- Corresponding Author:
- E. Şalva
Pharmaceutical Biotechnology, Faculty of Pharmacy, Pathology Laboratory, Vocational Health School, Marmara University, Tıbbiye Street, Haydarpasa Campuss, 34668, Istanbul, Turkey
E-mail: esalva@marmara.edu.tr
| Date of Submission | 2 January 2011 |
| Date of Revision | 12 March 2011 |
| Date of Acceptance | 20 March 2011 |
| Indian J Pharm Sci, 2011, 73 (2): 131-138 |
Abstract
Granulocyte macrophage colony stimulating factor, a potent hematopoietic cytokine, has been shown to stimulate production of white blood cells following chemotherapy. Therefore, the granulocyte macrophage colony stimulating factor gene is a potential candidate for the treatment of different pathological conditions. The purpose of this study is to investigate the suitability of chitosan as carrier for pORF-hGMCSF plasmid encoding granulocyte macrophage colony stimulating factor gene and also to study the effect of complexes on protein production and cell proliferation. Chitosan/pGM-CSF complexes were prepared using different (+/-) ratios (from 0.01/1 to 5/1). Complex formation was checked with agarose gel electrophoresis. The size and zeta potential values were measured. Enzyme and serum stability of complexes were studied. In vitro transfection properties of complexes were studied in HeLa cells. According to agarose gel electrophoresis, full complexation was obtained at 0.1/1 and higher chitosan/ pGM-CSF ratios. Complexes having about 132 nm size and +13.7 mV zeta potential value were obtained. Chitosan complexes protected plasmid against enzymatic and serum effects. The gene expression-dependent cell proliferation after transfection of chitosan/pGM-CSF complexes at 72 h was markedly increased in comparision with the level of control group. These results indicate that the effect of chitosan/pGM-CSF complexes on cell proliferation was changed with N/P ratio and time-dependently. For GM-CSF therapy, chitosan/pGM-CSF complexes may be used as alternative to conventional protein treatments. Chitosan may be a good carrier for pORF-hGMCSF. Further, in vivo study is ongoing.
Keywords
Chitosan, cell proliferation, MTT, pGM-CSF
Colony stimulating factors (CSFs) are produced by many different cells. They have high specificity to target cells (G-CSF for granulocyte and GM-CSF for granulocyte/monocyte cell line) and are active at low concentrations [1]. Lineage-specific growth factors, such as G-CSF (granulocyte colony stimulating factor) and GM-CSF (granulocyte macrophage colony stimulating factor), have pleitropic effects on both malignant and normal cells. They enhanced cell proliferation and survival, resulting in favorable differentiation and desired functional activity of myeloid cells [2]. GM-CSF is widely used in the neutropenia therapy emerging from cancer chemotherapy. It is one of the well-known hematopoietic cytokines [3], in addition to, it can also play diverse roles in wound healing [4]. GMCSF is secreted by keratinocytes in skin shortly after injury, which mediates epidermal cell proliferation in an autocrine manner and it is a pleiotropic cytokine, evoking complex process during wound repair [5]. Moreover, plasmid coding GM-CSF gene has adjuvant effect in vaccine [6]. Although recombinant human granulocyte macrophage colony stimulating factor (rHu-GM-CSF) has to be used effectively in clinic, it has been used as limited not only high cost but also the requirement for daily dosing due to its short halflife [7]. Therapy with cytokine gene may be a beneficial alternative [8]. A possible alternative route to multiple rHu-GM-CSF injections is GM-CSF expression by cells after gene transfection, i.e gene therapy [9]. As reported by Nemunaitis [10], recent preclinical studies suggested enhanced antitumor activity of plasmid GM-CSF in comparison with recombinant protein. However, delivery and in vivo transfection of pDNA encoding cytokines are inefficient, therefore, available carrier system is important for delivery [11-13]. DNA must delivered to the cell in a transcriptionally active form, hence the suitable carrier is a key factor in the gene therapy. One means to achieve long lasting expression of cytokine is to use a vector-based delivery system such as viral vectors and non-viral vectors. In general, viral vectors have been reported for pGM-CSF carrier [10,14]. However, safety concerns over the use of viral vectors and uncontrolled cytokine production are disadvantages. The success of cytokine gene treatment is largely dependent on the delivery vehicle which should be administered efficiently, safely and repeatedly. There is very limited information about the GM-CSF coding plasmid delivery system. Among the limited papers, Kim et al. [15] applied the gene therapy of GM-CSF for neural cell injury using water-soluble lipopolymer (WSLF) as gene delivery vehicles. Silica nanoparticles were used as a vehicle for GM-CSF gene transfer to dogs for white blood cell production and good results were obtained by Choi et al. [16]. Acid-terminated polyglycolide microparticles were prepared as sustained release GM-CSF for dendritic cells [17]. Cationic polymers have widely used in gene delivery applications [18]. These polymers generally serve as gene carriers though either complexation or physical entrapment for intracellular gene delivery. Among them chitosan is a biodegredable polysaccharide that has been widely studied for gene delivery [8,19-22]. It is also nonimmunogenic and low toxic [20,21,23]. Although a great number of studies on different pharmacological properties of chitosan are present, problems exist. Chitosan is cationic at acidic and neutral pH. Its cationic nature allows it to interact with negatively charged DNA and easily form complexes [24,25]. Gene delivery efficiency of chitosan is significantly influenced from the different factors such as formulation-related parameters [26]. In addition to, there is no information on the chitosan-based system used in GM-CSF coding plasmid delivery.
