of the extracted compound was studied using agar well diffusion method. Minimal inhibitory concentration of the compound for antibacterial activity was estimated using the plate dilution method. The isolate was characterized by morphological, biochemical and molecular taxonomy using 16S rDNA sequencing. Chloroform extract was prepared and purified by silica gel column chromatography. The nitrogenous fraction was identified with Dragondorff’s reagent. The structure of the pure compound was identified by gas chromatography coupled with mass spectrometry, Fourier-transform infrared spectroscopy and proton nuclear magnetic resonance spectroscopy. Crude extract of VITLGK012 showed antibacterial activity with a MIC of 0.5 mg/ml against Proteus vulgaris. VITLGK012 strain was identified to be a Streptomyces species based on 16S rDNA sequencing. Gas chromatography and mass spectrometry analysis showed the presence of more than 10 molecules in the crude extract. Based on spectroscopic analysis, the extracted compound was found to be nitrogenous in nature and was identified as gancidin W. The results of the study suggested that Streptomyces sp. VITLGK012 could be a good source for nitrogenous antibacterial secondary metabolites.
Keywords
Actinomycetes, Streptomyces sp. VITLGK012, molecular taxonomy, gancidin W
Actinomycetes are Gram-positive, filamentous, saprophytic bacteria, which belong to the order Actinomycetales and the family Actinomycetaceae. Among various prokaryotes, Actinoplanes, Amycolatopsis, Micromonospora, Saccharopolyspora and Streptomyces are economically important actinomycetes genera and also major producers of commercially important antibiotic molecules [1]. Numerous bioactive compounds and new chemical entities have been extracted from actinomycetes. Out of 23 000 bioactive compounds isolated from microorganisms, 10 000 bioactive compounds were isolated from actinomycetes alone, among which, 7600 were isolated from Streptomyces species [2]. These bioactive compounds were reported to have numerous activities like antibacterial, antifungal, antiviral, anticancer and antimalarial. However, in the recent past, the rediscovery of known bioactive compounds in terrestrial actinomycetes have increased and therefore researchers have shown increased interest in exploring marine actinomycetes. The marine ecosystem offers a rich diversity, which is vastly different from the terrestrial environment.
The incidence of pathogens developing resistance to current drugs has increased in the resent past and cancer cells also becoming resistant to existing class of cancer drugs; hence there is a pressing need to discover novel bioactive compounds capable of acting on drug resistant pathogens and cancer cells. Marine actinomycetes from unexplored or under-explored regions are capable of producing novel secondary metabolites and complex chemical structures. Rameswaram and Dhanushkodi are examples of unexplored or under-explored regions from India for actinomycetes diversity and secondary metabolites. Only very few studies are available on actinomycetes diversity [3]. In the present study, Rameswaram and Dhanushkodi marine soil samples were screened for potential antagonistic actinomycetes with the aim to isolate antibacterial secondary metabolites.
Materials and Methods
Isolation of marine actinomycetes
Marine soil samples were collected from Rameswaram and Dhanushkodi cost, Tamil Nadu, India at different locations. The top layer of the soil was removed using sterile tools and samples were collected in sterile air lock bags under aseptic conditions. The soil samples were serially diluted and 0.1 ml of each dilution was plated on composed-starch casein agar (composed- SCA) medium using spread plate technique. The plates were incubated at room temperature for a week and the colonies with dry powdery appearance were subcultured onto another fresh media plate to obtain a pure culture. Composed-SCA contained in g/l of, starch- 10 g, casein- 0.3 g, KNO3- 2 g, NaCl- 2 g, K2HPO4- 2 g, MgSO4.7H2O- 0.05 g, CaCO3- 0.02 g, FeSO4.7H2O- 0.01 g and agar- 18 g in 1 l of distilled water [4].
