- *Corresponding Author:
- S. S. Ray
Department of Biotechnology and Medical Engineering, National Institute of Technology, India
E-mail: dawn.foster@yale.edu
| Date of Submission | 16 March 2014 |
| Date of Revision | 24 January 2015 |
| Date of Acceptance | 03 August 2015 |
| Indian J Pharm Sci, 2015;77(4):439-445 |
This is an open access article distributed under the terms of the Creative Commons Attribution‑NonCommercial‑ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non‑commercially, as long as the author is credited and the new creations are licensed under the identical terms.
Abstract
Pueraria tuberosa is known for its therapeutic potentials in cardiovascular disorders, but its effect in angiogenesis has not been studied so far. In this study, a computational approach has been applied to elucidate the role of the phytochemicals in inhibition of angiogenesis through modulation of vascular endothelial growth factor receptors: Vascular endothelial growth factor receptor-1 and vascular endothelial growth factor receptor-2, major factors responsible for angiogenesis. Metabolite structures retrieved from PubChem and KNApSAcK – 3D databases, were docked using AutoDock4.2 tool. Hydrogen bond and molecular docking, absorption, distribution, metabolism and excretion and toxicity predictions were carried out using UCSF Chimera, LigPlot+ and PreADMET server, respectively. From the docking analysis, it was observed that puerarone and tuberostan had significant binding affinity for the intracellular kinase domain of vascular endothelial growth factor receptors-1 and vascular endothelial growth factor receptor-2 respectively. It is important to mention that both the phytochemicals shared similar interaction profile as that of standard inhibitors of vascular endothelial growth factor receptors. Also, both puerarone and tuberostan interacted with Lys861/Lys868 (adenosine 5'-triphosphate binding site of vascular endothelial growth factor receptors-1/vascular endothelial growth factor receptors-2), thus providing a clue that they may enforce their inhibitory effect by blocking the adenosine 5'-triphosphate binding domain of vascular endothelial growth factor receptors. Moreover, these molecules exhibited good drug-likeness, absorption, distribution, metabolism and excretion properties without any carcinogenic and toxic effects. The interaction pattern of the puerarone and tuberostan may provide a hint for a novel drug design for vascular endothelial growth factor tyrosine kinase receptors with better specificity to treat angiogenic disorders.
Keywords
Angiogenesis, VEGFR1, VEGFR2, phytochemicals, molecular docking, ADME and Tox
Angiogenesis is a complex and intricate process wherein the preexisting blood vessels give rise to the new ones [1,2]. These blood vessels distribute the nutrients; carry out gas exchange and transport waste, thus maintaining a balanced and healthy environment throughout the tissue [3]. Optimally regulated angiogenesis is essential for proper growth and development of tissue. On the other hand, a deregulated angiogenesis leads to various diseases such as cancer, obesity, diabetes, arthritis, uterine bleedings, Alzheimer’s, osteoporosis, stroke and atherosclerosis. There exist various chemical mediators like vascular endothelial growth factors (VEGF), fibroblast growth factor (FGF), epidermal growth factor (EGF), angiostatin and interferon, which regulate the angiogenic process [4]. Amongst all such mediators, VEGF is the most prominent inducer of angiogenesis. VEGF is essential in migrating, proliferating and forming of capillary like structures by endothelial cells. VEGF executes its actions by binding to its receptors (VEGFR1 and VEGFR2) on the endothelial cells, thus activating the cascade of many downstream signaling pathways essential for angiogenesis [5]. Chemicals that interrupt this signaling cascade act as angiogenesis inhibitors. Angiogenic inhibitors have potential therapeutic utility in cancer, age related macular degeneration and other diseases where vascular proliferation leads to pathogenesis. Recently, a number of plant derived natural compounds or phytochemicals have been reported possesing a potential antiangiogenic activity [6,7]. Schindler et al. and Bagli et al. reported that the flavonoids like genistein and luteolin have a profound inhibitory effect on VEGF secretion in the cancer cells, thus inhibiting the tumor growth [8,9]. Also, curcumin has been reported for its antiangiogenic potentials through down‑regulation of VEGF expression and inhibition of VEGFRs [10].
