- *Corresponding Author:
- K. B. Charuvil
Biotechnology and Bioinformatics division
Saraswathy Thangavelu Centre of Jawaharlal Nehru Tropical Botanic Garden and Research Institute Puthenthope
Thiruvananthapuram–695 586
Kerala
India
E-mail: drbijuck@gmail.com
| Date of Received | 29 January 2020 |
| Date of Revision | 08 February 2021 |
| Date of Acceptance | 09 May 2021 |
| Indian J Pharm Sci 2021;83(3):443-450 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Hepatitis C virus infection is the leading cause of chronic liver disease and hepatocellular carcinoma. There is no effective vaccine for hepatitis C virus prevention despite the fact that severalvaccinesare under development. Currently, the Unites States Food and drug administration approved combination drugs for all genotypes that would help to cure the infection more quickly and efficiently than ever before. However, the high costs, development of various side effects and emergence of drug resistant strains demand the need for new anti-viral to treat different stages of the hepatitis C virus life cycle. Focussing drug candidate from herbal ingredients is the novel approach of pharmaceutical science over the past few decades. In this perspective, the present study aimed to investigate the phytochemicals present in Acacia nilotica (L.) Delile against hepatitis C virus non-structural protein3-4A serine protease. The N-terminal Protease domain of non-structural protein3 along with non-structural protein4A protein is responsible for the cleavage of four polypeptide junctions’ viz., non-structural protein3-4A, non-structural protein4A-non-structural protein4B, non-structural protein4B-non-structural protein5A and non-structural protein5A-5B that are essential for viral genome replication. Hence targeting non-structural protein3-4A blocks the replication process. Here, in silico molecular docking study was executed to estimate the efficacy ofphytochemicals along with the two Food and drug administration approved hepatitis C virus non-structural protein3-4A inhibitors-Grazoprevir and simeprevir as reference compoundsagainst the selected target. Docking results revealed that about six phytochemicals (+)-Catechin 5-Gallate, Acacetin, (+)-Mollisacacidin, Catechin, Acalinol A and Chlorogenic acid are better than the reference compounds and hence selected as hits. Further, the hit molecules were filtered through analysing druglikeness properties, pharmacokinetics, medicinal chemistry friendliness including pan assay interference compounds and Brenk structural alerts, leadlikeness and finally prediction of potential toxicity and toxic substructure to ascertain a lead molecule. The results obtained in the current study propose Acacetin as the lead molecule for further in vitro and in vivo study.
Keywords
Acacetin, acacia, antipyretics, hepatitis C, phytochemicals, non-structural protein3-4A serine protease
Hepatitis C virus (HCV) is a member of the family Flaviviridae and the genus Flavivirus. It has a 9.6 kb positive-strand Ribonucleic acid (RNA) genome that encodes a polyprotein precursor of approximately 3000 amino acids which is proteolytically processed into four structural proteins viz. Capsid, Envelope proteins (E1, E2) and p7 with the aid of host proteases and six non-structural proteins (NS) namely NS2, NS3, NS4A, NS4B, NS5A and NS5B with the help of viral serine proteases namely NS2-NS3 and NS3-4A[1]. Among all the ten HCV proteins, the serine proteases (NS3-4A) and the RNA-dependent RNA polymerases (NS5B-RdRp) were considered as the most important target for anti-HCV drug development.
NS3-4A is a heterodimeric serine protease that belongs to the chymotrypsin family. The NS3 is a 631 amino acid residue multi-functional protein with N-terminal protease domain and C- terminal helicase domain. The C-terminal helicase domain is responsible for the unwinding of duplex RNA that is formed when the single-stranded RNA genome is copied. Also, it cleans out RNA binding proteins from viral RNA, assist translation and polyprotein processing[2]. The N-terminal protease domain (181aa) with the help of NS4A which acts as a cofactor is responsible for the cleavage of four polypeptide junctions namely NS3-4A, NS4A-NS4B, NS4B-NS5A and NS5A-5B. In the absence of NS4A, NS3 could only partially cleave at NS5A-5B but not the other sites (NS3-4A, NS4A-5A and NS5A-5B). NS4A is a 54 amino acid residue protein with residue 1-20 is hydrophobic and forms a transmembrane α helix, 21-34 is also hydrophobic and forms β strands and the remaining 20 residues are hydrophilic and forms helical conformation. The residues 21-34 are essential to fully activate the NS3 serine protease[1].