The aim of this study was to develop chitosan/GMCSF complexes for in vitro GM-CSF expression and to investigate the formation and stability of chitosanbased complexes formed with pGM-CSF, as well as the size, zeta potential, morphology and in vitro transfection efficiency were investigated.
Materials and Methods
Chitosan (Low MW; 150 kDa, DDA;75-85%) was supplied from Fluka (Germany), DNase I (10.000 U/ μg) and MTT cell proliferation kit were from Roche (Gemany). All of the cell culture media and reagents were purchased from Biological Industries (Israel). All the substance were of pharmaceutical or molecular grade.
Plasmid structure and isolation
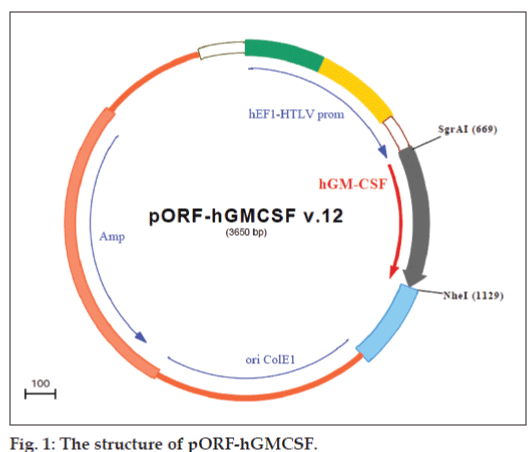
pORF-hGMCSF plasmid DNA (InvivoGen, San Diego, USA) has 3650-bp size, encodes human GM-CSF gene and contains elongation factor-1α (EF-1α)/human T cell leukemia virus (HTLV) hybrid promoter which is a composite promoter comprised from EF-1α (1) promoter and 5′ untranslated region of the HTLV; hGM-CSF gene from the ATG to the stop codon; Escherichia coli origin of replication; and ampicillin resistance gene was used (fig. 1). Plasmid was amplified in E. Coli GT100, extracted by the alkaline lysis method and purified by phenol/ chloroform extraction followed by PEG:NaCl extraction and ethanol precipitation. The quantity and quality of the purified plasmid DNA were assesed spectrophotometrically at 260-280 nm and also by electrophoresis in agarose gels.
Linear DNA was formed by using the restriction endonucleases EcoRI and Hind III, which cleave the plasmid in the CMV promoter region and in the ampicillin resistance region, respectively. The plasmid stock solution was incubated with enzyme for 2 h at 37º according to the instructions of the manufacturer (Fermentas, Germany, 50 units of enzyme per 26.6 μg DNA).
Preparation of Chitosan/pDNA complexes and gel retardation assay
Chitosan stock solutions (0.25% w/v, 0.25 mg/ ml) were prepared by dissolving chitosan in sterile 40 mM acetate buffer (pH 5.0) and then filtering (0.22 μm) the solutions under sterile conditions. DNA samples were dissolved in TE (Tris:EDTA, pH 8.0) buffer solution at 1-10 μg/ml. ChitosanpDNA complexes were formed by the addition of chitosan to the pDNA solution (in acetate buffer) by stirring on a vortex mixer (Ika-MS3, Germany). The mixed solution was vortexed rapidly for 30 s and incubated for another 30 min at room temperature for forming complexes completely [22]. Chitosan and pDNA solutions were mixed in a series of N/P ratios (The N/P ratio is defined as the molar ratio between the maximum number of protonable amines in chitosan and the number of negative phosphates on pDNA). Different charge ratios (+/-) of chitosan/ pDNA complexes (0.05/1-0.3/1) were produced at a constant pDNA concentration (13.3 μg/ml) for use in in vitro studies (particle size and zeta potential measurements, gel retardation assays and transfection experiments).
