- *Corresponding Author:
- A. P. Mishra
Department of Pharmaceutical Chemistry, H. N. B. Garhwal University, Srinagar Garhwal-246 174, India
E-mail: abhaypharmachemhnbgu@gmail.com
| Date of Submission | 14 September 2017 |
| Date of Revision | 10 December 2018 |
| Date of Acceptance | 03 April 2019 |
| Indian J Pharm Sci 2019;81(3):565-568 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The present study was designed to evaluate the antidiabetic potential of diospyrin isolated from Diospyros lotus, using protein tyrosine phosphatase 1B enzyme as the target. Molecular binding mode of diospyrin to protein tyrosine phosphatase 1B was essential to explore its molecular interactions. Molecular docking, the simulation technique used to model the interaction between two molecules were performed using Open Eye software. This compound exhibited significant protein tyrosine phosphatase 1B inhibitory activity (IC50 value: 27.59±0.03 μM). Molecular docking studies showed significant molecular interactions of the diospyrin with Gly 220, Tyr 46, Val 49 and Asp 48 inside the active site of protein tyrosine phosphatase 1B. The in silico result builds prospect that diospyrin can be further developed as a new lead compound targeting protein tyrosine phosphatase 1B inhibition.
Keywords
Diospyros lotus, Ebenaceae, diospyrin, PTP1B, molecular docking
The genus Diospyros belonging to family Ebenaceae is well distributed in the equatorial and subtropical areas of the world. Around 750 species of the genus are known around the world, of which 24 are native to India [1,2]. Diospyros lotus, also known as date plum, is native to the Himalayan province and grows up to a medium height [3]. Diospyros species are documented for medicinal uses in various traditional healing systems all over the world. All plant parts are used for therapeutic purposes such as treatment of lumbago, pain, fever, inflammation, and microbial infections [4]. In addition, Diospyros species are carminative, astringent, sedative and febrifuge [1,5,6]. Several antiinflammatory triterpenoids belonging to oleanane, lupane and ursane series have been isolated [7-9]. Furthermore, the presence of naphthalene derivatives, naphthoquinones and dimeric naphthoquinones with significant activities has been investigated in several species [10,11].
Protein tyrosine phosphatases (PTPs) play a key role in cellular processes like cell growth, proliferation, and differentiation. Several recent studies have revealed that PTPs are involved in various metabolic processes, immunological response, cell-cell adhesion and cellmatrix interactions [12-14]. In fact, PTP antagonizes protein tyrosine kinases in multiple signaling pathways. PTP1B is an intracellular non-receptor type PTP, validated as a target for many diseases, including diabetes and cancer [15]. Extensive biochemical and genetic investigations have shown that PTP1B is involved in the negative control of insulin- and leptinreceptor [16]. PTP1B-knockout mice have revealed an increased sensitivity to insulin, enhanced glycemic control, and resistance to diet-induced obesity [17]. Therefore, inhibition of PTP1B provides a novel approach in regulating type 2 diabetes and obesity, and many laboratories worldwide are currently focusing on novel PTP1B inhibitors from natural as well as synthetic sources. In this study, diospyrin, a bisnaphthoquinonoid from D. lotus, was screened for PTP1B inhibitory activity. Diospyrin and its analogs were found to possess antimycobacterial [18] properties. Diospyrin inhibited lipoxygenase [19], urease [20], and hyaluronidase [21], among other enzymes. In this investigation, through molecular docking simulation studies, it was shown, for the first time, the molecular mechanism of PTP1B inhibition by this dimeric naphthoquinone.
Diospyros lotus L. roots were collected from Toormang Razagram, Dir, KPK, Pakistan, in May 2009. The samples were identified in the Department of Botany, University of Peshawar, Peshawar, Pakistan and a voucher specimen (Bot. 20036) was deposited in the Herbarium of the same department. Shade-dried D. lotus roots (14 kg) were powdered and kept at room temperature in methanol for 6 days, with constant stirring by percolation method. Afterward, the extract was concentrated by evaporating solvent via rotary vacuum evaporator, under reduced pressure at a temperature below 55°. The process was repeated four times until the extraction was completed. Finally, the dark green residue (202 g) was obtained. Diospyrin was isolated from the methanol extract as previously reported [22].