Currently, only few drugs were approved for HCV treatment and most of them were given in the form of combination therapy to treat various HCV disorders. Specific direct acting antivirals (DAAs) targeting the NS3-4A protease, NS5B polymerase and NS5A protein are also available in the market. However, resistance-associated substitutions induce amino acid changes that can reduce the susceptibility to one or more antiviral drugs and hence leads to treatment failure. Hence the limitations provided by the existing treatment demonstrate the need for the development of more efficient new antivirals against HCV.
Over the past decades, there have been worthwhile studies on compounds extracted from plants that have shown activity against a range of microorganisms that cause human diseases. A variety of natural compounds have manifested antiviral activity worldwide, including anti-HCV activity[3] or hepatoprotective effect as described for Naringenin, (-)-Epigallocatechin gallate (EGCG), silymarin and caffeine[4-7]. In this connection, plant-derived compounds can provide an alternative approach to new antivirals. It has been estimated that almost 50 % of drugs approved since 1994 are based on natural products[8].
Therefore, in the present investigation, we have conducted a computer-based drug design strategy to investigate the efficiency of phytochemicals present in the plant Acacia nilotica (L.,) Delile (reported to be anti-HCV)[9] against HCV NS3-4A serine protease to identify a series of hit molecules through molecular interaction study and thereafter subjected to druglikeness profiling, pharmacokinetics analysis, medicinal chemistry friendliness as well as prediction of toxicity and toxic substructure to identify an appropriate lead candidate.
Materials and Methods
Retrieval and preparation of target Protein:
The 3D structure of the target protein NS3-4A was retrieved from protein data bank (PDB id: 2OC0). The protein structure was prepared using protein preparation wizard (pre-processed, optimized and minimized) in the Schrodinger software graphical user interface Maestro v11.9. The pre-processing stage exploits the options such as assign bond orders, add hydrogens, create disulphide bonds and delete water molecules beyond 5 A0 from any of the het groups including ions. The next stage is optimization of hydrogen-bonding network by reorienting hydroxyl and thiol groups, water molecules, amide groups of Asn and Gln and the imidazole ring in His and predicting protonation states of His, Asp and Glu and tautomeric states of histidine. These optimizations are necessary because the orientation of hydroxyl groups, the terminal amide groups in asparagine and glutamine and the ring of histidine cannot be determined from the X-ray structure. Finally a minimization was carried out in which the heavy atoms kept restrained. Hydrogen atoms are not restrained, which allows the optimized H-bond network to be refined. The minimization is done with a Root mean square deviation (RMSD) cut-off of 0.30 Ǻ[10].
Selection and preparation of ligand:
A total of 51 ligands were chosen here for the molecular docking study, out of which 49 are phytochemicals reported from the plant Acacia nilotica and the remaining two are Food and drug administration (FDA) approved drugs specifically used against NS3-4A target to treat HCV. Among 49 phytochemicals, the structure of 47 phytochemicals was retrieved from PubChem and the remaining 2 compounds were drawn using Chemsketch and their canonical smiles were generated. The canonical smiles were submitted to an online file format converter (Open Babel) to get the 2D structures in Spatial DataFile(SDF) format. The 2D structures of two anti-HCV drugs were retrieved from Drugbank 2.0. The ligands were subjected to ligand preparation by LigPrep module of the Maestro v11.9. The process of ligand preparation includes addition of hydrogen molecules, conversion of 2D structures to low energy 3D structures, correction of bond lengths and bond angles. Protonation states of ligands were generated by Epik[11] and pH was set to neutral pH 7.0. Finally various ligand conformers were generated as output in Maestro format.
Generation of Receptor Grid:
Receptor Grid Generation module was used to generate grid around the active site already occupied by the co-crystallized ligand of the receptor. Amino acid residues in the active site include His57, Asp81,Ile132, Leu135, Lys136, Ser139, Phe154, Arg155, Ala156, Cys159, Ala157, Gly137whereas His57, Asp81 and Ser139 forms the catalytic triad[1]. Vanderwaals scaling factor 1.00 and partial charge cut off 0.25 were default parameters used for grid generation.
Validation of docking procedure:
The most suitable method of evaluating the accuracy of a docking procedure is to determine how intimately the lowest energy pose predicted by the scoring function resembles an experimental binding mode as determined by X-ray crystallography. In the present study, the extra precision Glide docking procedure was validated by removing the natural ligand, Ketoamide Inhibitor SCH491762 from the binding site and re-docking it to the HCV NS3-4A Protein (PDB ID: 2OC0). The Root means square deviation (RMSD) between the predicted conformation and the observed X-ray crystallographic conformation of Ketoamide Inhibitor SCH491762 was found to be 1.0589.This indicates the reliability of the docking method in reproducing the experimentally observed binding mode.