Pueraria tuberosa or Indian Kudzu is widely known in traditional medicines to have many medicinal benefits. It has exhibited cardiotonic, aphrodisiac, antihyperglycemic, antilipidemic and galactogogic activities [11‑14]. Tubers of Pueraria tuberosa are rich in daidzin, puerarin, puerarone, genistein, puetuberosanol, tuberostan, tuberosin, and puerarin 4’,6’‑diacetate [15‑17]. Though it has much health beneficial effect but its effect on angiogenesis not been studied much.
In the current investigation, we have tried to evaluate the effect of various phytochemicals derived from aforesaid plant source on the Vascular Endothelial Growth Factor Receptors (VEGFRs) using in silico approach. The phytochemicals were screened for their toxic and mutagenic effects. Further, the nontoxic and nonmutagenic phytochemicals were docked onto kinase domain of VEGFR1 and VEGFR2.
Materials and Methods
Protein structure retrieval and active site predictions
The three‑dimensional structural models of intracellular kinase domain of VEGFR1 and VEGFR2 proteins were deducted from RCSB Protein Data Bank (http:// www.rcsb.org). Cleaning and optimizing the protein model geometry was carried out by the removal of miscellaneous ligands and other hetero‑atoms such as water and ions using the Argus Lab Software. Further, the protein models were used for the active site prediction using CASTp Calculations (Computed Atlas of Surface Topography of proteins) (http://stsfw. bioengr.uic.edu/castp/calculation.php). Amongst all predicted sites, the site having the catalytic amino acids was selected for docking. The clue about the amino acids involved in protein catalysis was gained from the UniProt Server (http://www.uniprot.org/).
Substrate selection
The three‑dimensional PDB structures of the phytochemicals present in the aforesaid plants and the standard FDA approved drug molecules were retrieved from KNApSAcK‑3D (http://knapsack3d. sakura.ne.jp/) and PubChem (http://pubchem.ncbi.nlm nih.gov/) databases using PRODRG Server (http:// davapc1.bioch.dundee.ac.uk/prodrg/). Further, energy minimization and ligand optimization were carried out using Argus lab software.
Molecular property predictions
The selected phytochemicals were examined for their molecular properties, absorption, distribution, metabolism and excretion (ADME) and toxicity using PreADMET (http://preadmet.bmdrc.org/). The phytochemicals, which passed the selection criteria were used for the docking analysis.
Molecular docking
The selected phytochemicals were docked into the active sites of the protein models using AutoDock4.2 software tool. In brief, polar hydrogen and Kollman charges were added to the protein models and docking was done by Lamarckian Genetic Algorithm using a standard protocol on the basis a population size of 150 randomly placed individuals; a maximum number of 2.5×107 energy evaluations, a mutation rate of 0.02 and a crossover rate of 0.8 [18]. Twenty independent docking runs were carried out and results were clustered according to the 1.0 Ǻ RMSD criteria. The grid maps representing the proteins were calculated using auto grid and grid size was set to 60×60×60 points with grid spacing of 0.375 Ǻ. Thereafter, the interaction pattern in the protein‑ligand complex was visualized using UCSF chimera [19] and LigPlot+ [20].
Results and Discussion
VEGFR1 and VEGFR2 are the transmembrane tyrosine kinase receptors with all the three domains: Extracellular domain, a transmembrane region, and an intracellular domain. Although VEGFR1 was earlier considered as a negative regulator of VEGFR2, but recently it was also found to accumulate the angioblast cells in the blood vessels. On the other hand, VEGFR2 plays a primary role in angiogenesis, vascular permeability and differentiation of the above mentioned angioblast cells [21].
