- *Corresponding Author:
- X. Li
School of Basic Medical Sciences and Forensic Medicine, Hangzhou Medical College, Hangzhou, Zhejiang 310053, China
E-mail: leexiang2004@126.com
| Date of Received | 10 January 2021 |
| Date of Revision | 24 October 2021 |
| Date of Acceptance | 16 May 2022 |
| Indian J Pharm Sci 2022;84(3):617-630 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Drug repositioning may be a promising way to find potential therapies against coronavirus disease 2019. Although chloroquine and hydroxychloroquine showed controversial results against the coronavirus disease 2019 disease, the potential common and diverging mechanisms of action are not reported and need to be dissected for better understanding them. An integrated strategy was proposed to systematically decipher the common and diverging aspects of mechanism of chloroquine and hydroxychloroquine against coronavirus disease 2019-disease network based on network pharmacology and in silico molecular docking. Potential targets of the two drugs and coronavirus disease 2019 related genes were collected from online public databases. Target function enrichment analysis, tissue enrichment maps and molecular docking analysis were carried out to facilitate the systematic understanding of common and diverging mechanisms of the two drugs. Our results showed that 51 chloroquine targets and 47 hydroxychloroquine targets were associated with coronavirus disease 2019. The core targets include tumor necrosis factor, glyceraldehyde 3-phosphate dehydrogenase, lymphocyte-specific protein-tyrosine kinase, beta-2 microglobulin, nuclear receptor coactivator 1, peroxisome proliferator-activated receptor gamma and glutathione disulfide reductase. Both chloroquine and hydroxychloroquine had good binding affinity towards tumor necrosis factor (affinity=-8.6 and -8.4 kcal/mol, respectively) and glyceraldehyde 3-phosphate dehydrogenase (-7.5 and -7.5 kcal/mol). Chloroquine and hydroxychloroquine both had good affinity with angiotensinconverting enzyme 2, 3-chymotrypsin-like protease and transmembrane serine protease 2. However, hydroxychloroquine manifested better binding affinity with the three proteins comparing with that of chloroquine. Chloroquine and hydroxychloroquine could have potential to inhibit over-activated immunity and inflammation. The potential tissue-specific regulation of the two drugs against severe acute respiratory syndrome coronavirus 2 infection may related with the lung, liver, brain, placenta, kidney, blood, eye, et al. In conclusion, our data systematically demonstrated chloroquine and hydroxychloroquine may have potential regulatory effects on coronavirus disease 2019 disease network, which may affect multiple organs, protein targets and pathways. Routine measurements of the chloroquine and hydroxychloroquine blood concentrations and tailored therapy regimen may be essential. But, further rigorous and high quality randomized controlled clinical trials are warranted to validate the antiviral effects of chloroquine and hydroxychloroquine against severe acute respiratory syndrome coronavirus 2. Our proposed strategy could facilitate the drug repurposing efforts for coronavirus disease 2019 treatment.
Keywords
Coronavirus disease 2019, chloroquine, hydroxychloroquine, network pharmacology, molecular docking, blood concentration
Coronavirus Disease 2019 (COVID-19), the novel coronavirus pneumonia, caused by the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) has devastated the world for nearly 2 y. The World Health Organization (WHO) declared COVID-19 as a pandemic on 11th March, 2020. According to the Johns Hopkins University, there are more than 243 million cases globally and over 4.9 million reported deaths in the world by 23rd October, 2021. Whereas there are huge investments towards the fight against COVID-19, there are still no specific drugs available against COVID-19. Veklury (remdesivir) was approved by the Food and Drug Administration (FDA) to treat COVID-19. Therefore, research towards the development of anti-COVID-19 drugs is still important.
Drug repositioning may be a promise way to find potential therapies against COVID-19. According to DrugBank database, Chloroquine (CQ) was first developed in 1940s for the treatment of malaria. Its current clinical indications include human immunodeficiency virus, systemic lupus erythematosus and rheumatoid arthritis. Hydroxychloroquine (HCQ), a CQ derivative, is a prescribed drug against uncomplicated malaria, rheumatoid arthritis, chronic discoid lupus erythematosus or systemic lupus erythematosus. Both CQ and HCQ are under investigation as potential treatment options for COVID-19. A multinational registry analysis failed to confirm the benefits conferred by CQ or HCQ in the treatment of COVID-19[1]. Besides, the WHO has halted the CQ or HCQ trials against COVID-19.