Molecular Docking Studies:
All the conformers obtained from the LigPrep-output were docked into the receptor active site residues enclosed in a grid box. Flexible ligand docking was employed for the current study and the best molecular docked complexes were evaluated using extra precision Glide score (XPG Score). The XPG score optimizes ligand binding energy based on force field parameters and docking penalties that had significant influences over the receptor-ligand binding[12].
Assessment of Druglikeness, Pharmacokinetics and Toxicity:
The concept of druglikeness and pharmacokinetics has been widely used in Pharmaceutical industry to ward off side effects caused by the drug like small molecules. In the current study, the best hit molecules procured through molecular interaction studies were further filtered to determine a lead candidate by evaluating physiochemical parameters using Molinspiration. Pharmacokinetics as well as medicinal chemistry friendliness parameters were studied using SwissADME[13]. Detection of Carcinogenicity through admetSAR 2.0[14] and ProTox-II[15]. Hepatotoxicity through admetSAR and pkCSM[16]. Detection of HERG (Human Ether-a-go-go-Related Gene) inhibition or non-inhibition through pkCSM and ADMETlab[17]. Cytotoxicity through ProTox-II and finally to identify potential toxic substructure in the selected molecules, ‘mcule-Toxicity checker’ has been used.
Results and Discussion
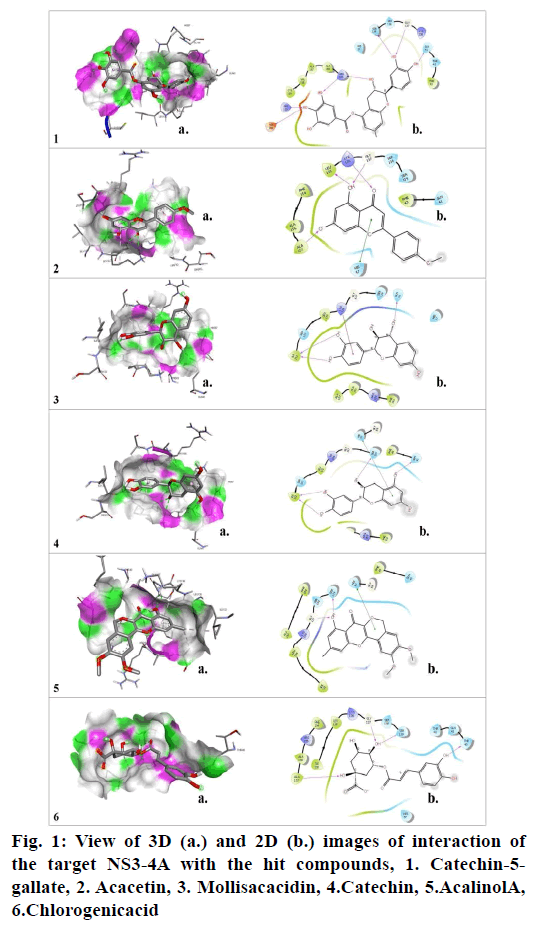
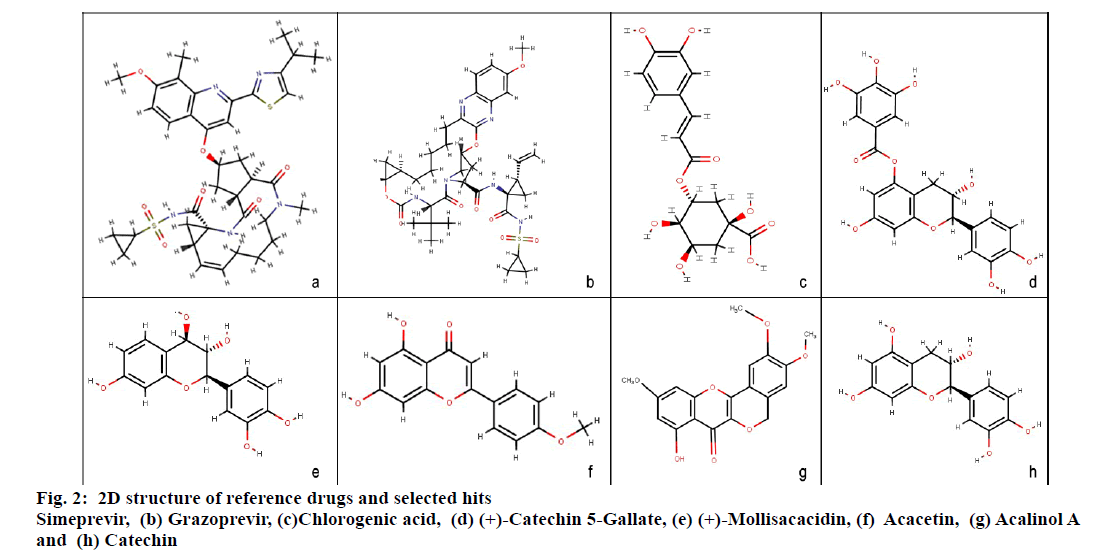
Molecular docking studies were carried out to identify potential hit molecules from Acacia nilotica against NS3-4A serine protease through GLIDE (Schrodinger suite). A total of 49 phytochemicals were subjected to molecular docking along with two FDA approved drugs for HCV–Grazoprevir (DB11575) and Simeprevir (DB06290) as reference molecules. Among the phytochemicals six compounds viz. (+)-Catechin5-Gallate, Acacetin,(+)-Mollisacacidin, Catechin, Acalinol A and chlorogenic acid were selected as top hits (fig.1) since they showed least XP-glide score as compared with the reference drugs (Table 1). Further upon analysing the interaction of the hits with NS3-4A, it was observed that all the hit compounds except (+)-Mollisacacidin specifically bound with the catalytic triad residues. Catechin, AcalinolA, (+)-Catechin-5-Gallate and Chlorogenic acid interact with one of the catalytic triad residues SER139. Besides that, Catechin, Acacetin and AcalinolA showed pi-pi stacking interaction with another residue, HIS57 in the catalytic