- *Corresponding Author:
- Monica R. P. Rao
AISSMS College of Pharmacy, Kennedy Road, Near RTO, Pune-411 001, India
E-mail: monicarp_6@hotmail.com
| Date of Submission | 7 November 2005 |
| Date of Revision | 9 January 2007 |
| Date of Acceptance | 2 March 2007 |
| Indian J Pharm Sci, 2007, 69 (2): 167-172 |
Abstract
Pharmacogenomics is an emerging arena which aims to improve the therapeutic efficacy of a drug based on the genetic profile of a patient. This technology tries to detect the link between the genetic blueprint of a person and the heterogenous response to a drug so as to use this information to maximize the efficacy of the drug. This article explores the genesis of this field along with its benefits and the various techniques used for the same.
Keywords
Genomics, pharmacogenomics, drug response, SNP’s, genetic profiling
Medicine and therapy are geared around taking statistical information about the general population and then applying it to the individual. Physicians are forced to take empirical decisions about types of treatment and drug dosage based on information that has been gathered on the basis of population averages, rather than individual profiles[1]. However, there is a great heterogeneity in the way individuals respond to drugs, in terms of both host toxicity and treatment efficacy. Potential causes for such variability in drug effects include the pathogenicity and severity of disease being treated, drug interactions and the individuals age, nutritional status, renal and liver function and concomitant disease.
Despite the potential importance of clinical variables, it isnow recognized that inherited differences in the metabolism and disposition of drugs and geneticpolymorphisms in the targets of therapy (such as receptors), can have even greater influence on the efficacy and toxicity of medications [2]. Clinical observations of such inherited differences in drug effects were first documented in the 1950s, exemplified by the relation between prolonged muscle relaxation after suxamethonium and an inherited deficiency of choline esterase [3], hemolysis after antimalarial therapy and inherited levels of erythrocyte glucose 6-phosphate dehdrogenase activity [4] and peripheral neuropathy of isoniazid and inherited differences in acetylation of this medication [5]. Such observations gave rise to the field of pharmacogenomics, which is the study of genetic variation underlying differential response to drugs. The term comes from the words pharmacology and genomics and is thus the intersection of pharmaceuticals and genetics. The distinction between terms pharmacogenomics and pharmacogenetics is considered arbitrary.
It combines traditional pharmaceutical sciences such as biochemistry with annotated knowledge of genes, proteins and single nucleotide polymorphisms (SNPs, pronounced ‘snips’) (http://www.oml.gov/hgmis/medicine/pharma.html). It applies the large-scale systematic approach of genomics to speed the discovery of drug response markers, to identify whether they act at a level of the drug target, drug metabolism or disease pathways. Pharmacogenetic studies have established the importance of polymorphic drug metabolizing enzymes such as CYP2D6, a member of cytochrome P450 family; in the differential response to drugs [6]. Recently, the genetic factors at the level of drug target or the disease pathway have been identified. For example, ApoE4, an allele at the apolipoprotien E locus, not only correlates with risk of developing Alzheimer’s disease but also predicts poor response to cholinesterase treatment [7-9].
This is an example of polymorphism within a disease related gene that is predictive of drug response. It is likely for many common disease like cancer, atherosclerosis and the neurodegenerative disorders, each represents a collection of separate conditions with a similar clinical endpoint, but they have distinct etiologies and therefore, distinct responses to therapy. That is the underlying hypothesis of pharmacogenomics.
Determinants of altered drug response
Pharmacogenomics has its roots in pharmacogenetics, a field that dates back to 1950s and studies the linkage of genetic differences (polymorphism) in drug metabolism with safety and efficacy of a therapeutic agent; as well as genetic differences in mechanism of drugs on its target.