Formation of complexes between DNA and chitosan was controlled using gel electrophoresis. The complexes were prepared as described above using constant DNA concentration and different chitosan/ DNA ratios. The complexes were electrophoresed on 0.7% (w/v) agarose gel in TBE buffer for 90 min at 80 V. The gel was stained with ethidium bromide (0.5 μg/ml) and illuminated on an UV illuminator (Vilber Lourmat, France).
Physicochemical characterization of complexes
The size and zeta potentials of the complexes were determined using the Malvern Nano ZS, (Malvern Instruments, UK). Measurements were done at pH 7.4 in phosphate buffered saline (PBS). The instrument is equipped with both a particle sizer and zetameter unit. The samples was measured in glass cuvettes at 25°C with a constant angle of 90°. Each measurement was done in triplicate.
Morphology of complexes
Complexes were examined by transmission electron microscopy (TEM, Jeol, Japan). Briefly, a small drop (10-20 μl) of sample solution (1 mg/ml pDNA of chitosan complexes prepared in PBS pH 7.4) was deposited on to a copper grid covered by a 0.2% polyvinyl formal (Vinylec K). Excess liquid was blotted away with filter paper. The grids were allowed to dry at room temperature and performed negative staining technique. The grid was stained with 20 μl of 0.2% ammonium molibdate solution and left to stand for 5 min at room temperature [27].
DNase I and serum stability of complexes
In order to study the stability of pDNA in chitosan/ pGM-CSF complexes against the DNase, free DNA and DNA complexes were incubated with DNase I. Chitosan/pDNA (2/1) complexes containing 1 μg pORF-GM-CSF were reacted with 1 μl DNase I (1U), 10 μl DNase I reaction buffer (10x). Reaction was performed at 37° and aliquots were taken at intervals 0, 15, 30 min, 1, 2, 24 and 48 h. For the stopping of reaction, 0.5 M EDTA was used. DNA degradation was analyzed with agarose gel electrophoresis [22].
The interaction between DNA and the serum components was studied by gel electrophoresis. Serum stability of complexes was studied by incubating the samples in 10% foetal bovine serum and 150 mM NaCl solution at 37°. Samples were taken at intervals 0, 15, 30 min, 1, 2, 24 and 48 h and studied as mentioned above.
Cell culture
The human epithelial carcinoma cell line (HeLa) was obtained from the ATCC (ATCC CCL-2). Cells were cultured in DMEM supplemented with foetal bovine serum and 0.1% antibiotic solution in humidified atmosphere [(5% CO2, 95% air) Sanyo, USA] at 37°. The cells, which formed a single layer were trypsinized with trypsin solution (0.05% trypsin and 0.05% EDTA) for 5 min, collected by centrifugation (Hettich, Germany) at 3000 rpm at 3 min, resuspended in the medium and counted using a hemocytometer.
In vitro transfection and MTT assay
HeLa cells (5´103 cells/well) were seeded in 96 well tissue culture plates 24 h prior to the transfection experiments. Transfections were performed on cells that were approximately 70% confluent. Prior to transfection, the DMEM was removed and the cells were rinsed once with PBS. The wells were refilled (50 μl) with serum-free media. The cells were incubated with complexes in different ratios (N/P; 0.5/1, 1/1, 2/1, 5/1 and 10/1) for 6 h. Medium was removed and the cells were incubated with 10% FBS-containing culture medium for 24, 48 and 72 h.
The effect of different ratios of chitosan/pGMCSF complexes on cell proliferation and viability was determined by MTT assay (Cell Proliferation Kit I, Roche Diagnostic, Germany) [2,28]. HeLa cell proliferation and cell viability were determined by measuring the activity of dehydrogenase-enzyme as marker for the biological activity. After the incubation period, 10 μl of MTT was added to each well for three incubation times and incubated overnight. The purple formazan salt crystals were formed. MTT containing medium was removed and 100 μl solubilization buffer (10% SDS in 0.01 M HCl) was added to the formazan crystal formed by live cells. Plates were incubated for overnight and absorbances were measured at 550 and 600 nm using UV spectrophotometer (Shimadzu, Japan).
Statistical analysis
Data are expressed as mean value±SEM (standard error of the mean) and gene expression levels were estimated using Student’s t-test. p value of <0.05 was considered as statistically significant.