triad. So it is evident from the analysis that interaction of the above hits with the catalytic triad residues would lead to the stronger inhibition of the NS3-4A protease. In contrast with the phytochemicals, none of the reference drugs interact with any of the residues in the catalytic triad. Moreover both the hits and reference molecules interact with active site residues and also displayed acceptable range of Hydrogen–Acceptor distance (1.5-2.5Å), Donor-acceptor distance (2.4-3.5Å) and Donor-H-acceptor angle (Θ: 120º–180º)[18]. The 2D structure of the selected hits and reference drugs were shown in fig. 2.
| Name of Compound | XP GS |
H- bond interaction |
HAD | DAD | DHAA | Hydrophobic interaction |
pi-pi interaction ( Å) |
|---|---|---|---|---|---|---|---|
| Grazoprevir* | -4.69 | Gly137:N-H--O:Lig Ala157:N-H--O:Lig |
1.93 2.46 |
2.92 3.31 |
167.5 141.3 |
Phe43,Phe154, Ile132, Leu135, Ala156,Ala157, Val158, Cys159 |
- |
| Simeprevir * | -2.36 | Ala157:N-H--O:Lig | 2.30 | 3.17 | 143.4 | Phe154, Ala156, Ala157, Ile132, Cys159, Val78, Tyr56, Val158 | - |
| (+)-Catechin5-Gallate | -7.193 | Gly137:N-H--O:Lig Arg155:N-H--O:Lig Arg123:N-H--O:Lig Lig:O-H--O: ASP168 Lig:O-H--O: Ser139 Lig:O-H--O: GLN41 Lig:O-H--O:ARG155 |
1.97 2.21 2.09 1.90 2.05 1.97 2.16 |
2.87 2.92 3.01 2.92 3.00 2.90 2.87 |
146.7 126.6 149.7 164.2 153.3 164.5 130.6 |
Phe43, Ala156, Ala157,Val158, Cys159 |
- |
| Acacetin | -7.092 | Lys136:N-H--O:Lig Gly137:N-H--O:Lig Ala157:N-H--O:Lig Lig:O-H--O: Leu135 |
2.15 1.85 1.82 2.01 |
3.50 2.72 2.70 2.95 |
146.5 132.6 143.5 158.0 |
Phe43, Phe154, Leu135, Ala157, Ala156. |
His57(5.02) |
| (+)-Mollisaca- cidin | -6.472 | Lig:O-H --O:Ile132 Lig:O-H--O:Ile132 Lig:O-H--O: Gln41 |
2.58 1.83 1.88 |
2.74 3.43 2.82 |
149.8 159.0 171.0 |
Leu135,Ile132, Ala156, Ala157, Phe154 |
|
| Catechin | -6.289 | Lig:O-H--O:Ser139 Lig:O-H--O:Gln41 Lig:O-H --O:Ile132 Lig:O-H --O:Ile132 |
2.43 1.83 2.65 1.79 |
3.34 2.72 3.33 2.74 |
161.1 153.7 130.2 170.1 |
Phe43, Leu135, Ile132,Ala156, Ala157,Phe154. |
His57(4.78) |
| AcalinolA | -5.216 | Ser139:O-H--O:Lig Gly137:N-H--O:Lig |
1.95 2.04 |
2.82 2.95 |
134.6 148.5 |
Phe43, Leu135, Ile132,Ala157, Phe154 |
His57(4.77) |
| Chlorogenic acid | -4.973 | Lig:O-H--- O:Thr40 Lig:O-H--O:Ser139 Gly137:N-H--O:Lig Ala157:N-H--O:Lig |
1.74 1.92 2.13 2.50 |
2.91 2.82 2.76 3.49 |
163.7 154.0 172.7 165.1 |
Ala156,Ala157, Phe154,Ile132, Leu135 |
(*=reference drugs; XPGS=Extra precision Glide Score; HAD=Hydrogen-Acceptor distance; DAD= Donor-Acceptor distance; DHAA=Donor-HAcceptor Angle).