Pharmacokinetic variations
With the advent of molecular biology, the isolation and sequencing of DNA clones of drug metabolizing enzymes also became possible. This allowed definition of the catalytic specificity and activity of many individual drug-metabolizing enzymes. Some of the best studied ones being the cytochrome P450 isoenzymes, N-acetyl transferase (NAT) isoenzymes, the UDP glucuronoyl transferase and the methyl transferases. Of these enzymes, the cytochrome P450 bear the brunt of the load, metabolizing drug into products that are more readily excreted into urine and faeces. For most of the activity of CYP 450 determines how much and how long a drug remains in the body. In humans, six forms of CYP 450 viz; CYP1A2, CYP2C9, CYP2C19, CYP2D6, CYP2E1 and CYP3A4 are largely responsible for eliminating drugs5. For CYP2C9, CYP2C19, and CYP2D6, polymorphisms account for the majority of inter individual variability. Individuals who are poor or slow metabolisers have enzyme deficiency related polymorphisms and are at an increased risk of concentration related toxicity. While enhance enzyme activity/ levels; they are characterized others have polymorphism (e.g., gene amplification) that as extensive or ultra rapid metabolisers and can be resistant to therapy. Quotable examples include poor metabolism of antidepressants, anti psychotics, b-blockers, antiarrhythmics, and others that leads to systemic accumulation and toxicity linked with polymorphisms in CYP2C19 and glucuronosyl transferase locus UGT2B7; poor metabolisers of psychotropic drugs as S-mephenytoin suffer from drowsiness or more serious side effects associated with CYP2D6 and CYP3A4. In patients associated with CYP2D6 and CYP3A4. In patients associated with CYP2D6 and CYP3A4. polymorphic for poor metabolisers forms of CYP2D6, terfenadine competes with erythromycin for CYP3A4, which slows the breakdown, leading to concentration-related toxicity [1].
Pharmacodynamic variations
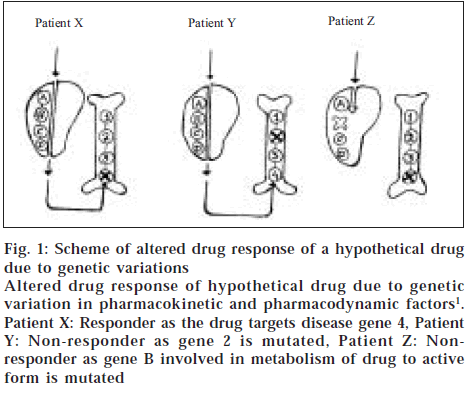
The mechanism of action of drug is also important in determining the differences in therapeutic efficacy and side effects between individuals. However, genetic variations in receptor function have been relatively rare in healthy individuals, this is because metabolizing enzymes are low affinity systems and receptors are high affinity systems with stringent structural requirements. Mutations in receptors may therefore be incompatible with life or cause severe disease. Differences in sequences of receptors subtypes for dopamine, serotonin and catecholamines may result in individual differences in behaviour and drug responses [3]. For example, large variations in efficacy of the psychotropic drug sumitriptan have been attributed to single amino acid substitutions in 5-hydroxy tryptamine (5HT) receptors [10]. Thus genetic variation in both pharmacokinetic and pharmacodynamic factors attributes to drug response. Some patterns do not respond to a given drug because it is not processed efficiently, others do not respond because the disease gene defect or its pathway is not targeted by the drug [1] (fig. 1).
Figure 1: Scheme of altered drug response of a hypothetical drug due to genetic variations
Altered drug response of hypothetical drug due to genetic variation in pharmacokinetic and pharmacodynamic factors1. Patient X: Responder as the drug targets disease gene 4, Patient
Y: Non-responder as gene 2 is mutated, Patient Z: Non-responder as gene B involved in metabolism of drug to active form is mutated
Identification of pharmacogenetic markers for predicting drug response
The methodology of genome wide DNA genotyping as applied to pharmacogenomic studies evolved from linkage and association studies of complex disease. Linkage studies involve genotyping families with micro satellite-markers and the goal is to correlate inheritance of a particular chromosomal region with inheritance of disease. However, because drug response data can rarely be obtained from multiple members of a family linkage studies are almost impractical in pharmacogenetics.
On the other hand, association studies correlate the presence of chromosomal region and a trait (disease or drug response) in unrelated individuals of a population. Because the common ancestry of unrelated individuals in an open population is much more distant than that of family members, the shared chromosomal region are much smaller, 100 kilo bases or less. Thus in order to perform association studies in an open population, 1,00,000 markers or more is required. Such dense maps are not yet available. However, rapid pace of DNA marker discovery together with novel genotyping techniques will soon permit genome-wide association studies.