Results and Discussion
In this study, we have evaluated the availability of chitosan for delivery of GM-CSF plasmid in vitro. Chitosan/pGM-CSF complexes were prepared by self-assembled method [22]. The complex formation occurs because of ionic interaction between the positively charged amino groups of chitosan and the negatively charged phosphate groups of DNA. The molecular weight of chitosan is an important factor that affecting complex size, stability and cell uptake efficiency. The appropriate balance between protection and release of pDNA from complexes for biological functionality is needed [26]. High molecular weight chitosans have superior stability of complexes which is beneficial for the DNA protection in cells. Whereas, this situation could limited the release of DNA from chitosan complexes, hence low or delayed gene expression was obtained [29]. In contrast, complexes formed with rather lower MW chitosan (<10 kDa) are not sufficiently stable and can lead to weaker association between DNA and chitosan. It can not provide effective protection for DNA due to early dissociation and therefore show little or no transgene expression [26]. However, complexes prepared with 150 kDa molecular weight chitosan are shown higher stabilities and transfection efficiency [8,22]. Therefore, we selected in chitosan 150 kDa wt, not to rather low.
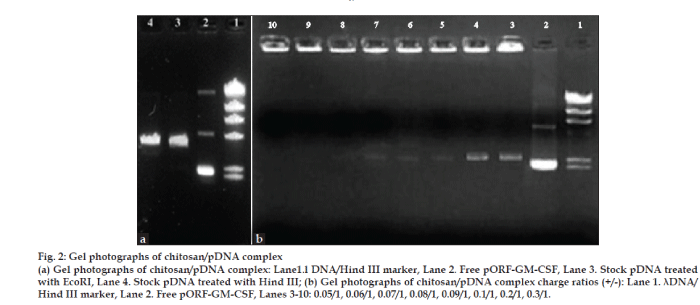
In order to determine the optimal complexation conditions between chitosan and pGM-CSF, it was necessary to evaluate the degree of binding between chitosan and plasmid DNA. Before complex formation, for the control of plasmid DNA, DNA was treated with the restriction endonucleases EcoRI and Hind III, which cleaved the plasmid in the CMV promoter region and in the ampicillin resistance region. These treatments resulted in pure linear plasmid forms as shown by agarose gel electrophoresis (fig. 2a). As seen in this figure, pGM-CSF was highly in supercoiled form. This is important for nucleic acid medicine.
Figure 2: Gel photographs of chitosan/pDNA complex
(a) Gel photographs of chitosan/pDNA complex: Lane1.l DNA/Hind III marker, Lane 2. Free pORF-GM-CSF, Lane 3. Stock pDNA treated with EcoRI, Lane 4. Stock pDNA treated with Hind III; (b) Gel photographs of chitosan/pDNA complex charge ratios (+/-): Lane 1. λDNA/
Hind III marker, Lane 2. Free pORF-GM-CSF, Lanes 3-10: 0.05/1, 0.06/1, 0.07/1, 0.08/1, 0.09/1, 0.1/1, 0.2/1, 0.3/1.
We first confirmed the complex formation between plasmid DNA and chitosan by electrophoresis on an agarose gel. When DNA is mixed with chitosan, electrostatic interactions mainly drive the formation of complexes. The migration of DNA on the agarose gel was retarded because of the charge neutralization and/ or an increase in the molecular size of the complexes (fig. 2b). When the charge was greater than 0.1/1 ratio, migration of DNA was completely retarded.
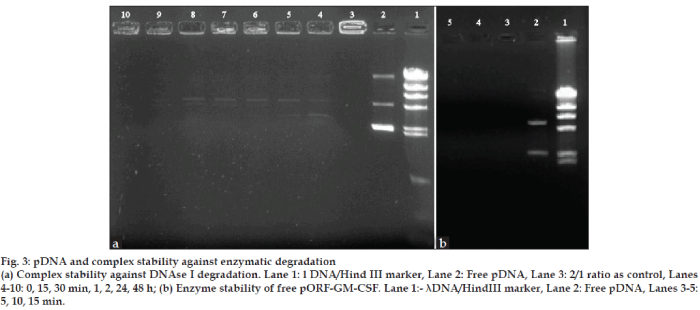
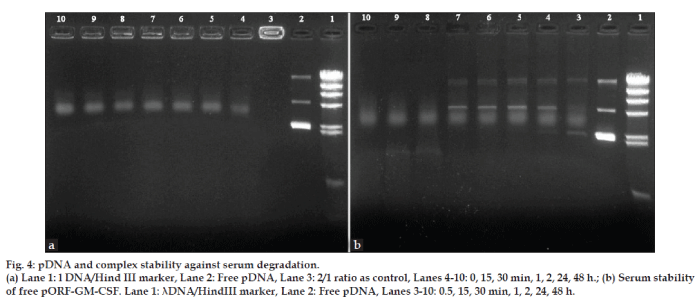
The development of gene delivery system that protect DNA against serum and enzyme is very important for the in vitro and in vivo applications [30,31]. The effect of DNase I and serum on DNA stability was studied and chitosan-based complexes protected GM-CSF gene against the serum and DNase I attacks. This data was confirmed by earlier report [8].