Table 1: Interaction Details of Hits and Reference Molecules with Ns3-4a
Properties such as log P, Molecular weight, Topological polar surface area (TPSA), number of Hydrogen bond donors (HBD), number of Hydrogen bond Acceptors(HBA) and number of rotatable bonds (Table 2) were considered here for calculating druglikeness of the hits. According to Lipinski’s rule of Five (RO5), a drug molecule tends to show good oralbioavailability, smoothmembrane permeabilityand highgastrointestinal absorptionin human gut when its log P≤5; MW≤500 Da; HBAs≤10 and HBDs≤5[19]. Veber’s rule proposes that a compound possess high oral bioavailabilitywhen its TPSA≤140 Å and number of rotatable bonds ≤10[20]. ‘GSK 4/400 rule’ suggests that the propensity of a drug tends to be toxic when its log P>4 and MW>400 Da[21]. Physiochemical properties of the four hit molecules Catechin, (+)-Mollisacacidin, Acalinol A and Acacetin were found perfect conformity with RO5 and Veber’s rule which signifies their considerable drug like properties and also with GSK 4/400 rule indicating their nontoxic nature.
| Compound name | Mol.wt. (Da) | Log P | TPSA | H-bond donors |
H-bond acceptors | Rotatable bonds |
|---|---|---|---|---|---|---|
| (+)-Catechin 5-Gallate | 442.38 | 1.99 | 177.13 | 7 | 10 | 4 |
| Acacetin | 284.27 | 3.00 | 79.90 | 2 | 5 | 2 |
| (+)-Mollisacacidin | 290.27 | 0.46 | 110.37 | 5 | 6 | 1 |
| Catechin | 290.27 | 1.37 | 110.37 | 5 | 6 | 1 |
| AcalinolA | 340.33 | 3.26 | 78.14 | 1 | 6 | 2 |
| Chlorogenic Acid | 354.31 | -0.45 | 164.74 | 6 | 9 | 5 |
(TPSA=Topological polar surface area; Logp=Logarithm of partial coefficient; Mol.wt=Molecular weight)
Table 2: Physiochemical Properties of the Hits
Majority of the drug failure in pharmaceutical industries is mainly due to lack of proper ADME analysis. Some important pharmacokinetic properties such as water solubility, gastrointestinal (GI) absorption, blood brain barrier (BBB) permeation, P-gp (P-glycoprotein) substrate and CYP450 enzymes inhibition were computed and depicted in Table 3. Estimation of water solubility revealed that all the concerned hit molecules except Acalinol A were soluble. In case of GI absorption the compounds except (+)-Catechin 5-Gallate and Chlorogenic acid showed high absorption. Recent research findings suggested that, in common with HIV infection, HCV may cross the blood brain barrier leading to neuro-inflammation[22]. None of the selected hits except Acalinol A cross the BBB. Assessment of P-gp substrate indicates that all the selected hits except Catechin act as non-substrates. P-gp plays a major role in limiting cellular uptake of drugs resulting in therapeutic failure because the drug concentration would be lower than expected[23,24]. The interaction of small molecules with various Cytochrome P450 isoforms CYP1A2, CYP2C19, CYP2C9, CYP2D6 and CYP3A4 is a significant factor in drug elimination through metabolic biotransformation[25]. Inhibition of these isoenzymes is an important cause of pharmacokinetics-related drug-drug interactions due to the accumulation of metabolites/drugs[26]. The compounds (+)-Catechin5-Gallate, (+)-Mollisacacidin, Catechin and Chlorogenic acid are non-inhibitors of all the five Cytochrome P450 isoforms whereas Acalinol A inhibits four and Acacetin inhibits 3 isoforms. Further, medicinal chemistry friendliness parameters-structural alerts (PAINS and Brenk) and leadlikeness were predicted. Identification of compounds with structural alerts is an imperative screening step of drug development. PAINS (Pan Assay Interference Compounds) are commonly referred to as toxicophores with alarming chemical groups that have been reported to interfere with biological assays, interact and damage DNA/proteins [27]. Brenkis another structural alert which warns about allegedly toxic, metabolically unstable and chemically reactive fragments present in the structure[28]. Here Acalinol A and Acacetin were not found to possess PAINS and Brenkalerts. Regarding leadlikeness criteria proposed by Teague[29] all the compounds except Chlorogenic acid and (+)-Catechin5-Gallate conceded leadlikeness and hence they were suitable for initiating further optimization.