Usually, the FDA approved antiangiogenic drugs are targeted at the intracellular domain of the VEGFRs, including the adenosine 5'-triphosphate (ATP) binding domain. ATP binding domain gains a special attention while considering receptor tyrosine kinases as ATP hydrolysis plays a crucial role in their activation. Thus, inhibition of this domain may potentially inhibit the signaling pathways. Keeping this perspective in mind, we evaluated the inhibitory potentials of phytochemicals by blocking the ATP binding domain of VEGFRs. Recently, interaction between juxtamembrane domain and catalytic domain of VEGFR2 has gained a significant attention. Juxtamembrane domain connects the transmembrane domain with the catalytic intracellular domain in receptor tyrosine kinases [22]. Solowiej et al. have reported that the presence of the Juxtamembrane domain of VEGFR2 has a substantial effect on inhibitor binding to the catalytic domain of VEGFR2. It was observed that the presence of juxtamembrane domain enhanced the affinity of Axitinib for the catalytic domain of VEGFR2 [23]. However, absence of an intact X‑Ray crystallographic structure for the same, limits it use for the present in silico investigation. Thus, three‑dimensional structures of catalytic domain of VEGFR1 (3HNG) and VEGFR2 (3VHE) were taken for the analysis from RSCB PDB (Table 1). For in silico analysis of the ligand‑protein interaction, the knowledge of active site of the protein is essential because ligands usually target the active sites. The active site of proteins was predicted using CASTp calculations.
| Proteins | Notations | PDB ID | Organism | References |
|---|---|---|---|---|
| Vascular endothelial | VEGFR1 | 3HNG | Homo | [24] |
| growth factor receptor 1 | Sapiens | |||
| Vascular endothelial | VEGFR2 | 3VHE | Homo | [25] |
| growth factor receptor 2 | Sapiens |
Table 1: Targets To Check The Effect On Angiogenesis
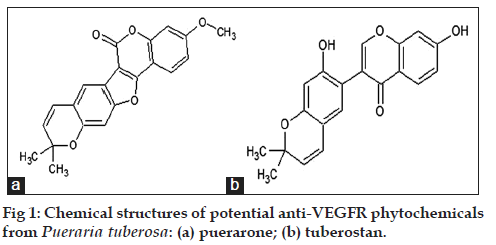
The three‑dimensional structure of phytochemicals was isolated from KNApSacK‑3D and PubChem database. Screening of the phytochemicals was carried out using PreADMET server and was based on their molecular properties including drug likeness, carcinogenicity and mutagenicity. The ligands which passed the selection criteria were further subjected to docking analysis. 18 out of 140 molecules were found to be nontoxic and were used for docking study using AutoDock4.2. Nontoxic 18 molecules were docked with the intracellular domain of VEGFRs, within which puerarone and tuberostan gave best binding with VEGFR1 and VEGFR2. Further study was continued with these two molecules. The origin and chemical structures of these two molecules is mentioned in Table 2 and fig. 1.
| KNApSAcK ID | Name of the Phytochemical | Origin | Ref |
|---|---|---|---|
| C00009871 | Puerarone | Puerariatuberosa(vidarikanda) | [26] |
| C00010049 | Tuberostan | Puerariatuberosa(vidarikanda) | [27] |
Table 2: Name And The Origin Of The Selected Phytochemicals
Puerarone and tuberostan, both were found to follow Lipinski’s rule of five which permit to assess the pharmacokinetics of the drug, including absorption, distribution, metabolism and excretion (ADME). Furthermore, they were recognized as nontoxic, nonmutagenic and noncarcinogenic in nature (Table 3).