As seen in figs. 3 and 4, free pDNA was rapidly degraded with enzyme and serum but the formation of complexes with chitosan efficiently protected the plasmid DNA from degradation up to 48 h from enzyme and serum effects.
Figure 3: pDNA and complex stability against enzymatic degradation
(a) Complex stability against DNAse I degradation. Lane 1: l DNA/Hind III marker, Lane 2: Free pDNA, Lane 3: 2/1 ratio as control, Lanes 4-10: 0, 15, 30 min, 1, 2, 24, 48 h; (b) Enzyme stability of free pORF-GM-CSF. Lane 1:- λDNA/HindIII marker, Lane 2: Free pDNA, Lanes 3-5:
5, 10, 15 min.
Figure 4: pDNA and complex stability against serum degradation.
(a) Lane 1: l DNA/Hind III marker, Lane 2: Free pDNA, Lane 3: 2/1 ratio as control, Lanes 4-10: 0, 15, 30 min, 1, 2, 24, 48 h.; (b) Serum stability
of free pORF-GM-CSF. Lane 1: λDNA/HindIII marker, Lane 2: Free pDNA, Lanes 3-10: 0.5, 15, 30 min, 1, 2, 24, 48 h.
The N/P ratio of complexes is an important parameter affected complex properties and transfection efficiency. The N/P ratio is defined as the ratio between chitosan nitrogen per DNA phosphate [32]. Therefore, we next investigated changes of the size and zeta potential of the complexes at various charge ratios. The particle size and zeta potential of complexes are an important factor for the cell uptake in in vitro and in vivo studies [29]. Their sizes and surface charges increased with an increase of the charge ratio (Table 1). This data indicated similarity with earlier reports [26]. This is likely due to the intermolecular cross-linking between DNA strands, a phenomenon typically observed with either high DNA concentrations or an excess amount of polycations [25]. It was considered that a positive zeta potential value could benefit the complexes as a gene delivery system because the cell membrane has a net negative zeta potential value.
| Chitosan/ pDNAnanoplex | Zeta potential | Mean particle size |
|---|---|---|
| (+/-) | (mV±SD) | (nm±SD) |
| 0.5/1 | 6.8±2.3 | 113±5.8 |
| 1/1 | 13.7±3.7 | 132±2.7 |
| 2/1 | 24.5±5.1 | 143±6.9 |
| 5/1 | 26.0±7.2 | 173±12.3 |
| 10/1 | 29.0±6.5 | 199±10.8 |
The results are expressed as the mean±SD (n = 5).
Table 1: Zeta potential and particle size of chitosan/pgm-csf complexes
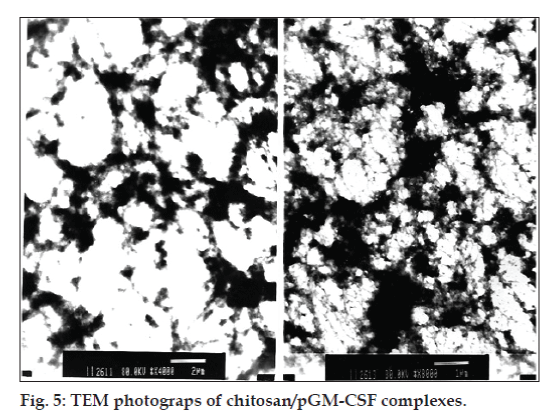
The shape of the chitosan/pGM-CSF complexes was investigated using transmission electron microscopy. Different topological conformations including spherical, annular, toroidal and globular morphologies were observed by changing of N/P ratios of the complexes [33-35]. In our study, the TEM photograps revealed that chitosan/pGM-CSF complexes has globular structure (fig. 5).