On the other hand, China has achieved first stage success on the combat with SARS-CoV-2 under the great efforts of the nation. The China plan, combining with Western medicine and traditional Chinese medicine, has played a major role in combating COVID-19[2]. The latest diagnosis and treatment guidelines on COVID-19 (8th version, 18th August, 2020) in China[3] recommended CQ phosphate trials while evaluating its efficacy. In addition, our study highlighted contradicting data concerning the clinical efficacies of CQ and HCQ against COVID-19 patients. We noted that CQ or HCQ serum or blood concentrations need further investigation, as the post-treatment serum drug concentration is directly associated with antiviral efficacy. Studies showed that the steady-state whole blood concentration of CQ was at least 16 μM, which should suffice in its antiviral effects with minimal toxicity[4]. “Expert consensus on CQ phosphate for the treatment of novel coronavirus pneumonia of China” recommended a therapy regimen of 500 mg twice daily for 7 d for patients weighing over 50 kg and 500 mg twice a day for the first 2 d and once daily for the rest of the 5 d for patients weighing ≤50 kg[5]. Besides, HCQ shows a wide variability among individuals[6], in terms of the efficacy and side effects.
Therefore, whereas CQ or HCQ may still harbor potential efficacies against the COVID-19 disease, their mechanisms of action need to be dissected.
However, the interactions between drug and disease are often non-linear and complex, traditional pharmacology investigations could not meet the relationships deciphering requirement. Network pharmacology[7], the next paradigm in drug discovery, can be applied to visualize and analyse the intrinsic information contained in drug-disease interaction network. Herein, an integrated strategy was proposed to explore the mechanism of CQ and HCQ action against COVID-19 based on both network pharmacology and in silico molecular docking. We applied network pharmacology to construct drug (CQ or HCQ)-target networks while network analysis was used to obtain functional sub-networks, then Gene Ontology (GO) biological processes and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were obtained with gene function enrichment analysis. Finally, the top ranked target genes were obtained and drug-target molecular docking was conducted. In addition, SARS-CoV-2 infection related target proteins such as Angiotensin- Converting Enzyme 2 (ACE2), 3-Chymotrypsin- Like protease (3CLpro) and Transmembrane Serine Protease 2 (TMPRSS2) were also used to carry out molecular docking with CQ and HCQ.
In summary, this study proposed an integrated strategy to systematically decipher the modes of action of CQ and HCQ from the molecular network view and explored the common and diverging aspects of these two drugs on the modulation of COVID-19 disease network. Our study provides an informative basis for further design and interrogation of CQ and HCQ on the treatment of COVID-19. However, more rigorous and high quality randomized controlled clinical trials are needed to provide definitive and reliable data. The strategy proposed herein could facilitate drug repurposing studies for COVID-19 treatment.
Materials and Methods
Collecting and sorting of drug targets:
Online public databases such as Drugbank (https:// www.drugbank.ca/), Therapeutic Target Database (TTD) (http://db.idrblab.net/ttd/), TargetNet (http:// targetnet.scbdd.com/), Data Recovery and Repair- Cloud Provider Interface (DRAR-CPI) (https://cpi. bio-x.cn/drar/), SwissTargetPrediction (http://www. swisstargetprediction.ch/) or PharmMapper (http:// lilab-ecust.cn/pharmmapper/) were used to collect and identify potential CQ or HCQ targets. We used an Area Under Curve (AUC) ≥0.7 and probability >0.9 as a target selection criteria in TargetNet. For DRAR- CPI, SwissTargetPrediction and PharmMapper, we used Z-score <-0.5, probability=1 and fit value >3.8 as the criteria. UniProt (https://www.uniprot.org/) and Database for Annotation, Visualization and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/) were used to convert and unify the target gene names. We filtered and removed duplicate genes and then used the rest to construct networks for further analysis.
Target network construction and analysis:
Coronavirus pneumonia was used as a key word to retrieve disease related genes and adopted databases such as PubMed Gene (https://www.ncbi.nlm.nih. gov/gene/), GeneCards (https://www.genecards. org/), Coats Disease (CTD) (http://ctdbase.org/) and Online Mendelian Inheritance in Man (OMIM) (https://omim.org/). Co-expressed ACE2 genes were downloaded from paper of Wang et al.[8] and GeneMANIA prediction server. The collected COVID-19 associated genes were standardized and converted into official gene symbols in Homo sapiens species. The common genes between COVID-19 and CQ targets were identified and then used String (https://string-db.org/) to retrieve Protein-Protein Interaction (PPI) networks. Cytoscape 3.7.2 was applied to construct and analyse CQ regulation networks. The same procedure was applied for HCQ.
Target function enrichment analysis:
DAVID was used to conduct functional enrichment analysis on common drug and disease targets and KEGG pathways with p<0.05 were obtained. We then applied the grammar of graphics plot2 (ggplot2) in R 3.6.1 to draw bubble diagrams.