The PTP1B inhibition assay was carried out in 96-well plates using 3,3-dimethyl glutarate buffer (pH 7.0). The reaction mixture contained various concentrations of test compounds and standard compound ursolic acid (Sigma), p-nitrophenol phosphate (pNPP, 1 mM) and PTB1B (10 mM). After incubation at 27° for 40 min, the absorbance of the released pNPP was measured at 405 nm. Assay was carried out in triplicates and IC50 values were calculated.
Open Eye Docking 3.0.0. software was used to dock the OMEGA pre-generated multi-conformer library [23]. Fast Rigid Exhaustive Docking (FRED), a docking program, which docks molecules from a multi-conformer database into a receptor site, using an exhaustive search algorithm, was used, which thoroughly docks/scores all conceivable positions of each ligand in the binding site [24]. The comprehensive search is based on rigid rotations and transformations of each conformer within the binding site well-defined by a box. FRED program filters the poses ensemble, by refusing the ones that clash with PTP1B, or that has not adequate contacts with the protein. Final poses can then be scored or re-scored by using one or more scoring functions. In the current study, the smooth shape-based Gaussian scoring function (shapegauss) was selected to analyze the shape complementarity between individual ligand and the binding pocket [24]. Default FRED protocol was used except for the size of the box defining the binding sites. In an effort to improve the docking-scoring performance, wideranging docking was performed with shapegauss, applying the optimization mode. The latter involves an organized solid body optimization of the top graded poses from the exhaustive docking. Three diverse boxes were explored for PTP1B (PDB: 2BGD) and 3 different imitations were carried out with an added value of 8 Å, around the reference ligand.
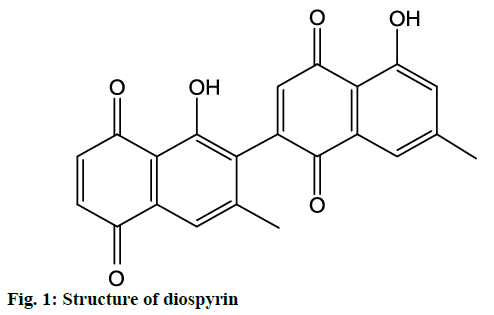
In the present study, molecular docking analysis was used to reveal the favorable binding interactions between PTP1B enzyme and diospyrin (fig. 1). The latter showed significant enzyme inhibitory activity (IC50 27.59±0.03 μM). The activity of ursolic acid (positive control) recorded in the present study was IC50 3.21±0.02 μM, which was in agreement with that previously reported for the compound [18,19]. It appears that this is the first report on the identification of potentially antidiabetic phytochemical from D. lotus.
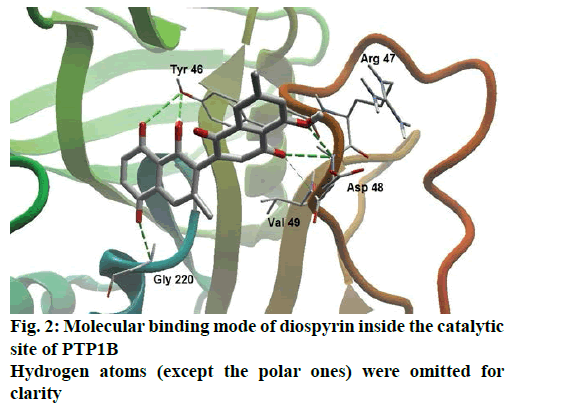
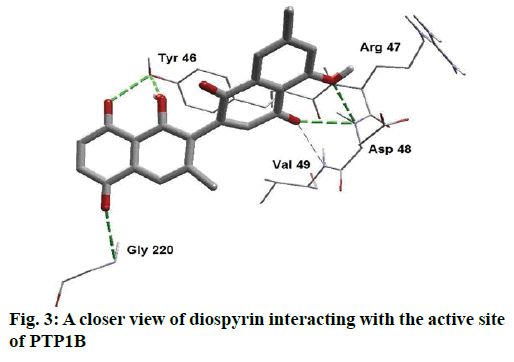
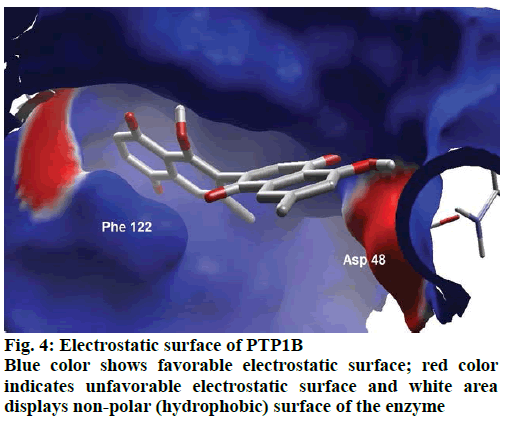
Detailed computational and structural insights revealed promising interactions between the PTP1B enzyme and diospyrin. Various moieties of diospyrin showed significant and multiple binding interactions with important amino acid residues of PTP1B (figs. 2 and 3). Interestingly, both phenolic and adjacent carbonyl groups on one side of the quinone dimer revealed simultaneous hydrogen bonding interactions with Tyr 46 at the active site of PTP1B, at the distance of 3.08 and 