Preparation of crude extract
All actinomycetes isolates were screened for antibacterial activity against selected test pathogens. Based on the antibacterial activity, the isolate VITLGK012 was selected for further analysis. The isolate VITLGK012 (3 ml seed culture) was inoculated aseptically on International Streptomyces Project (ISP) No.1 broth media and incubated at room temperature (+/–37°) for 7 d, in a rotary incubator at 110 rpm. After incubation, the culture was filtrated using mesh cloth to remove the biomass and the cell free culture supernatant was mixed with equal volumes of chloroform. The solvent and supernatant were mixed vigorously and allowed to mix further by leaving on a rotary shaker overnight. The solvent phase was separated and condensed in a rotatory vacuum evaporator. The procedure was repeated till sufficient quantity (5 g) of the crude was obtained [5].
Assay of antibacterial activity
Bacterial pathogens, Escherichia coli MTCC-1886, Proteus vulgaris MTCC-7299, Bacillus cereus MTCC- 1168, and Staphylococcus aureus MTCC-7405, were freshly cultured in a nutrient broth as inoculum. Lawn culture of these pathogens was made on Muller-Hinton agar (MHA) HiMedia, plate. Wells were punched on these MHA plates using a well borer. To these wells, various concentrations of crude extract were added and incubated overnight at 37°. After which, the zone of inhibition (ZOI) was observed and measured using ruler compared with Streptomycin (positive control) [6,7].
16S rDNA sequencing
The genomic DNA of VITLGK012 was isolated by phenol:chloroform method and was subjected to PCR amplification with 16S rDNA universal primers, i.e., 27F and 1492R. The obtained product was then labeled using fluorescent primers, i.e., 785F and 907R and the fluorescent-labeled PCR products are sequenced using an ABI-3730xl sequencer (Applied Biosystems). The obtained sequence was searched in the NCBI database for similarity using BLAST search. The MEGA 6.06 software was used to construct the phylogenetic tree of the sequence, using neighbor-joining method [8].
Gas chromatography-mass spectrometry (GC-MS) analysis
The GC-MS analysis (Jeol GCMATE II GC-MS, Agilent Technologies 6890N Network GC system for gas chromatography) was performed and the instrument is attached to a mass spectrum (MS) component. GC column (fused silica) HP5 (50 m×0.25 mm) was used. Column temperature was maintained at 100° for 20 min, 235° for 3 min and injector temperature 240°, Helium was used as carrier gas, and split ratio was 5:4. The results from the MS analysis are cross-matched with the NIST library and the closest match is identified [9].
Fourier-transform infrared (FTIR) and nuclear magnetic resonance (NMR) analysis
The FTIR spectrum of the pure compound was analysed on a PerkinElmer Spectrum 1 FTIR spectrometer at a resolution of 1 cm−1 at a scan range of 450 to 4000 cm−1 [10]. H1NMR analysis was done in AV500 FT-NMR spectrometer to identify the functional group present in the compound [11].
Structure elucidation
The molecular weight of the pure compound was determined based on the GC-MS and mass spectral analysis. The functional groups present in the molecule were identified using FTIR spectrum. 1H-NMR spectrum revealed the positioning of the hydrogen atoms in the molecule. By combining all the spectral data, the structure of the pure compound was elucidated and identified.
Results and Discussion
Rameswaram and Dhanushkodi soil samples yielded 100 actinomycetes isolates. All isolates were screened for antibacterial activity, an isolate designated as VITLGK012 was found to be antibacterial in nature [12]. The appearance of VITLGK012 on a composed-SCA is shown in Figure 1. Gram staining of the isolate showed that it was Gram-positive. Based on antibacterial activity (preliminary screening), VITLGK012 was chosen for further studies.
The chloroform extract prepared from VITLGK012 had demonstrated significant antibacterial activity against the selected bacterial pathogens. At 20 mg/ml concentration, the crude extract demonstrated the highest activity against P. vulgaris with a 24 mm ZOI. The antibacterial activities exhibited by chloroform extract against tested pathogens are given in Table 1. The chloroform extract showed the least MIC value of 0.5 mg/ml against P. vulgaris.