Tissue enrichment maps:
The 51 CQ target genes which overlapped with COVID-19 genes were imported to DAVID 6.8 to carry out tissue enrichment analysis. We applied the function of “Tissue_Expression (UP_TISSUE) and Functional Annotation Chart” (p<0.05) with the background set as Homo sapiens. Similarly, the 47 HCQ target genes were conducted following the same protocol of CQ.
Molecular docking analysis:
In order to validate the potential CQ or HCQ target genes and explore their potential modes of action against COVID-19 disease, we employed AutoDock Vina (http://autodock.scripps.edu/) to verify and dissect the potential interaction modes. We retrieved the CQ (PubChem CID: 2719) or HCQ (PubChem CID: 3652) structures from PubChem (https:// pubchem.ncbi.nlm.nih.gov/). On the other hand, we downloaded the three-dimensional structures of the target proteins from Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB) database (https://www.rcsb.org/). The top ranked CQ or HCQ target proteins and the proteins associated with SARS-CoV-2 infection, including ACE2 (PDB: 1R42, X-ray diffraction, 2.20 Å resolution) and 3CLpro (PDB: 6LU7, X-ray diffraction, 2.16 Å resolution), were obtained from the RCSB PDB database. We employed SWISS-MODEL database (https://swissmodel.expasy.org/repository/ uniprot/O15393?csm=C05B5531C8A311C7) to obtain the HUMAN_Homology model TMPRSS2. Docking energy scores were used to evaluate the binding affinity between the drugs and the targets.
Results and Discussion
Our analysis showed a total of 153 and 116 targets for CQ and HCQ, respectively. 5713 genes were associated with COVID-19. 51 CQ and 47 HCQ target genes were related with COVID-19 disease.
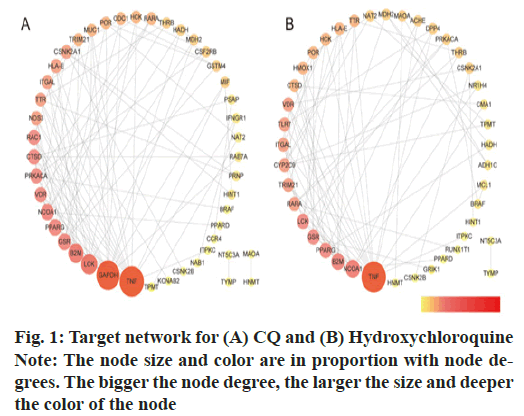