3.00 Å, respectively. One the same monomer, the second carbonylic oxygen held Gly 220 (3.00 Å) tightly on opposite side with respect to Tyr 46. Such bidirectional interactions indicate that diospyrin might closely and effectively fill the binding pocket, thus blocking the biochemical catalysis of substrate molecules. Asp 48 is the critical and terminal amino acid lining the outer rim of the binding pocket of PTP1B. The phenolic group showed a promising contact with Asp 48 via hydrogen bonding at a distance of 3.00 Å. This favorable interaction of Asp 48 was further reinforced by the simultaneous, but strong and favorable hydrogen bonding interaction of adjacent carbonylic oxygen from the second monomer with Asp 48 (2.85 Å) and Val 49 (3.22 Å). Phe 122 is an amino acid surrounding another side of the outer surface of the active site, which showed favorable steric interactions with diospyrin (fig. 4). The rotatable bond between both quinone monomers afforded effective molecular penetration of diospyrin inside the active site, which was supported by favorable electrostatic and steric interactions. This study indicates the promising potential of diospyrin to be further explored and developed as a new lead compound, targeting PTP1B, for diabetes and cancer alleviation.
Recently, the enzyme inhibitory activity of diospyrin was reported on DNA gyrase of Mycobacterium tuberculosis [20]. Its derivatives and analogs have demonstrated anticancer and antiparasitic activities [2,21]. In vivo antidiabetic effects of different species belonging to the same genus, namely Diospyros kaki L., Diospyros lotus L. and Diospyros peregrina (Gaertn) Gürke, have been previously demonstrated [22-24]. In conclusion, these results appear to be the first report on the antidiabetic potential of diospyrin and D. lotus. Further studies based on in vivo models are needed to elucidate this relevant biological activity.
Acknowledgements
Authors thank the Higher Education Commission for financial support. Authors also thank Dr. Abdur Rashid, plant taxonomist, Department of Botany, University of Peshawar, Peshawar, Pakistan for identifying the plant material collected.
Conflicts of interest:
The authors declare that there is no conflict of interest.
References
- Rauf A, Uddin G, Raza M, Patel S, Bawazeer S, Ben Hadda T, et al. Urease inhibition potential of Di-naphthodiospyrol from Diospyros lotus roots. Nat Prod Res 2017;31:1214-8.
- Uddin G, Rauf A, Siddiqui BS, Muhammad N, Khan A, Shah SU. Anti-nociceptive, antiinflammatory and sedative activities of the extracts and chemical constituents of Diospyros lotus L. Phytomedicine. 2014;21:954-59.
- Rauf A, Uddin G, Siddiqui BS, Khan H. In vivo sedative and muscle relaxants activity of Diospyros lotus L. Asian Pac J Trop Biomed 2015;5:277-80.
- Rauf A, Hadda TB, Uddin G, Cerón-Carrasco JP, Peña-García J, Pérez-Sánchez H, et al. Sedative-hypnotic-like effect and molecular docking of di-naphthodiospyrol from Diospyros lotus in an animal model. Biomed Pharmacother 2017;88:109-13.