In order to investigate the carrier availability of chitosan for GM-CSF plasmid, in vitro gene transfection assay at different ratios of chitosan/ pGM-CSF complexes (0.5/1, 1/1, 2/1, 5/1 and 10/1) were performed in HeLa cell line. Cell proliferation and viability assays are importance for routine applications (the capability of the cells to incorporate a radioactively labeled substance [3H]- thymidine), or to release a radioisotope such as [51Cr] after cell lysis, the incorporation of 5-bromo-2’-deoxyuridine (BrdU) in place of thymidine for DNA synthesis and cellular proliferation in immuno-histo and cytochemistry, ELISA and FACS analysis (Cell Proliferation Kit I, MTT, Roche Applied Science, Mannheim, Germany). Among cell proliferation and viability assays, the MTT assay provides a rapid and versatile method for assessing cell viability, cytotoxicity and cell proliferation [36]. This test is a quantitative colorimetric method to determine cell proliferation and it utilizes the yellow tetrazolium salt [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide] which is metabolized by mitochondrial succinic dehydrogenase activity of proliferating cells to yield a purple formazan reaction product which is largely impermeable to cell membranes, thus resulting in its accumulation within healthy cells. Solubilization of the cells results in the revelation of the product that can readily be detected using a simple colorimetric assay. MTT also provides an indication of the mitochondrial integrity and activity [37].
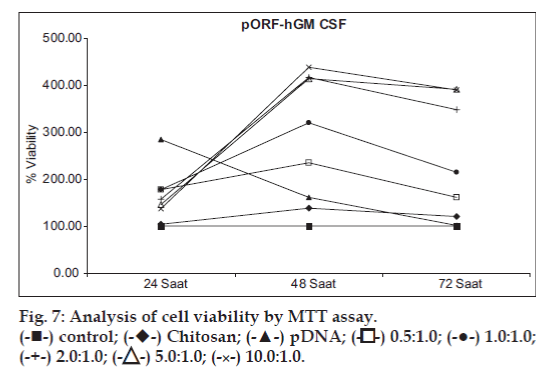
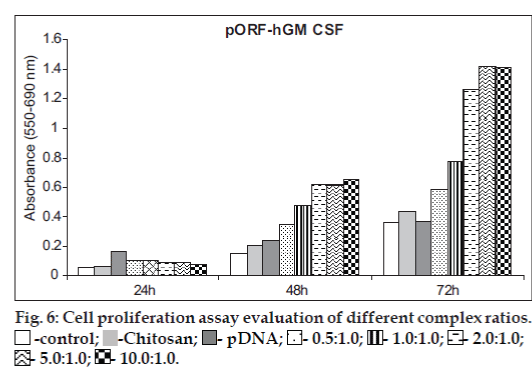
The effect of chitosan/pGM-CSF complexes on the proliferation of HeLa cells was assayed by MTT technique (hGM-CSF proliferation assay, MTT, Roche Applied Science, Mannheim, Germany) [2,28,38,39]. Changes of gene expressiondependent cell proliferation and cell viability during the experiment are given in figs. 6 and 7. Transfection efficiency of complexes was followed in the different incubation times. As seen in fig 6, after 24 h, free pDNA transfer showed higher efficiency than complexes (P>0.05). However, transfection efficiency of chitosan/pGM-CSF complexes was very low at 24 h. At 48 h, GM-CSF expression increased gradually in all the complexes except free plasmid (fig. 6). Highest gene expression were obtained with complexes after 72 h of transfection, but free plasmid showed lower values than 24 h. In general, differences between the values obtained at 48 and 72 h are statistically significant. (P<0.05). In addition to, expression increased as the chitosan/ pGM-CSF ratio increase, however there is no statistically significant difference between 2/1 and 5/1 ratios at 48 h or 5/1 and 10/1 ratios at 72 h (P>0.05). Similar data were reported by Ishii et al [32]. They reported that the transfection activity of chitosan/DNA complexes increased at a stoichiometries of complex. For all type of chitosan, gene expression varied with the stoichiometry of the complexes as given by the N/P ratio. In general, expression increased with an increasing N/P ratio to a certain point before declining [40]. Moreover, the cell uptake efficiency of the complexes were found to be in correlation with transfection activities by them [41]. Wang et al. [28] showed significantly enhanced the proliferation of endothelial progenitor cells of GM-CSF by MTT tecnique. In our study, MTT assay also demonstrated that the effect of chitosan/pGMCSF complexes on cell proliferation is dependent on N/P ratios and time. Furthermore, MTT data confirmed the accelerating effect of GM-CSF on the proliferation of HeLa cells.
According to these results, it can be said that chitosan has good carrier ability for GM-CSF coded gene, long and sustained GM-CSF release can be obtained with these complex forms. The useful potential of chitosan/ pGM-CSF complexes for the wound healing in terms of cell proliferation effect of GM-CSF is promising. The in vivo study is ongoing.
Acknowledgements
This study was supported by TUBITAK (SBAG-HD-155, 106S232). The authors wish to thank to TUBITAK for this financial support.