These technical considerations favor the use of SNP’s rather than micro satellite markers used for linkage studies [6]. SNP’s are simple base substitutions that occur within and outside genes [11-13]. SNP’s can be used as a diagnostic tool to predict drug response. For SNP’s to be used in this way a person’s DNA must be sequenced for the presence of specific SNP’s. The traditional gene sequencing technology, which is very slow and expensive however, has impacted the widespread use of SNP’s as a diagnostic tool.
DNA micro arrays (or DNA chips) are an evolving technology that should make it possible for doctors to examine their patients for the presence of specific SNP’s quickly and affordably. A single micro array can now be used to screen 1,00,000 SNP’s found in patient’s genome in a matter of hours. As DNA micro array technology is developed further SNP screening in doctor’s office to determine patient’s response to a drug prior to drug prescription will be commonplace (http://www.oml.gov/hgmis/medicine/pharma.html). Pharmacogenomic applications of array based transcript profiling include analysis of patient tissues in response to therapy during the clinical trials. Expression based studies prove to be appropriate in cancers, because RNA can be obtained from biopsies and surgical specimens. This technology readily detects the somatic changes associated with the development of some tumors and their response to chemotherapy. These changes linked to therapeutic outcomes include the amplification of the oncogene erbB2, which predicts good response to cyclophosphomide-methotrexate-5 flurouracil (CMF) adjuvant therapy of breast cancer [14]. Current areas of technology development in transcript profiling include RNA amplification protocols that permit the use of low quantity of starting materials [15]. Laser capture microdissection (LCM) which facilitates isolation of individual’s cells from contaminating material in heterogeneous clinical samples, and continuing development of arrays and associated imaging systems to improve sensitivity [16]. In 1997, Cohen proposed a new and ambitious approach to mapping disease and drug response genes. This was brought into practice with the help of Abbott Labs; which developed and marketed diagnostic kits for stratifying patients to drug response. To find relevant genes the use of linkage disequilibrium studies was made. This is a relatively unproven approach, which relies on the detection of recombinatorially conserved regions around an ancestral mutation. Thus this study is an attempt beyond traditional linkage and association genetics [17].
Pharmacogenomics and its applications
To maximize the benefits of pharmacogenetics to drug discovery and the provision of better health care it is imperative to apply this science to identify targets and discover new medicines that will stop or prevent disease processes and discuss how pharmacogenetics will impact the pharmaceutical industry and the provision of health care. A limited number of molecular target families have been identified, including the receptors and enzymes, for which high through put screening is now possible. A good target is one against which many compounds can be screened rapidly to identify active molecules (hits). These hits can be developed into optimized molecules (leads), which have properties of well-tolerated and effective medicines. The best-validated targets are those that have medicines. The best-validated targets are those that have already produced well-tolerated and effective medicines in human (precedented targets). Many targets are chosen on the basis of scientific hypotheses and do not lead to effective medicines because the initial hypotheses are subsequently disproved [18,19].
Technologies such as differential gene expression, transgenic animal models, proteomics, in-situ hybridization and immuno- histo chemistry are used to imply relationships between a gene and a disease process [20] However, screening with these powerful tools has yet to lead to a specific target for a drug candidate with proven efficacy in humans or to a marketed drug.