| Properties | Puerarone | Tuberostan |
|---|---|---|
| Molecular weight (g/mol) | 336.33804 | 348.34874 |
| Log P | 5.09 | 5.87 |
| HB donor | 2 | 0 |
| HB acceptors | 5 | 5 |
| Ames test | Nonmutagen | Nonmutagen |
| Carcinogenicity | Noncarcinogen | Noncarcinogen |
| Human intestinal absorption (%) | 93.881 | 95.894 |
HB: Hydrogen bond
Table 3: Molecular Properties Of The Selected Phytochemicals - Puerarone And Tuberostan
Puerarone (KNApSAcK_3D ID C00009871) displayed best affinity towards the intracellular domain of VEGFR1 with a binding energy of ‑ 9.91 kcal/mol (Ki: 0.182 μM). On the other hand, tuberostan (KNApSAcK_3D ID C00010049) showed highest affinity towards the intracellular domain of VEGFR2 with a binding energy of ‑ 9.32 kcal/Mol (Ki: 0.148 μM). When compared with all the FDA approved drugs as mentioned in the Table 4, it was found that puerarone showed better binding affinity with VEGFR1 intracellular domain except pazopanib and vatalanib where it showed comparable results. Similarly, tuberostan showed better results than all the drugs except pazopanib and axitinib where it showed almost equal results. In Table 4, values in bold font represent best affinity of the test molecules with the intracellular domain of VEGFR1 and VEGFR2.
| PDB ID | Ligands | Minimum binding energy (kcal/mol)VEGFR1 | Ki (µM) | Hydrogen Bonds | Interacting amino acids |
|---|---|---|---|---|---|
| 3HNG | Test molecule | ||||
| Puerarone | −9.91 | 0.182 | 0 | ‑ | |
| Tuberostan | −8.44 | 0.646 | 0 | ‑ | |
| Standard inhibitors | |||||
| Sunitinib | −7.44 | 7.0 | 3 | Cys912 (2), Cys1039 | |
| Pazopanib | −10.19 | 0.034 | 3 | Lys861, His1020, Asp1040 | |
| Axitinib | −9.86 | 0.059 | 3 | Cys1018, Asp1040 (2) | |