References
- Rodak BF, Fritsma GA, Doig K. Hematology: Clinical principles and applications. In: Andrew A, editor. Hematopoesis. 3rd ed. New York: Saunders Elsevier; 2007. p. 69-70.
- Ural AU, Avcu F, Zerman M, Yılmaz MI, Pekel A, Beyzadeoglu M. Differentian Effect of Thalidomide and GM-CSF Combination on HL-60 Acute PromyelocyticLeukemia Cells. ExpOncol 2006;28:216-9.
- Armitage JO. Emerging application of recombinant human granulocytemacrophage colony stimulating factor. Blood 1998;92:4491-508.
- Imokawa G, Yada Y, Kımura M, Morisaki N. Granulocyte⁄macrophage colony-stimulating factor is an intrinsic keratinocyte-derived growth factor for human melanocytes in UVA-induced melanosis. Biochem J 1996;313:625-31.
- Fang Y, Gong SJ, Xu YH, Hambly BD, Bao S. Impaired cutaneous wound healing in granulocyte⁄macrophage colony-stimulating factor knockout mice. Br J Dermatol 2007;157:458-65.
- Ojima T, Iwahashi M, Nakamura M, Matsuda K, Nakamori M. Benefits of gene transduction of granulocyte macrophage colony-stimulating factor in cancer vaccine using genetically modified dendritic cells. Int J Oncol 2007;31:931-9.
- Borrello I, Pardoll D. GM-CSF-based cellular vaccines: A review of the clinical experience. Cytokine Growth Factor Rev 2002;13:185-93.
- Akbuga J, Turan SO, Erdogan N. Plasmid-DNA loaded chitosan microspheres for in vitro IL-2 expression. Eur J Pharm Biopharm 2004;58:501-7.
- Meuli M, Liu Y, Liggitt D, Kashani-Sabet M, Knauer S, Meuli-Simmen C, et al. Efficient Gene Expression in Skin Wound Sites Following Local Plasmid Injection. J Invest Dermatol 2001;116:131-5.
- Nemunaitis J. GVAX (GMCSF gene modified tumor vaccine) in advanced stage non small cell lung cancer. J Control Release 2003;91: 225-31.
- Tangney M, Casey G, Larkin JO, Collins CG, Soden D. Non viralin vivo immune gene therapy of cancer: Combined strategies for treatment of systemic disease. Cancer ImmunolImmunother 2006;55:1443-50.
- Desire L, Mysiakine E, Bonnafous D, Couvreur P. Sustained delivery of growth factors from methylidenemalonate 2.1.2- based polymers. Biomaterials 2006;27:2609-20.
- Scheerlinck JP, Casey G, Mcwaters P, Kelly J, Woollard D, Lightowlers MW, et al. The immune response to a DNA vaccine can be modulated by co-delivery of cytokine genes using a DNA prime-protein boost strategy. Vaccine 2001;19:4053-60.
- Kass E, Panicali DL, Mazzara G, Schlom J, Greiner JW. Granulocyte/ Macrophage-Colony Stimulating Factor produced by recombinant Avian Poxviruses enriches the regional lymph nodes with antigen-presenting cells and acts as an immunoadjuvant. Can Res 2001;61:206-14.
- Kim JM, Lee M, Kim KH, Ha Y, Choi JK, Park SR, et al. Gene therapy of neural cell injuries in vitro using the hypoxia-inducible GM-CSF expression plasmids and water-soluble lipopolymer (WSLP). J Control Release 2009;133:60-7.
- Choi EW, Shin IS, Chae YJ, Koo HC, Lee JH, Chung TH, et al. Effects of GM-CSF gene transfer using silica nanoparticles as a vehicleon white blood cell production in dogs. ExpHematol 2008;36:807-15.
- Shalaby WS, Yeh H, Woo E, Hendren S, Corbett JT. Development of novel substrates for tumor immunotherapy. J Control Release 2003;91:209-24.
- Kawakami S, Higuchi Y, Hashida M. Nonviral approaches for targeted delivery of plasmid DNA and oligonucleotide. J Pharm Sci 2008;97:726-45.
- Borchard G. Chitosans for gene delivery. Adv Drug Del Rev 2001;52:145-50.
- Köping-Höggård M, Vårum KM, Issa M, Danielsen S, Christensen BE, Stokke BT, et al. Improved chitosan-mediated gene delivery based on easily dissociated chitosan polyplexes of highly defined chitosan oligomers. Gene Ther 2004;11:1441-52.
- Özbaş-Turan S, Aral C, Kabasakal L, Keyer-Uysal M, Akbuğa J. Coencapsulation of two plasmids in chitosan microspheres as a nonviral gene delivery vehicle. J Pharm PharmSci 2003;6:27-32.