- Ganapaty S, Thomas PS, Karagianis G, Waterman PG, Brun R. Antiprotozoal and cytotoxic naphthalene derivatives from Diospyros assimilis. Phytochemistry 2006;67:1950-56.
- Rauf A, Uddin G, Siddiqui BS, Muhammad N, Khan H. Antipyretic and antinociceptive activity of Diospyros lotus L. in animals. Asian Pac J Trop Biomed 2014;4:S382-6.
- Fazl-i-Sattar, Ullah Z, Rahman A, Rauf A, Tariq M, Tahir AA, et al. Phytochemical, spectroscopic and density functional theory study of Diospyrin, and non-bonding interactions of Diospyrin with atmospheric gases. Spectrochim Acta A Mol Biomol Spectrosc 2015;141:71-79.
- Ullah Z, Rahman A, Fazl-i-Sattar, Rauf A, Yaseen M, Hassan W, et al. Density functional theory and phytochemical study of 8-hydroxyisodiospyrin. J Mol Struct 2015;1095:69-78.
- Rauf A, Uddin G, Khan H, Siddiqui BS, Arfan M. Antihyperalgesic activity of crude extract and 7-methyljuglone of Diospyros lotus roots. Nat Prod Res 2015;29:2226-29.
- Rauf A, Uddin G, Siddiqui BS. Isolation and structure elucidation of a new dimeric naphthoquinone from Diospyros lotus. Chem Nat Compd 2015;51:1049-51.
- Rauf A, Uddin G, Khan H, Arfan M, Siddiqui BS. Bioassay-guided isolation of antibacterial constituents from Diospyros lotus roots. Nat Prod Res 2016;30:426-28.
- Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol 2006;7:833-46.
- Fischer EH, Charbonneau H, Tonks NK. Protein tyrosine phosphatases: A diverse family of intracellular and transmembrane enzymes. Science 1991;253:401-6.
- Hunter T. Protein kinases and phosphatases: The Yin and Yang of protein phosphorylation and signaling. Cell 1995;80:225-36.
- Comeau AB, Critton DA, Page R, Seto CT. A focused library of protein tyrosine phosphatase inhibitors. J Med Chem 2010;53:6768-72.
- Liu G, Trevillyan JM. Protein tyrosine phosphatase 1B as a target for the treatment of impaired glucose tolerance and type II diabetes. Curr Opin Investig Drugs 2002;3:1608-16.
- Owen C, Czopek A, Agouni A, Grant L, Judson R, Lees EK, et al. Adipocyte-specific protein tyrosine phosphatase 1b deletion increases lipogenesis, adipocyte cell size and is a minor regulator of glucose homeostasis. PLoS One 2012;7(2):e32700.
- Sagar S, Green IR. Synthesis of quinonoid analogues of diospyrin. Synthesis 2009;6:935-40.
- Uddin G, Rauf A, Arfan M, Waliullah, Rehman TU, Khan AZ, et al. Molecular docking of Diospyrin as a LOX inhibitory compound. J Saudi Chem Soc 2016;20:S448-S450.
- Uddin RG, Siddiqui BS, Khan A, Farooq U, Khan FA, Bukhari SM, et al. Bioassay-guided isolation of novel and selective urease inhibitors from Diospyros lotus. Chin J Nat Med 2017;15:865-70.
- Pinho BR, Sousa C, Valentão P, Oliveira JMA, Andrade PB. Modulation of basophils’ degranulation and allergy-related enzymes by monomeric and dimeric naphthoquinones. PLoS One 2014;9:e90122.
- Ebalunode JO, Ouyang Z, Liang J, Zheng W. Novel approach to structure-based pharmacophore search using computational geometry and shape matching techniques. J Chem Inf Model 2008;48:889-901.
- McGann M. FRED pose prediction and virtual screening accuracy. J Chem Inf Model 2011;51:578-96.
- McGann MR, Almond HR, Nicholls A, Grant JA, Brown FK. Gaussian docking functions. Biopolymers 2003;68:76-90.