| Vandetanib | −8.54 | 0.5493 | 0 | ‑ | |
| Vatalanib | −12.18 | 0.0018 | 3 | Glu878, Cys912, Asp1040 | |
| Sorafenib | −9.44 | 0.120 | 3 | Cys912 (2), Asp1040 | |
| VEGFR2 | |||||
| 3VHE | Test molecule | ||||
| Puerarone | −7.91 | 1.58 | 1 | Cys1045 | |
| Tuberostan | −9.32 | 0.148 | 0 | ‑ | |
| Standard inhibitors | |||||
| Sunitinib | −6.26 | 51.98 | 4 | Asp814, Lys868, Ala 881, Leu1049 | |
| Pazopanib | −9.58 | 0.095 | 1 | Asp1046 | |
| Axitinib | −9.94 | 0.052 | 1 | Lys868 | |
| Vandetanib | −7.36 | 4.01 | 0 | ‑ | |
| Vatalanib | −8.78 | 0.367 | 1 | Asp1046 | |
| Sorafenib | −7.7 | 2.26 | 1 | Lys868 |
Values in bold face indicate the test molecule with a better minimum binding energy. VEGFRs: Vascular endothelial growth factor receptors
Table 4: Binding Interactions Of Phytochemicals Along With Inhibitors With Vegfrs−Docking And Chimera Analysis
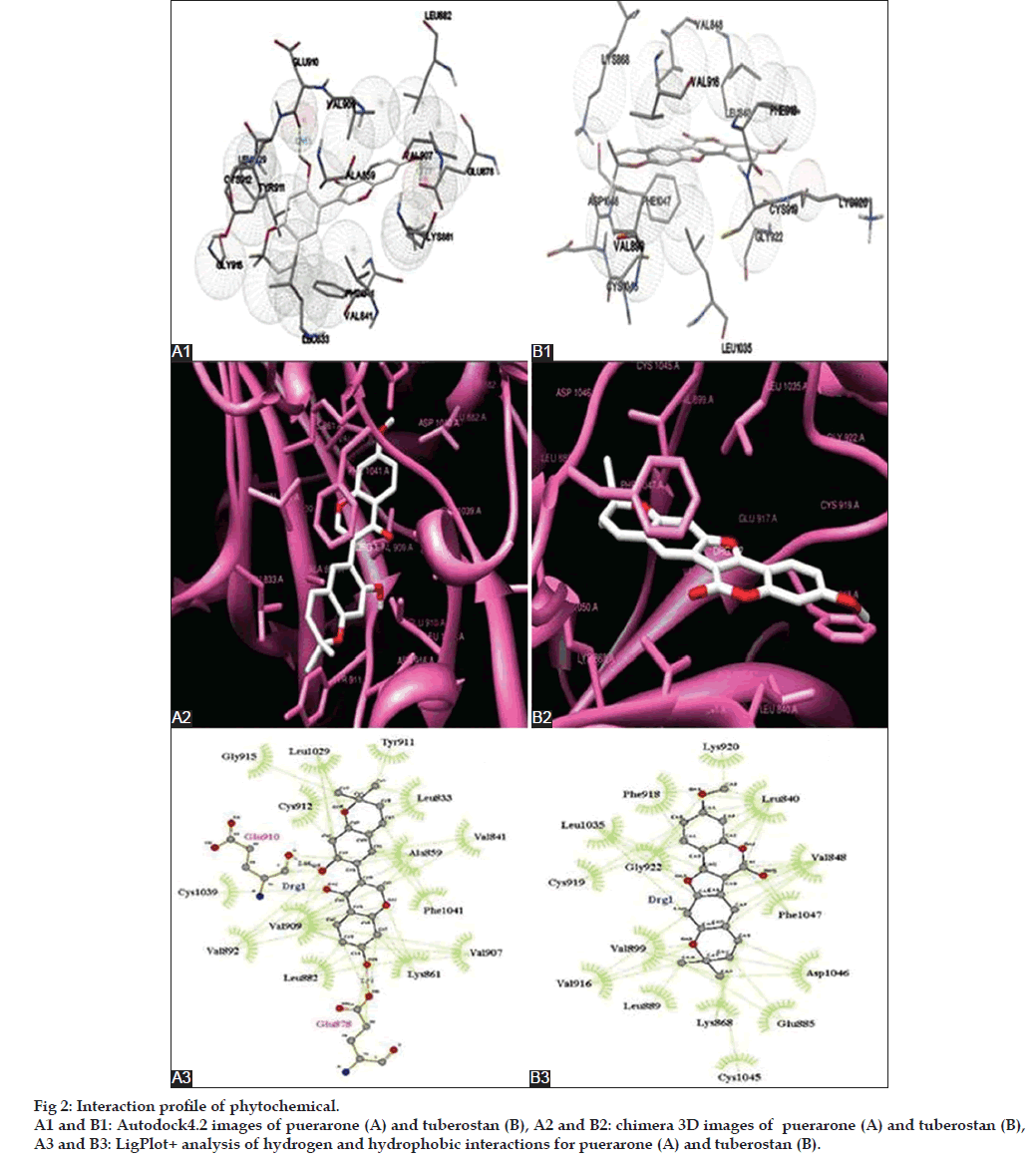
For an effective binding of a ligand to a protein receptor, the affinity of the ligand towards the protein as well as the stability of the protein ligand complex, are some of the important benchmarks. The above mentioned binding energies along with the interaction profile (hydrogen bonds and hydrophobic interactions) provide an important indication for the affinity and stability of ligand with the protein. The hydrogen bonds and hydrophobic interactions formed between proteins and phytochemicals were illustrated through LigPlot+ (Table 5). Interaction profiles were figured out only for the best possible orientation of the selected ligand molecule. For LigPlot+ analysis, an illustrative figure was generated for each ligand, describing bonded as well as nonbonded interactions between the ligands and protein.