- Özgel G, Akbuğa J. In vitro characterization and transfection of IL-2 gene complexes. Int J Pharm 2006;315:44-51.
- Lavertu M, Methot S, Tran-Khanh N, Buschmann MD. High efficiency gene transfer using chitosan/DNA nanoparticles with spesific combinations of molecular weight and degree of deacetylation. Biomaterials 2006;27:4815-24.
- Duceppe N, Tabrizian M. Factors influencing the transfection efficiency of ultra low molecular weight chitosan/hyaluronic acid nanoparticles. Biomaterials 2009;30:2625-31.
- Lee KY, Kwon IC, Jo WH, Jeong SY. Complex formation between plasmid DNA and self-aggregates of deoxycholic acid-modified chitosan. Polymer 2005;46:8107-12.
- Mao S, Sun W, Kissel T. Chitosan-based formulations for delivery of DNA and siRNA. Adv Drug Del Rev 2010;62:12-27.
- MacLaughlin FC, Mumper RJ, Wang J. Chitosan and depolymerized chitosan oligomers as condensing carriers for in vivo plasmid delivery. J Control Release 1998;56:259-72.
- Wang QR, Wang F, Zhu WB, Lei J, Huang YH. GM-CSF accelerates proliferation of endothelial progenitor cells from murine bone marrow mononuclear cells in vitro. Cytokine 2009;45:174-8.
- Köping-Höggard M, Tubulekas I, Guan H, Edwards K, Nilsson M. Chitosan as a non viral gene delivery system: Structure-property relationships and characteristics compared with polyethylenimineinvitro and after lung administration in vivo. Gene Ther 2001;8:1108-21.
- Barry ME, Pinto-Gonzalez D, Orson FM, McKenzie GJ, Petry GR, Barry MA. Role of Endogenous Endonucleases and Tissue Site in Transfection and CpG-Mediated Immune Activation after Naked DNA Injection. Hum Gene Ther 1999;10:2461-80.
- Huang M, Fong C-W, Khor E, Lim LY. Transfection efficiency of chitosan vectors: Effect of polymer molecular weight and degree of deacetylation. J Control Release 2005;106:391-406.
- Ishii T, Okahata Y, Sato T. Mechanism of cell transfection with plasmid/chitosan complexes. BiochimBiophysActa-Biomembranes 2001;1514:51-64.
- Liu W, Sun S, Cao Z, Zhang X, Yao K, Lu WW, et al. An investigation on the physicochemical properties of chitosan/DNA polyelectrolyte complexes. Biomaterials 2005;26:2705-11.
- Köping-Höggard M, Melnikova YS, Varum KM, Lindman B, Artursson P. Relationship between the physical shape and the efficiency of oligomeric chitosan as a gene delivery system in vitro and in vivo. J Gene Med 2003;5:130-41.
- Erbacher P, Zou S, Bettinger T, Steffan AM, Remy JS. Chitosan based vector/DNA complexes for gene delivery: Biophysical characteristic and transfection ability. Pharm Res 1998;15:1332-9.
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J Immunol Methods 1983;65:55-63.
- Mao S, Shuai X, Unger F, Simona M, Bi D, Kissel T. The depolymerization of chitosan: Effects on physicochemical and biological properties. Int J Pharm 2004;281:45-54.
- Fogolín MB, Eberhardt MO, Kratje R, Etcheverrigaray M. Choice of the adequate quantification method for recombinant human GMCSF produced in different host systems. Electron J Biotechnol 2002;5:243-50.
- Chan WK, Cheung CC, Law HK, Lau YL, Chan GC. Ganodermalucidum polysaccharides can induce human monocyticleukemia cells into dendritic cells with immuno-stimulatory function. J HematolOncol 2008;1:1-12.
- Strand SP, Lelu S, Reitan NK, Davies CL, Artursson P, Vårum KM. Molecular design of chitosan gene delivery systems with an optimized balance between polyplex stability and polyplex unpacking. Biomaterials 2010;31:975-87.
- Kim TH, Jiang HL, Jere D, Park IK, Cho MH, Nah JW, et al. Chemical modification of chitosan as a gene carrier in vitro and in vivo.ProgPolymSci 2007;32:726-53.
 -control;
-control;
 -Chitosan;
-Chitosan;
 - pDNA;
- pDNA;
 - 0.5:1.0;
- 0.5:1.0;
 - 1.0:1.0; -
- 1.0:1.0; -
 2.0:1.0; -
2.0:1.0; -
 5.0:1.0;
5.0:1.0;
 - 10.0:1.0.
- 10.0:1.0.