| Hits | Pharmacokinetics Analysis | Medicinal Chemistry Friendliness | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Water solubility | GIAb | BBBp | P-gps | CYP450 inhibition | ||||||||
| CYP1A2 | CYP2C19 | CYP2C9 | CYP2D6 | CYP3A4 | PAINS | Brenk | LL | |||||
| (+)-Catechin5-Gallate | soluble | low | NP | NS | no | no | no | no | no | 1 alert | 2 alerts | no |
| Acacetin | soluble | high | NP | NS | yes | no | yes | yes | no | no alert | no alert | yes |
| (+)-Mollisacacidin | soluble | high | NP | NS | no | no | no | no | no | 1 alert | 1 alert | yes |
| Catechin | soluble | high | NP | S | no | no | no | no | no | 1 alert | 1 alert | yes |
| Acalinol A | moderate | high | P | NS | yes | no | yes | yes | yes | no alert | no alert | yes |
| chlorogenic acid | soluble | low | NP | NS | no | no | no | no | no | 1 alert | 2 alert | no |
(GIAb=Gastro Intestinal absorption; BBBP=Blood Brain Barrier Permeation; P-gps=P-glycoprotein substrate; PAINS=Pan Assay Interference Compounds; LL=Leadlikeness; NP=Non permeable; P=Permeable; NS=Non-substrate, S=Substrate)
Table 3: Details of Pharmacokinetic Properties and Medicinal Chemistry Friendliness of the Hits
Forecasting the toxicity of a small molecule is one of the most crucial aspects in effective drug development. Drug toxicity is one of the major causes of rejection of a large number of therapeutic components at a later stage of drug development. In the present study, in silico assessment of mutagenicity in correlation with Ames mutagenicity through pkCSM and admetSAR revealed that (+)-Mollisacacidin exhibited mutagenicity. Hepatotoxic effects of the compounds were investigated through pkCSM and ProTox-II web servers. None of the compounds were predicted to be hepatotoxic. Carcinogenicity detection of the phytochemicals through admetSAR and ProTox-II revealed the carcinogenic effect of (+)-Mollisacacidin. Regarding the inhibitory effect of selected hits towards HERG (Human Ether-a-go-go-Related Gene) through pkCSM and ADMETlab server admit them as non-HERG blockers. Subsequently prediction of cytotoxicity through ProTox-II revealed that none of the compounds exhibit cytotoxic effect. Additionally they have also been subjected to mcule-Toxicity checker and found that among the hits, Acacetin does not possess any potential toxic substructure. Details of the predicted results were depicted in Table 4.