| PDB ID | Ligands | Hydrogen Bonds | Hydrophobic Interactions |
|---|---|---|---|
| VEGFR1 | |||
| 3HNG | Test molecules | ||
| Puerarone | Glu878, Glu910 | Leu833, Val841, Ala859, Lys861, Leu882, Val892, Val907, Val909, | |
| Tyr911, Cys912, Gly915, Leu1029, Cys1039, Phe1041 | |||
| Tuberostan | ‑ | Leu833, Val841, Ala859, Lys861, Glu878, Ile881, Leu882, Ile885, | |
| Val892, Val907, Val909, Cys1018, Leu1029, Ile1038, Cys1039, Asp1040 | |||
| Standard inhibitors | |||
| Sunitinib | Cys912 (2), Cys1039 | Leu833, Val841, Ala859, Glu878, Leu882, Val907, Val909, Glu910, | |
| Tyr911, Gly915, Asn916, Leu1029, Asp1040, Phe1041 | |||
| Pazopanib | Asp1040 | Leu833, Val841, Lys861, Glu878, Val892, Val909, Tyr911, Cys912, | |
| His1020, Leu1029, Cys1039, Phe1041 | |||
| Axitinib | Asp1040 (3) | Asp807, Lys861, Glu878, Ile881, Leu882, Vaal892, Cys1018, Ile1019, | |
| His1020, Arg1021, Ile1038, Cys1039 | |||
| Vandetanib | ‑ | Val841, Glu878, Ile881, Leu882, Ile885, Val891, Val892, Val909, | |
| Leu1013, Leu1029, Ile1038, Cys1039, Asp1040, Phe1041 | |||
| Vatalanib | Glu878, Cys912, Asp1040 (2) | Leu833, Ala859, Lys861, Leu882, Val891, Val907, Val909, Glu910, | |
| Tyr911, Leu1013, Leu1029, Ile1038, Cys1039, Phe1041 | |||
| Sorafenib | Cys912, Asp1040 | Leu833, Val841, Ala859, Lys861, Glu878, Ile881, Leu882, Val892, | |
| Val909, Glu910, Tyr911, Gly915, Leu1013, Cy1018, Cys1039 | |||
| VEGFR2 | |||
| 3VHE | Test molecules | ||
| Puerarone | Glu885, Cys1045 | Leu840, Val848, Ala866, Lys868, Lue889, Val916, Gly922, Asn923, | |
| Lue1035, Asp1046, Phe1047 | |||
| Tuberostan | ‑ | Lue840, Val848, Lys868, Glu885, Leu889, Val899, Val916, Phe918, | |
| Cys919, Lys920, Gly922, Cys1045, Asp1046, Phe1047 | |||
| Standard inhibitors | |||
| Sunitinib | Asp814, Lys868, Ala 881, Leu1049 | Cys817, Leu882, Ser884, Glu885, Ile888, Asp1046, Gly1048 | |
| Pazopanib | Asp1046 | Val848, Ala866, Val867, Lys868, Glu885, Ile888, Leu889, Val899, | |
| Val914, Val916, Cys919, Gly922, Leu1035, Cys1045, Phe1047 | |||
| Axitinib | Asp1046 | Leu840, Val848, Ala866, Lys868, Glu885, Leu889, Val899, Val916, | |
| Phe918, Cys919, Gly922, Leu1035, Cys1045, Phe1047 | |||
| Vandetanib | Lys868, Glu885, Ile888, Leu889, Val899, Val916, Leu1035, Cys1045, | ||
| Asp1046, Phe1047, Gly1048 | |||
| Vatalanib | Val848, Lys868, Glu885, Leu889, Val899, Val914, Val916, Leu1035, | ||
| Cys1045, Asp1046, Phe1047 | |||
| Sorafenib | Lys868 | Leu840, Val848, Ala866, Glu885, Leu889, Val899, Val916, Glu917, | |
| Cys919, Phe918, Leu1035, Cys1045, Asp1046, Phe1047(2), Gly1048 | |||
Presence of bold faced amino acid indicates the interaction of the phytochemical at ATP binding site of the VEGFRs. VEGFRs: Vascular endothelial growth factor receptors
Table 5: Molecular Interactions Of The Selected Phytochemicals And Standard Inhibitors With Vegfrs − Ligplot+ Analysis
The hydrophobic amino acids of the intracellular domain of VEGFR1, which engage in the hydrophobic interactions with puerarone, are Leu833, Val841, Ala859, Lys861, Leu882, Val892, Val907, Val909, Tyr911, Cys912, Gly915, Leu1029, Cys1039, and Phe1041. Within the commercial inhibitors, vatalanib bound to the VEGFR1 intracellular domain more strongly than the others. Interestingly, the interaction profile of vatalanib interacted hydrophobically with Leu833, Ala859, Lys861, Leu882, Val907, Val909, Tyr911, Leu1029, Cys1039, and Phe1041, and formed hydrogen bonds with Glu878 which was similar to that of puerarone. It is important to mention that Asp1040 residue of VEGFR1 was involved in hydrogen bond formation with pazopanib, axitinib, vatalanib and sorafenib. Lys861 forms a major amino acid residue of ATP binding sites in intracellular domain VEGFR1 and interestingly, both, puerarone and most of the inhibitors interacted with it. This provides us the clue that puerarone may block the ATP binding site of VEGFR1, thus enforcing their inhibitory effect.