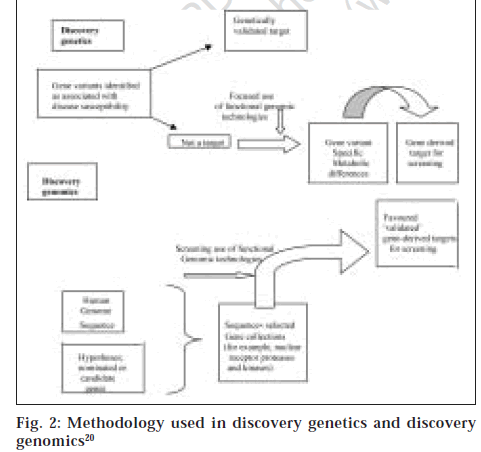
Nevertheless the differential gene expressions and proteomics are screening technologies that detect different levels and/or patterns of gene and protein expression intissues [21,22]. The identification of disease susceptible genes and study of the function of the susceptibility gene variants can be used to identify targets that will be related to the disease in patients and will therefore be validated. This process termed discovery genetics identifies fewer targets than the approach used in discovery genomics of data mining of sequences from the human genome project and similar programs with powerful bioinformatic tools that identifies gene families by locating domains that possess similar sequences [20] (fig. 2). Some of the successes of pharmacogenomics include the use of alosetron, a drug approved for treatment of female patients with diarrhoea-predominant irritable bowel syndrome (IBS) in the US [23,24].
Since the diagnosis of the IBS can be imprecise, a simple medicine response profile is done to determine whether the patients’ symptoms will be alleviated by alosetron[25].
Herceptin [Genentech, San Francisco], a monoclonal antibody that targets the protein product of the HER2 oncogene [also known as ERbB2] expressed in a subpopulation of breast cancer patients and Gleevec (Novartis, Basel, Switzerland) a drug designed to treat patients with chronic myeloid leukemia (CML) resulting from the Philadelphia chromosome translocation and a test linking hypersensitivity reactions to the HIV/AIDS drug Abacavir (Glaxo Smith Kline, Brentford UK) to the HLA-B* 5701 halotype are some of the treatments based on pharmacogenomics[26,27].
Use of pharmacogenetic technologies will help in identifying medicines profiles during the phase II clinical trials. This can be used in the selection of patient groups in the phase III studies. This is likely to make the trials smaller faster and more efficient [28]. Post approval surveillance for detection of adverse drug reactions (ADRs) can be streamlined by documenting and characterizing the DNA of patients experiencing an ADR and comparing with DNA from control patients who received the drug but did not experience any ADR. This would enable abbreviated SNP profiles for patients susceptible to the ADR to be determined [29] .
The other anticipated benefits of pharmacogenomics include more accurate methods of determining appropriate drug dosages. The ability of a person’s body to process a drug that is his genetic profile will form the basis for dosage calculations instead of the weight and age of the patient. This will maximize the therapy’s value and decrease the likelihood of overdose. Knowledge of the genetic characteristics of an individual will allow prospective suggestions of altered lifestyle with the view to avoid or suppress the disease. Besides this, knowledge of particular disease susceptibility will allow careful monitoring and treatments can be introduced at an appropriate stage to maximize therapy.
Economic and regulatory concerns
Although the impact of pharmacogenomics on various aspects of drug development can never be overestimated, one of the major concerns for pharmaceutical companies to venture into this field is its economic viability. Two types of stratification have been envisioned, viz, patient stratification and disease stratification. The features of patient stratification include differential dosing based on patient genotype, which could lead to increased market size. Disease stratification on the other hand involves giving different drugs based on patient genotype, which would decrease the market size for an individual drug [30] For example, people with an ultra rapid metabolizing allele of Cytochrome P450 i.e., CYP2D6 will require increased drug dosages to ensure that the drug is not deactivated too soon to gain a clinical effect [31]. This patient stratification could increase the market size and revenue but would not require the same R and D investments as a new drug.
However disease stratification involves subdividing diseases and prescribing different medicines to patients with similar symptoms based on their genetic profile. For example mutations in BRCA1 or BRCA2 genes can lead to ovarian cancer in women, treatment for which will be different than these with mutations in hereditary nonpolyposis colorectal cancer genes [which affect the risk of developing endometrial cancer]. Though this will increase the efficacy of the drug and the cost, it will restrict the market size and if a single company is not able to develop drugs for all segments of its existing market, the revenue loss will be considerable.