| Bacterial pathogens | Zone of crude extract (20 mg) |
Inhibition (mm) Streptomycin (10 μg) |
MIC (mg/ml) |
|---|---|---|---|
| Escherichia coli [MTCC: 1886] | 20.0 | 11.0 | 2.0 |
| Staphylococcus aureus [MTCC: 7405] | 21.0 | 13.0 | 2.0 |
| Proteus vulgaris [MTCC: 7299] | 24.0 | 10.0 | 0.5 |
| Bacillus cereus [MTCC: 1168] | 19.0 | 11.0 | 2.0 |
Table 1: Antibacterial activity of Streptomyces sp. VITLGK012 crude extract
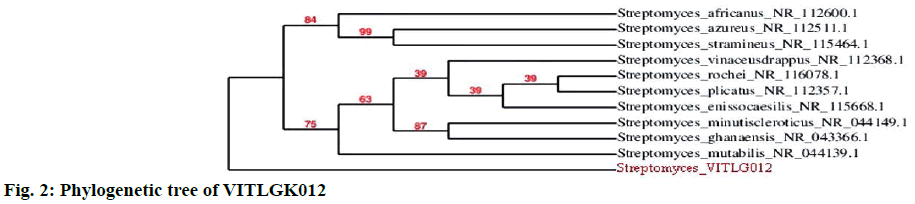
The optimal growth of VITLGK012 was studied using seven different ISP culture media. The isolate, grew well on all the media, except in ISP-6, where the growth was very low. The morphological characteristics of VITLGK012 observed on the different ISP growth media are given in Table 2. The 16S rDNA sequencing of VITLGK012, yielded 1403 nucleotides, it was searched through the NCBI database for similarity and the isolate showed 93 % similarity towards Streptomyces rochei. Since it showed only 93 % similarity with S. rochi, VITLGK012 could be a novel strain. The phylogenetic tree of VITLGK012 is shown in Figure 2. The 16S rDNA nucleotide sequence was submitted to the GenBank under the accession ID: KT597921.ID.
| Media | Description | |
|---|---|---|
| ISP 1 | Mycelium | Aerial: Creamy white Substrate: Creamy white |
| Morphology | Translucent, dry | |
| ISP 2 | Mycelium | Aerial: Pale white Substrate: Dirty white |
| Morphology | Translucent, dry | |
| ISP 3 | Mycelium | Aerial: Light grey Substrate: Amber |
| Morphology | Translucent, dry, powdery | |
| ISP 4 | Mycelium | Aerial: Grey Substrate: Khakhi |
| Morphology | Translucent, Dry, chalky | |
| ISP 5 | Mycelium | Aerial: Grey Substrate: Grey |
| Morphology | Translucent, powdery, dry | |
| ISP 6 | Morphology | Poor growth |
| ISP 7 | Mycelium | Aerial: Grey Substrate: Creamy white |
| Morphology | Translucent, dry, powdery | |
Table 2: Growth of Streptomyces sp. VITLGK012 on different ISP Culture Media
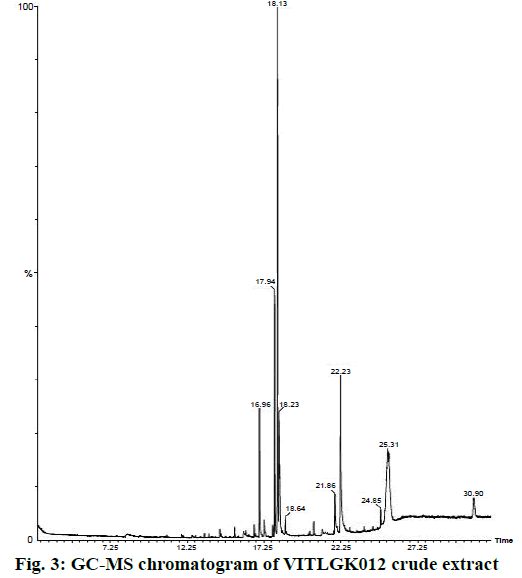
The chloroform extract of VITLGK012 was analysed using GC-MS, which demonstrated the presence of more than 10 different compounds, with a single major peak at a retention time of 18.13 min in the gas chromatogram. The GC-MS chromatogram of the chloroform extract of VITLGK012 is shown in Figure 3. The NIST library matching of GC-MS chromatogram for the major peak (18.13 min) showed more than 95 % similarity with 3-(2-methylpropyl)-octahydropyrrolo[1,2-a] piperazine-1,4-dione (gancidin W).