Equivalently, tuberostan was found to interact more effectively with the intracellular kinase domain of VEGFR2 through eleven hydrophobic amino acids, including Leu833, Lue840, Val848, Lys868, Glu885, Leu889, Val899, Val916, Phe918, Cys919, Lys920, Gly922, Cys1045, Asp1046, and Phe1047. Among the inhibitors, axitinib showed the highest affinity for intracellular domain of VEGFR2. Both tuberostan and axitinib were found to share their hydrophobic interactions with Leu840, Val848, Lys868, Glu885, Leu889, Val899, Val916, Phe918, Cys919, Gly922, Cys1045, and Phe1047. Also, tuberostan formed hydrophobic interaction with Asp1046, which on the contrary, was involved in the hydrogen bonding with axitinib. Similar to VEGFR1, Lys868 forms a crucial residue for ATP binding in the case of VEGFR2 which was recognized to show hydrophobic bonding with tuberostan and commercial inhibitors. Both the proteins and all the inhibitors showed hydrophobic interaction at the ATP binding site, but tuberostan, additionally, showed maximum hydrophobic interaction and highest binding affinity as per the LigPlot+ and Chimera analysis. Thus, making it a better candidate for inhibition of intracellular domain of VEGFR2. The interaction profile of the phytochemicals with the VEGFRs using Autodock4.0, Chimera and LigPlot+, can be viewed in fig. 2.
In this complete study in silico was done to explore the therapeutic potentials of the phytochemicals derived from Pueraria tuberosa. The overall analysis suggested that puerarone and tuberostan could show a high affinity towards the critical proteins VEGFR1 and VEGFR2, respectively. The study provides a hint for the design of novel drug leads against VEGFRs for the cure of multiple angiogenic diseases; exploiting the pharmacological aspects of these compounds. Further, in vitro and in vivo validation of the estimated mechanism as well as the therapeutic potentials of the selected phytochemicals is required.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- Laschke MW, Harder Y, Amon M, Martin I, Farhadi J, Ring A, et al. Angiogenesis in tissue engineering: Breathing life into constructed tissue substitutes. Tissue Eng 2006;12:2093‑104.
- Dai J, Rabie AB. VEGF: An essential mediator of both angiogenesis and endochondral ossification. J Dent Res 2007;86:937‑50.
- Carmeliet P. Angiogenesis in health and disease. Nat Med 2003;9:653‑60.
- Polverini PJ. The pathophysiology of angiogenesis. Crit Rev Oral Biol Med 1995;6:230‑47.
- Olsson AK, Dimberg A, Kreuger J, Claesson‑Welsh L. VEGF receptor signalling – In control of vascular function. Nat Rev Mol Cell Biol 2006;7:359‑71.
- Saha S, Islam MK, Shilpi JA, Hasan S. Inhibition of VEGF: A novel mechanism to control angiogenesis by Withaniasomnifera’s key metabolite withaferin A. In SilicoPharmacol 2013;1:11.
- Kadioglu O, Seo E, Efferth T. Targeting Angiogenesis By Phytochemicals. Med Aromat Plants 2013;2:1‑8.