The stratification effect of pharmacogenomics adds complexity to the regulatory requirements, as provision will have to be made for genetic tests, which is a prerequisite for enjoying the benefits of this field. The International Conference on Harmonization of technical requirements for registration of pharmaceuticals for human use, which brings together regulatory officers from Japan, Europe, and the United States has recommended that additional studies be required when drugs are submitted for licensing in a new jurisdiction to better define the clinical characteristics of the drug in the original patient population [32,33]. Also because of the intricacies of drug response profiling on the basis of genetic variability, exclusive national regulation of new medicines will be an inadequate means of appraisal in the future. The impact of pharmacogenomics on the legislation of orphan drugs will also be considerable. The US-FDA defines an orphan drug as one, which is used for treatment of diseases that occur in less than 2,00,000 patients [orphan disease] (http://www.news.bmn.com/hmsbeagle/). In the US,orphan products are fast tracked to approval because of the life threatening nature of some orphan diseases, a lack of other effective treatments and the reduced trial size required to license a drug that is badly needed by a small population [34]. Pharmacogenomics studies could lead to an increase in the number of orphan drug applications as parallel trials for a multitude of drugs targeted to various subgroups of a disease population need to be conducted and this will increase the expenditure [35].
Another significant outcome will be the reevaluation of drugs that previously failed during development because of serious ADR’s in a small number of patients or were withdrawn post approval [36]. An example of this is GlaxoSmithKline’s Lotronex, a drug initially prescribed for the treatment of IBS, which was withdrawn from the US market since it caused serious intestinal complications in a small number of patients. However it was reapproved with restrictions that it should be prescribed only those patients for whom existing therapies had failed [37].
Ethical issues
Social and ethical barriers towards any scientific advancement have been the norm since ages. With respect to pharmacogenomics these barriers stem from the inaccurate vernacular usage creating a confused public perception of genetic testing. More clarity in language will ensure that pharmacogenetics/ genomics is not confused with gene therapy, genetically modified foods, genetic engineering or with cloning of humans or other organs. Precise differentiation of the associated terminologies will clear any doubts regarding this exciting field.
Conclusion
From the precluding discussions, it is obvious that pharmacogenomics portends fascinating implications for the medical and pharma sector. Its impact on various aspects of drug development will be tremendous. Reaping the dividends of this field requires a clear focus on technologies and a synchronized multidisciplinary effort. At the same time, it is imperative to frame comprehensive regulatory policies at an international level, considering the vast domain of the field. In developing countries this field is still nascent and awareness negligible. Though the benefits are profound, the cost factor will be a major deterrent not only for the layman but also for the pharmaceutical companies. A concerted effort needs to be taken by the governments, regulatory bodies and pharma majors to ensure that high efficacy drugs reach the common man at a feasible low cost.
Acknowledgements
The authors would like to acknowledge the regular counsel and guidance of Dr. K. G. Bothara, Principal, AISSMS College of Pharmacy, Kennedy Road, Pune- 411 001.
References
- Marshall A. Laying the foundation for personalized medicines. Nat Biotech 1997;15:954-7.
- Evans WE, Relling MV . Pharmacogenomics: Translating functional genomics into the rational therapeutics. Science 1999;286:487-91.
- Kalow W. Familial incidence of low pseudomonas cholinesterase level. Lancet 1956;211:576.
- Carson PE, Flanagan CE, Alving AS, Ickes CE Enzymatic deficiency in primaquine-sensitive erythrocytes. Science 1956;124:484-5.
- Evans DA, Manley KA, McKusick VA. Genetic control of isoniazide metabolism in man. Br Med J 1960;2:485-91.
- Kleyn PW, Veesell ES. Genetic variation as a guide to drug development. Science 1998;281:1820-1.
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al . Gene dose of Apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science 1993;261:921-3.
- Poirier J, Davignon J, Bouthillier D, Kogan S, Bertrand P, Gauthier S. Apolipoprotein E polymorphism and Alzheimer's disease. Lancet 1993;342:697-9.
- Poirier J, Delisle M, Quirion R, Aubert I, Farlow M, Lahiri D, et al . Apolipoprotein E4 allele as a predictor of cholinergic deficits and treatment outcome in Alzheimer's disease. Proc Natl Acad Sci USA 1995;92:12260-4.
- Lichter J, McNamara D. What is in a gene: Using genetic information for the design of clinical trials. Curr Opin Biotechnol 1995;6:715-7.