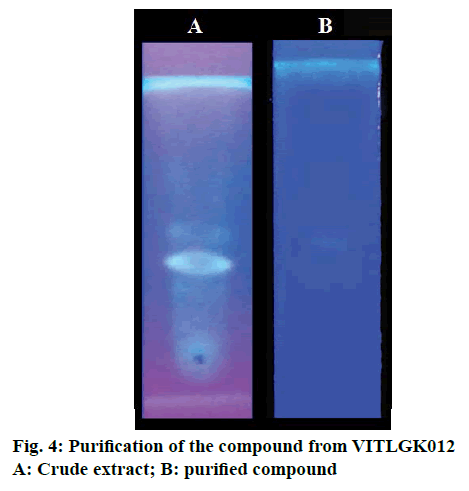
Individual peaks were obtained in TLC plate with chloroform:carbinol (9:1 v/v) solvent system (Figure 4). The chloroform extract was fractionated using silica gel 60-120, column chromatography, with the same solvent system. A total of 28 fractions were collected at a flow rate of 1 ml/min, with a volume of 5 ml each. Fractions 16-20 was positive for Dragondorff’s test and the pure compound separated using column chromatography are shown in Figure 4.
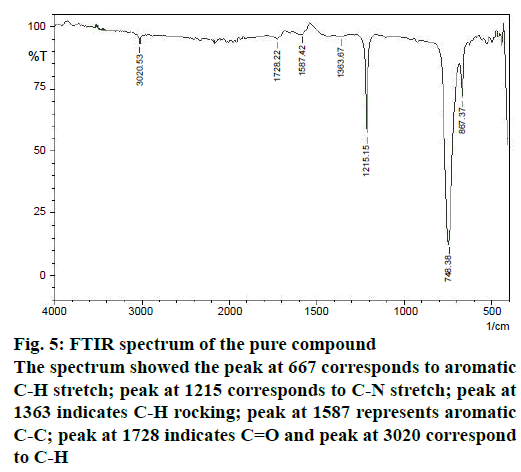
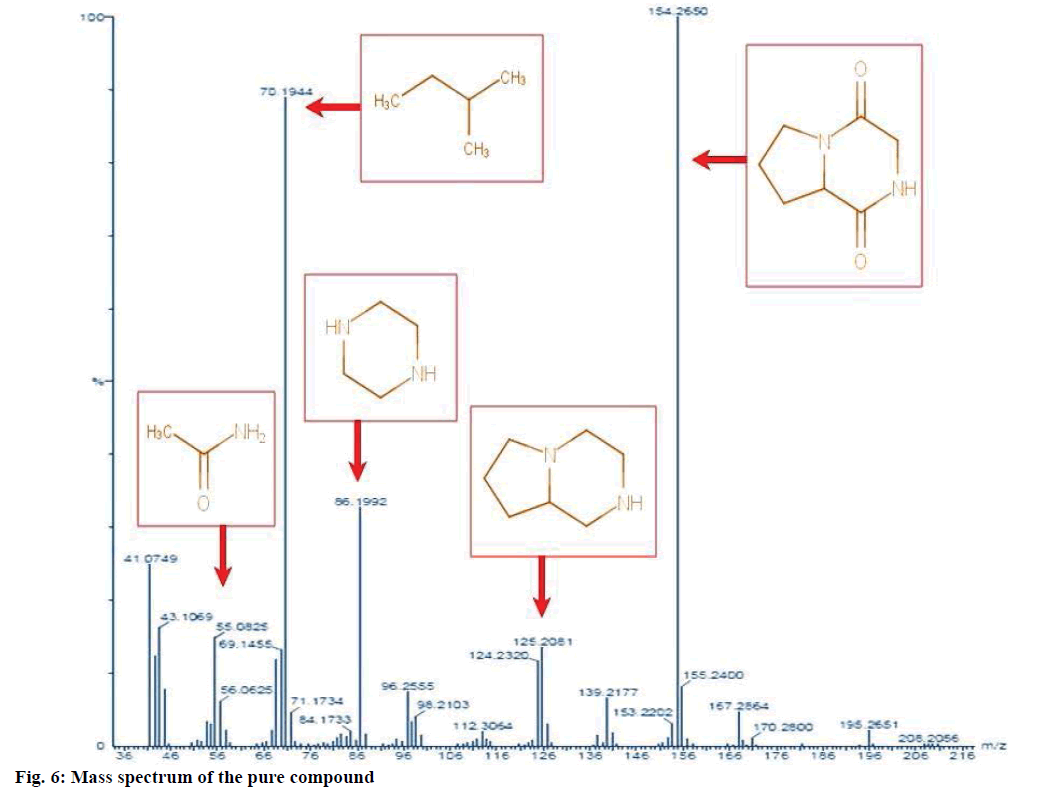
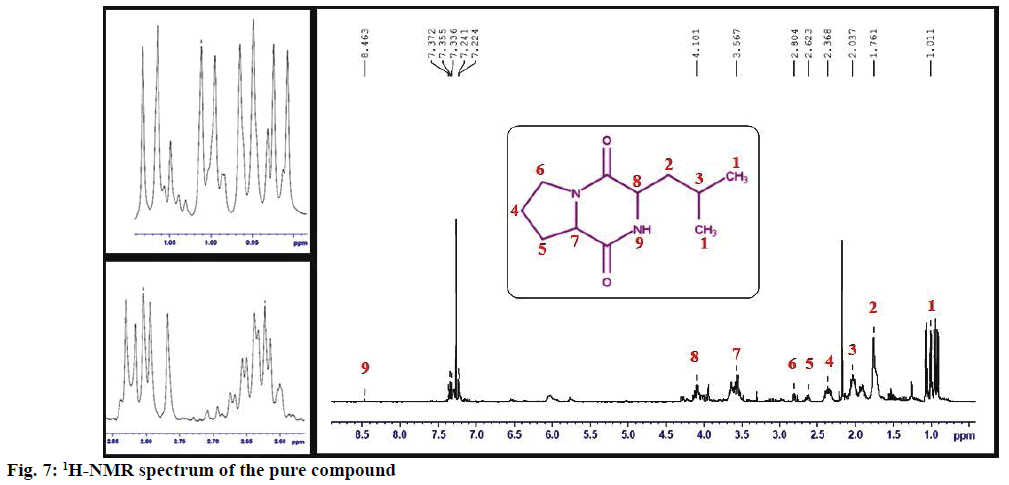
The structure of the pure compound was elucidated using FTIR, GC-MS, and 1H-NMR analysis. FTIR spectrum showed the peak at 667 corresponds to aromatic C-H stretch; peak at 1215 corresponds to C-N stretch; peak at 1363 indicates C-H rocking; peak at 1587 represents aromatic C-C; peak at 1728 indicates C=O and peak at 3020 correspond to C-H (Figure 5). The MS of the pure compound is shown in Figure 6. The 1H-NMR spectrum of the pure compound is shown in Figure 7. Combining these spectral data, the structure of the pure compound was identified as 3-(2-methylpropyl)- octahydropyrrolo[1,2-a]piperazine-1,4-dione and with a common name gancidin W. The molecular formula of the compound is C11H18N2O2 and the molecular weight is 210.27 g/mol.