- Schindler R, Mentlein R. Flavonoids and vitamin E reduce the release of the angiogenic peptide vascular endothelial growth factor from human tumor cells. J Nutr 2006;136:1477‑82.
- Bagli E, Stefaniotou M, Morbidelli L, Ziche M, Psillas K, Murphy C, et al.Luteolin inhibits vascular endothelial growth factor‑induced angiogenesis; inhibition of endothelial cell survival and proliferation by targeting phosphatidylinositol 3’‑kinase activity. Cancer Res 2004;64:7936‑46.
- Anand P, Thomas SG, Kunnumakkara AB, Sundaram C, Harikumar KB, Sung B, et al. Biological activities of curcumin and its analogues (Congeners) made by man and mother nature. BiochemPharmacol 2008;76:1590‑611.
- Hsu FL, Liu IM, Kuo DH, Chen WC, Su HC, Cheng JT. Antihyperglycemic effect of puerarin in streptozotocin‑induced diabetic rats. J Nat Prod 2003;66:788‑92.
- Tanwar Y, Goyal S, Ramawat K. Hypolipidemic effects of tubers of Indian kudzu (Puerariatuberosa). J Herb Med Toxicol 2008;2:21‑5.
- Gupta R, Sharma R, Sharma A, Choudhary R, Bhatnager A, Joshi Y. Antifertility effects of Puerariatuberosa. Root extract in male rats. Pharm Biol 2005;42:603‑9.
- Shukla S, Jonathan S, Sharma A. Protective action of butanolic extract of Puerariatuberosa DC. Against carbon tetrachloride induced hepatotoxicity in adult rats. Phytother Res 1996;10:608‑9.
- Khan RA, Agrawal PK, Kapil RS. Puetuberosanol, an epoxychalcanol from Puerariatuberosa.Phytochemistry 1996;42:243‑4.
- Joshi BS, Kamat VN. Tuberosin, a new pterocarpan from PuerariatuberosaDC. J ChemSoc Perkin 1 1973;9:907‑11.
- Koul A, Sumbali G. Detection of zearalenone, zearalenol and deoxynivalenol from medicinally important dried rhizomes and root tubers. Afr J Biotechnol 2008;7:4136‑9.
- Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, et al. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J ComputChem 1998;19:1639‑62.
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera – A visualization system for exploratory research and analysis. J ComputChem 2004;25:1605‑12.
- Laskowski RA, Swindells MB. LigPlot+: Multiple ligand‑protein interaction diagrams for drug discovery. J ChemInf Model 2011;51:2778‑86.
- Neufeld G, Cohen T, Gengrinovitch S, Poltorak Z. Vascular endothelial growth factor (VEGF) and its receptors. FASEB J 1999;13:9‑22.
- Roskoski R Jr. VEGF receptor protein‑tyrosine kinases: Structure and regulation. BiochemBiophys Res Commun 2008;375:287‑91.
- Solowiej J, Bergqvist S, McTigue MA, Marrone T, Quenzer T, Cobbs M, et al. Characterizing the effects of the juxtamembrane domain on vascular endothelial growth factor receptor‑2 enzymatic activity, autophosphorylation, and inhibition by axitinib. Biochemistry 2009;48:7019‑31.
- Seo EJ, Kuete V, Kadioglu O, Krusche B, Schröder S, Greten HJ, et al. Antiangiogenic activity and pharmacogenomics of medicinal plants from traditional Korean medicine. Evid Based Complement Alternat Med 2013;2013:1‑13.
- 5‑methyl‑ 4‑phenoxy‑5H‑pyrrolo[3,2 ‑d] pyrimidine derivatives: Novel VEGFR2 kinase inhibitors binding to inactive kinase conformation. Bioorg Med Chem 2010;18:7260‑73.
- Khan RA, Kapil RS. A facile synthesis of biogenetic precursor, puerarone, isolated from Pueraria sp. J HeterocyclChem 2001;38:1007‑9.
- Pandey N, Tripathi YB. Antioxidant activity of tuberosin isolated from Pueraria tuberose Linn. J Inflamm (Lond) 2010;7:47.