| Compound name | Ames mutagenicity | Hepatotoxicity | Carcinogenicity | HERG inhibition | Cytotoxicity | Toxic substructure | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| pkCSM | ADMETlab | pkCSM | Protox-II | admetSAR | ProTox-II | pkCSM | ADMETlab | ProTox-II | ||
| (+)-Catechin 5-Gallate | no | no | no | no | no | no | no | no | no | Present |
| Acacetin | no | no | no | no | no | no | no | no | no | Absent |
| (+)-Mollisacacidin | yes | yes | no | no | yes | yes | no | no | no | Present |
| Catechin | no | no | no | no | no | no | no | no | no | Present |
| Acalinol A | no | no | no | no | no | no | no | no | no | Present |
| Chlorogenic acid | no | no | no | no | no | no | no | no | no | Present |
Table 4: In Silico Assessment of Potential Toxicity and Toxic Substructure of the Hits
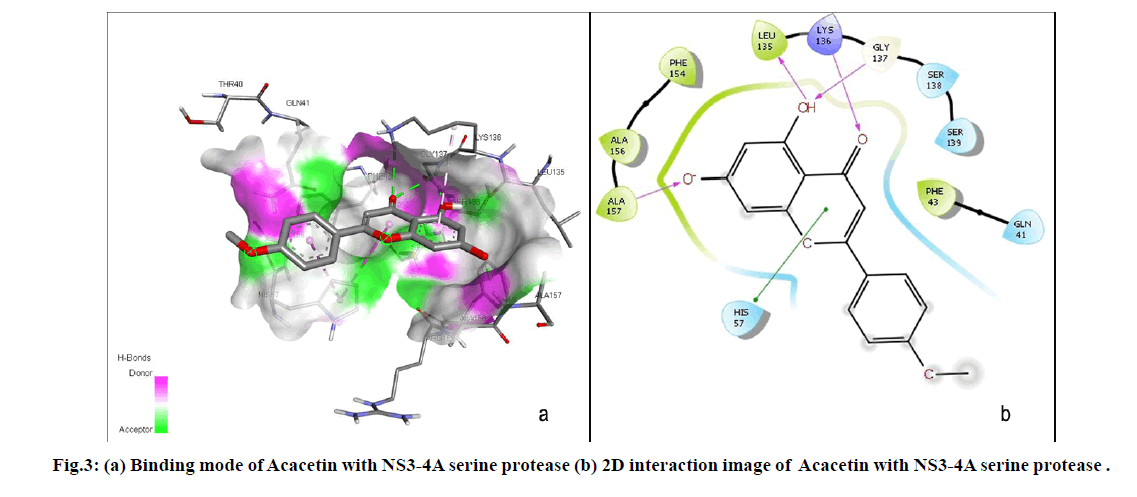
The current study resulted that Acacetin, a naturally occurring flavonoid was the lead molecule among the six hit compounds as it strongly binds with active site residues (GLY137, ALA157, LYS136 and LEU136) including the catalytic triad residue HIS57. Binding mode of Acacetin with NS3-4A serine protease was shown in fig. 3. Acacetin also satisfied Druglikeness properties, not exhibited any structural alerts and act as non-inhibitor of CYP3A4 which is the most abundant and significant isoform of CYP450 enzymes that contributes to metabolise 30.2 % of drugs[30]. Considering Toxicity prediction, Acacetin does not possess toxicity and toxic substructure. An in vitro study by Chien et al. indicated that Acacetin did not exert cytotoxicity[31]. Moreover, the result substantiates the Hepatoprotective effect of Acacetin which is experimentally proved by Cho et al.[32]. It also possess various other pharmacological activities including neuroprotective, cardioprotective, anticancer, anti-inflammatory, antidiabetic, anti-arthritic, anti-alzheimer’s, Antipyretic, Anti-ageing, Anti-oxidant, Antimicrobial, Anti-allergic and immunomodulatory activities[33]. However, further in vitro and in vivo studies are essential to propose Acacetin as a potential drug candidate against HCV.
Acknowledgements
The authors are thankful to Director, KSCSTE-JNTBGRI for providing the facilities and encouragements and thank KSCSTE, Govt. of Kerala for financial support.
Conflict of interests
The authors declared no conflicts of interest.
References
- Tan SL. Hepatitis C viruses: genomes and molecular biology. Norfolk (UK): Horiz Biosci 2006.
- Frick DN. HCV helicase: structure, function, and inhibition. Hepatitis C Viruses: Genomes and Molecular Biology 2006.
- Munir S, Saleem S, Idrees M, Tariq A, Butt S, Rauff B, et al. Hepatitis C treatment: current and future perspectives. Virol J 2010;7(1):1-6.
- Assini JM, Mulvihill EE, Burke AC, Sutherland BG, Telford DE, Chhoker SS, et al. Naringenin prevents obesity, hepatic steatosis and glucose intolerance in male mice independent of fibroblast growth factor 21. Endocrinology 2015;156(6):2087-102.
- Tipoe GL, Leung TM, Liong EC, Lau TY, Fung ML, Nanji AA. Epigallocatechin-3-gallate (EGCG) reduces liver inflammation, oxidative stress and fibrosis in carbon tetrachloride (CCl4)-induced liver injury in mice. Toxicology 2010;273(1-3):45-52.
- Zhang W, Hong R, Tian T. Silymarin’s protective effects and possible mechanisms on alcoholic fatty liver for rats. Biomol Ther 2013;21(4):264.
- Khalaf N, White D, Kanwal F, Ramsey D, Mittal S, Tavakoli-Tabasi S, et al. Coffee and caffeine are associated with decreased risk of advanced hepatic fibrosis among patients with hepatitis C. ClinGastroenterol Hepatol 2015;13(8):1521-31.