- Zhao LP, Aragaki C, Hsu L, Quiaoit F. Mapping of complex traits by single-nucleotide polymorphism. Am J Hum Genet 1998;63:225-40.
- Brookes AJ. The essence of SNP's. Gene 1999;234:177-86.
- Hacia JG, Fan JB, Ryder O, Jin L, Edgemon K, Ghandour G, et al . Determination of ancestral alleles for human single nucleotide polymorphism using high-density oligonucleotide arrays. Nat Genet 1999;22:164-7.
- Muss HB, Thor AD, Berry DA, Kute T, Liu ET, Koerner F, et al . c-erb-2 expression and response to adjuvant therapy in women with node-positive early breast cancer. N Engl J Med 1994;330:1260-6.
- Van Gelder RN, von Zastrow ME, Yool A, Dement WC, Barchas JD, Eberwine JH. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci 1990;87:1663-7.
- Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, et al . Laser capture microdissection. Science 1996;274:998-1001.
- Marshall A. Genset-Abbott deal heralds Pharmacogenomic era. Nat Biotech 1997;15:829-30.
- Curran ME. Potassium ion channels and human disease: Phenotypes to drug targets? Curr Opin Biotech 1998;9:565-72.
- Marton MJ, DeRisi JL, Bennett HA, Iyer VR, Meyer MR, Roberts CJ, et al . Drug target validation and identification of secondary drug target effects using DNA microarrays. Nat Med 1998;4:1293-301.
- Roses AD. Pharmacogenetics and the practice of medicine. Nature 2000 ; 405:857-65.
- Blackstock WP, Weir MP. Proteomics quantitative and physical mapping of cellular proteins. Trends Biotechnol 1999;17:121-7.
- Kozian DH, Kirschbaum BJ. Comparative gene expression analysis. Trends Biotechnol 1999;17:73-8.
- Talley NJ. Irritable bowel syndrome: Disease definition and symptom description. Eur J Surg 1998;583:24-8.
- Paterson WG. Recommendations for the management of irritable bowel syndrome in family practice. CMAJ 1999;161:154-60.
- Hamm LR, Sorrells SC, Harding JP, Northcutt AR, Heath AT, Kapke GF, et al . Additional investigations fail to alter the diagnosis of irritable bowel syndrome in subjects fulfilling the Rome criteria. Am J Gastroenterol 1999;94:1279-82.
- Lindpaintner K. Genetics in drug discovery and development: Challenge and promise of individualizing treatment in common complex disease. Br Med Bull 1999;55:471-91.
- Lindpaintner K. The impact of pharmacogenetics and Pharmacogenomics on drug discovery. Nat Rev Drug Discov 2002;1:463-9.
- Bonnie A, Fijal MS, Hall JM, Witte JS. Clinical trials in the genomics era. Effects of protective genotypes on sample size and duration of trial 2000. Controll Clin Trials 2000;21:7-20.
- Lazarou J, Pomeranz BH, Corey PN. Incidence of adverse drug reactions in hospitalized patients: A meta-analysis of prospective studies. J Am Med Assoc 1998;279:1200-5.
- Shah J. Economic and regulatory considerations in Pharmacogenomics for drug licensing and healthcare. Nat Biotech 2003;21:747-53
- Wolf CR, Smith G. Pharmacogenetics. Br Med Bull 1999;55:366-86.
- Hodgson J, Marshall A. Pharmacogenomics: Will the regulators approve? Nat Biotechnol 1998;16:243-6.
- Shah RR. Regulatory aspects of integration of pharmacogenetics into drug development. Int J Pharmaceut Med 2001;15:67-9.
- Milne CP. Orphan products-pain relief for clinical development headache. Nat Biotech 2002;20:780-4.
- Lesko LJ, Woodcock J. Pharmacogenetics-guided drug development: Regulatory perspective. Pharmacogenomics J 2002;2:20-4.
- Branca M. FDA fosters Pharmacogenomics. Bio-ITWorld 2002;1:45.
- McCarthy M. FDA allows controversial bowel drug back on to market. Lancet 2002;359:2095.