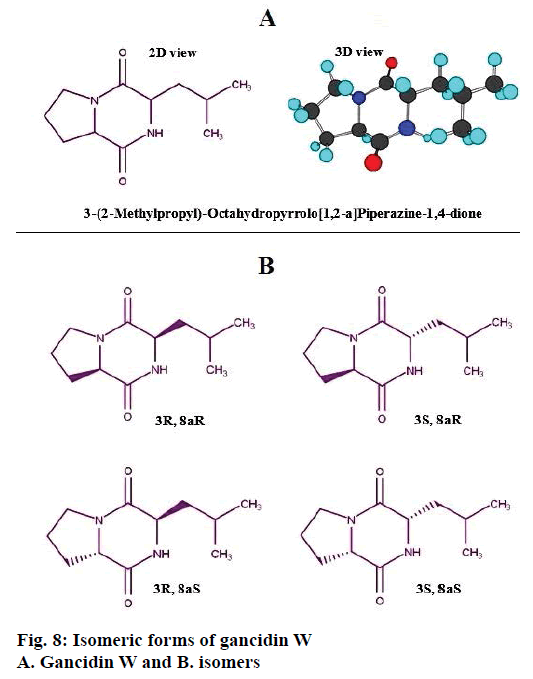
The identified compound contains two chiral carbons in its structure, hence, it has four isomeric forms, 3R, 8aR; 3S, 8aR; 3R, 8a and 3S, 8aS, which are represented in Figure 8. Based on the structure of the compound, it was identified to be a dipeptide molecule, made up of two aminoacids, proline and leucine.
VITLGK012 is a marine actinomycetes strain, isolated from marine soil samples of Rameswaram and Dhanushkodi, Tamil Nadu, India. Based on molecular phylogeny and taxonomy (16S rDNA gene sequencing), the genus of the isolate was identified as Streptomyces and designated as Streptomyces sp. VITLGK012. It showed 93 % similarity with S. rochi (MTCC 10109) and it was reported to possess broad spectrum of antagonistic activity against S. aureus and E. coli [13]. A terpenoid compound from S. rochei M32, 19-bis (3-hydroxyazetidin-1-yl) nonadeca-5,14- diene-1,8,12,19-tetraone having antagonistic activity against drug-resistant bacterial pathogens was recently reported [14]. The biosynthesized silver nanoparticles (AgNPs) from S. rochei MHM13 significantly inhibited the growth of medically important pathogenic bacteria (Vibrio fluvialis, Pseudomonas aeruginosa, Salmonella typhimurium, V. damsela, E. coli, B. subtilis, S. aureus and B. cereus) [15]. Actinomycin D produced by Streptomyces parvulus RSPSN2 (S. rochei) was very effective against bacterial pathogens P. mirabilis, P. putida and B. cereus [16].
Bioactivity-guided extraction and purification resulted in the identification of the nitrogenous compound, gancidin W. Isolation of gancidin W from other Streptomyces species was already been reported [17-19]. Gancidin W from Streptomcyes No: AAK-84 was reported as an antibiotic secondary metabolite in 1977 [20]. Initially, it was referred as cyclopeptide molecule made up of proline and leucine. It was reported that S. gancidicus produces cyclo(leucylpropyl) and the gancidin-complex [21]. It was also produced by a marine microbe Corollospora pulchella [21]. Antimalarial activity of gancidin W extracted from endophytic Streptomyces SUK10 was reported recently [22]. Our results confirmed the antibacterial potential of gancidin W extracted from Streptomyces sp. VITLGK012.
In summary, a novel actinomycetes strain, Streptomyces sp.VITLGK012 was isolated from Rameswaram and Dhanushkodi coastal soil sample were found to be a good source for antibacterial secondary metabolites. The above sampling sites could be explored further for actinomycetes diversity and bioactive secondary metabolites.
Acknowledgements
The authors thank the VIT University, for providing facilities to do this study.
Conflict of interest
There are no conflicts of interest.
Financial support and sponsorship
Nil.
References
- Chaudhary HS, Soni B, Shrivastava AR, Shrivastava S. Diversity and versatility of actinomycetes and its role in antibiotic production. J Appl Pharm Sci 2013;3(8):S83-94.
- Sharma M, Dangi P, Choudhary M. Actinomycetes: Source, identification and their applications. Int J Curr Microbiol Appl Sci 2014;3(2):801-32.
- Radhakrishnan M, Suganya S, Balagurunathan R, Kumar V. Preliminary screening for antibacterial and antimycobacterial activity of actinomycetes from less explored ecosystems. World J Microbiol Biotechnol 2010;26(3):561-6.
- Peela S, Bapiraju Kurada VVSN, Terli R. Studies on antagonistic marine actinomycetes from the Bay of Bengal. World J Microbiol Biotechnol 2005;21(4):583-5.