- Bhutani KK, Gohil VM. Natural products drug discovery research in India: status and appraisal. IndianJExpBiol 2010;48:199-7.
- Tavakkol-Afshari J, Meshkat Z, Zibaie S, Yousefi M, Hoseini MA, Nikpoor AR, et al. Natural Products for the Treatment of Hepatitis C. J Res Med Den Sci 2018;6:196-201.
- Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des 2013;27(3):221-34.
- Shelley JC, Cholleti A, Frye LL, Greenwood JR, Timlin MR, Uchimaya M. Epik: a software program for pK a prediction and protonation state generation for drug-like molecules. J Comput Aided Mol Des 2007;21(12):681-91.
- Aamir M, Singh VK, Dubey MK, Meena M, Kashyap SP, Katari SK, et al. In silico Prediction, Characterization, Molecular Docking and Dynamic Studies on Fungal SDRs as Novel Targets for Searching Potential Fungicides against Fusarium Wilt in Tomato. Front Pharmacol 2018;9:1038.
- Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 2017;7(1):1-3.
- Yang H, Lou C, Sun L, Li J, Cai Y, Wang Z, et al. admetSAR 2.0: web-service for prediction and optimization of chemical ADMET properties. Bioinformatics 2019;35(6):1067-9.
- Banerjee P, Eckert AO, Schrey AK, Preissner R. ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res 2018;46(W1):W257-63.
- Pires DE, Blundell TL, Ascher DB. pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J Med Chem 2015;58(9):4066-72.
- Dong J, Wang NN, Yao ZJ, Zhang L, Cheng Y, Ouyang D, et al. ADMETlab: a platform for systematic ADMET evaluation based on a comprehensively collected ADMET database. J Cheminform 2018;10(1):1-1.
- Raschka S, Wolf AJ, Bemister-Buffington J, Kuhn LA. Protein–ligand interfaces are polarized: discovery of a strong trend for intermolecular hydrogen bonds to favor donors on the protein side with implications for predicting and designing ligand complexes. J Comput Aided Mol Des 2018;32:511-28.
- Lipinski CA. Lead-and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol 2004;1:337-41.
- Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem 2002;45(12):2615-23.
- Gleeson MP. Generation of a set of simple, interpretable ADMET rules of thumb. J Med Chem 2008;51(4):817-34.
- Yarlott L, Heald E, Forton D. Hepatitis C virus infection, and neurological and psychiatric disorders - A review. J Adv Res 2017;8:139(2)-48.
- Levin GM. P-glycoprotein: why this drug transporter may be clinically important. Curr Psychiatr 2012;11(3):38-40.
- Lin JH, Yamazaki M. Role of P-glycoprotein in pharmacokinetics: Clinical implications. Clin Pharmacokinet 2003;42(1):59-98.
- Testa B, Kramer SD. The biochemistry of drug metabolism-an introduction: part 5. Metabolism and bioactivity. Chem Biodivers 2009;6(5):591-84.
- Hollenberg PF. Characteristics and common properties of inhibitors, inducers, and activators of CYP enzymes. Drug Metab Rev 2002;34(1-2):17-35.
- Baell JB, Holloway GA. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem 2010;53(7):2719-40.
- Brenk R, Schipani A, James D, Krasowski A, Gilbert IH, FrearsonJ, et al. Lessons learnt from assembling screening libraries for drug discovery for neglected diseases. Chem Med Chem 2008;3(3):435-44.
- Teague SJ, Davis AM, Leeson PD, Oprea T. The design of lead like combinatorial libraries. Angew ChemInt Ed Engl 1999;38(24):3743-8.
- Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther 2013;138(1):103-41.
- Cho HI, Park JH, Choi HS, Kwak JH, Lee DU, Lee SK, et al. Protective mechanisms of acacetin against D-galactosamine and lipopolysaccharide-induced fulminant hepatic failure in mice. J Nat Prod 2014;77(11):2497-503.
- Chien ST, Lin SS, Wang CK, Lee YB, Chen KS, Fong Y, et al. Acacetin inhibits the invasion and migration of human non-small cell lung cancer A549 cells by suppressing the p38α MAPK signaling pathway. Mol Cell Biochem 2011;350(1):135-8.
- Semwal RB, Semwal DK, Combrinck S, Trill J, Gibbons S, Viljoen A. Acacetin- A simple flavone exhibiting diverse pharmacological activities. Phytochem Lett 2019;32:56-65.