- Saraswathi M, Mallikarjuna N. Antibacterial activity of actinomycetes against bacterial pathogens of diabetic foot ulcers. J Appl Nat Sci 2013;5(2):335-7.
- Subashini E, Kannabiran K. Isolation and identification of anti-ESBL (extended spectrum β -lactamase) compound from marine Streptomyces sp. VITSJK8. J Adv Sci Res 2014;5(3):13-8.
- Astalakshmi A, Thangapandian V, Lingakumar K. Isolation, characterization of actinomycetes from the soil of Devathanam-A Foot-hill of Western Ghats. IJPRR 2014;3(1):15-20.
- Abirami M, Gopal JV, Kannabiran K. Extraction and identification of antibacterial compound from marine Streptomyces sp. VITAK1 isolated from the coast of Andaman and Nicobar Islands, India. Appl Biochem Microbiol 2015;51(4):406-10.
- Gnanavel V, Saral Mary A. GC-MS analysis of petroleum ether and ethanol leaf extracts from Abrus precatorius linn. Int J Pharma Bio Sci 2013;4(3):37-44.
- Suneetha SC, Raghupathy BP, Suresh PK. Physicochemical characterization and cytotoxicity screening of a novel colloidal nanogold-based phenytoin conjugate. Sci Pharm 2014;82:857-72.
- Badrinathan S, Shiju TM, Suneeva Sharon Christa A, Arya R, Pragasam V. Purification and structural characterization of sulfated polysaccharide from Sargassum myriocystum and its efficacy in scavenging free radicals. Indian J Pharm Sci 2012;549-55.
- Ravi L, Kannabiran K. Bioactivity-Guided Extraction and Identification of Antibacterial Compound from Marine Actinomycetes Strains Isolated from Costal Soil Samples of Rameswaram and Dhanushkodi, Tamil Nadu, India. Asian J Pharm 2016;10(4):S504-S509.
- Reddy NG, Ramakrishna DPN, Raja Gopal SV. A morphological, physiological and biochemical studies of marine Streptomyces rochei (MTCC 10109) showing antagonistic activity against selective human pathogenic microorganisms. Asian J Biol Sci 2011;4:1-14.
- Pazhanimurugan R, Radhakrishnan M, Shanmugasundaram T, Gopikrishnan V, Balagurunathan R. Terpenoid bioactive compound from Streptomyces rochei (M32): taxonomy, fermentation and biological activities. World J Microbiol Biotechnol 2016;32(10):161.
- Abd-Elnaby HM, Abo-Elala GM, Abdul-Raouf UM, Hamed M. Antibacterial and anticancer activity of extracellular synthesized silver nanoparticles from marine Streptomyces rochei MHM13. Egypt J Aquat Res 2016;42(3):301-12.
- Shetty PR, Buddana SK, Tatipamula VB, Naga YV, Ahmad J. Production of polypeptide antibiotic from Streptomyces parvulus and its antibacterial activity. Braz J Microbiol 2014;45(1):301-12.
- Wakaki S, Marumo H, Tomioka K, Shimizu M, Kato E, Kamada H, et al. Purification and isolation study on gancidins. J Antibiot 1958;11(4):150-5.
- Aiso K, Arai T, Suzuki M, Takamizawa Y. Gancidin, an antitumor substance derived from Streptomyces sp. J Antibiot 1956;9(3):97-101.
- Jain TC, Dingerdissen JJ, Weisbach J. Isolation and structure elucidation of Gancidin W. Heterocycles 1977;7:341-6.
- Bycroft BW, Payne DJ. Dictionary of antibiotics and related substances. 2nd ed. Boca Raton, Florida, United States: CRC Press; 2013.
- Farooq Biabani MA, Laatsch H. Advances in chemical studies on low-molecular weight metabolites of marine fungi. J Prakt Chem 1998;340:589-607.
- Zin NM, Baba MS, Zainal-Abidin AH, Latip J, Mazlan NW, Edrada-Ebel R. Gancidin W, a potential low-toxicity antimalarial agent isolated from an endophytic Streptomyces SUK10. Drug Des Devel Ther 2017;11:351-63.