Using STRING, we obtained the PPIs for the drug- disease common genes, with which the COVID-19 disease regulation networks for CQ or HCQ were constructed (Cytoscape 3.7.2; fig. 1). CQ-target network contained 46 nodes and 95 edges with an average node degree of 4.13 (16 key targets degree >4.13). HCQ- target network had 42 nodes and 76 edges with the average node degree of 3.62 (18 key targets degree >3.62) as shown in Table 1.
| CQ targets | Degree | HCQ targets | Degree |
|---|---|---|---|
| TNF | 18 | TNF | 18 |
| GAPDH | 17 | NCOA1 | 8 |
| LCK | 10 | B2M | 7 |
| B2M | 9 | PPARG | 6 |
| NCOA1 | 7 | LCK | 6 |
| PPARG | 7 | GSR | 6 |
| GSR | 7 | RARA | 5 |
| PRKACA | 6 | VDR | 5 |
| VDR | 6 | TRIM21 | 5 |
| RAC1 | 6 | TLR7 | 5 |
| CTSD | 6 | CYP2C9 | 5 |
| HLA-E | 5 | ITGAL | 5 |
| CSNK2A1 | 5 | HLA-E | 4 |
| ITGAL | 5 | CTSD | 4 |
| NOS3 | 5 | HCK | 4 |
| TTR | 5 | POR | 4 |
| HMOX1 | 4 | ||
| TTR | 4 |
Note: Nodes with degree values over the corresponding average values, PRKACA: Protein Kinase CAMP-Activated Catalytic Subunit Alpha; VDR: Vitamin D Receptor; RAC1: RAS-related C3 botulinum toxin substrate 1; CTSD: Cathepsin D; HLA-E: Major Histocompatibility Complex E; CSNK2A1: Casein Kinase 2 Alpha 1; ITGAL: Integrin Subunit Alpha L; NOS3: Nitric Oxide Synthase 3; TTR: Transthyretin; RARA: Retinoic Acid Receptor Alpha; TRIM21: Tripartite Motif Containing 21; TLR7: Toll Like Receptor 7; CYP2C9: Cytochrome P450 Family 2 Subfamily C Member 9; ITGAL: Integrin Subunit Alpha L; HCK: HCK Proto-Oncogene; POR: Cytochrome P450 Oxidoreductase and HMOX1: Heme Oxygenase 1
Table 1: The Targets and Node Degree Values for CQ and Hydroxychloroquine
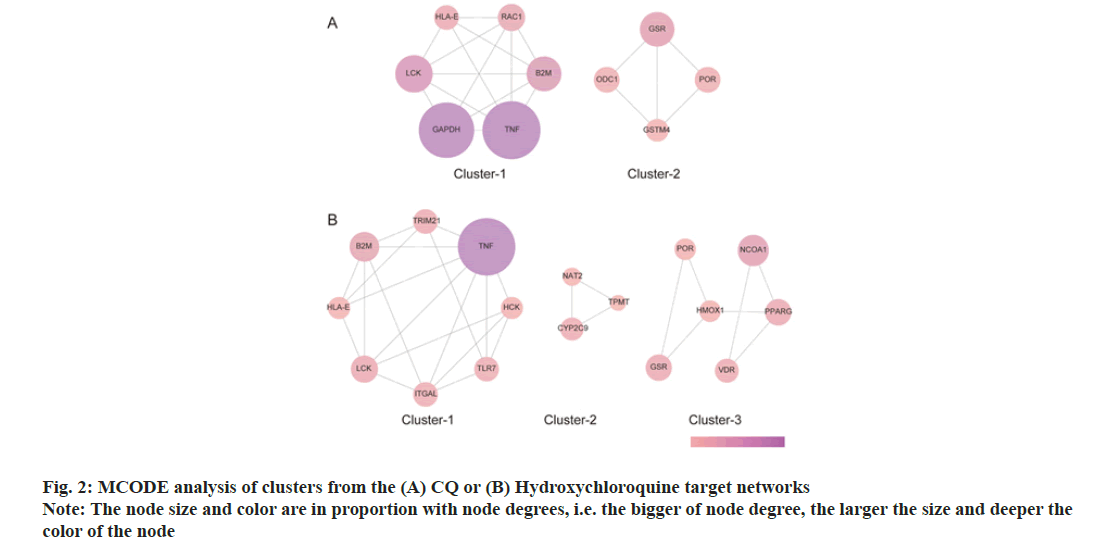
The Molecular Complex Detection (MCODE) analysis was conducted to analyze clusters. CQ had 2 clusters, while HCQ had 3 clusters as shown in fig. 2.
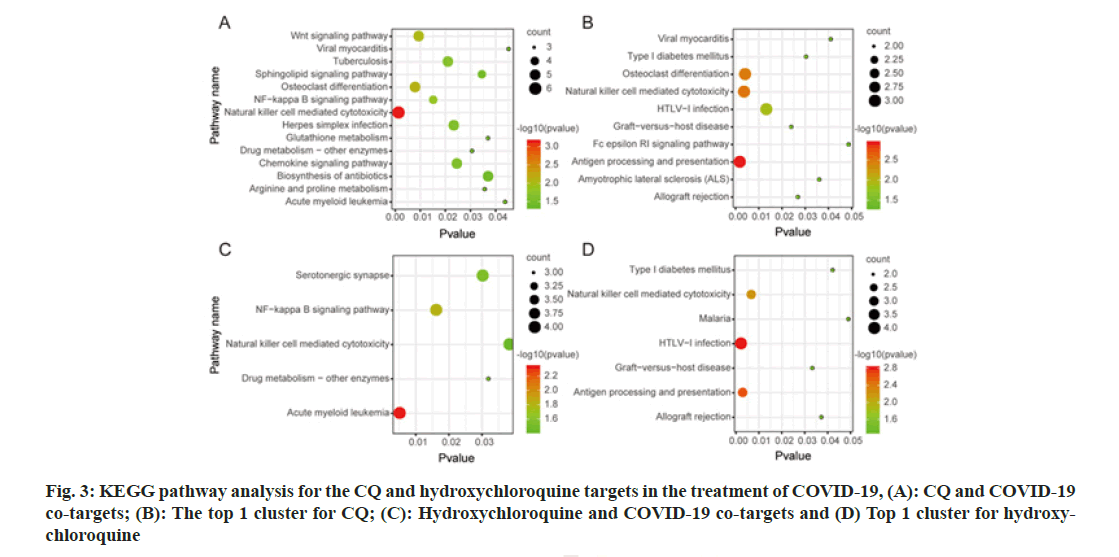
The common targets for CQ and COVID-19, and those for HCQ and COVID-19 were imported into DAVID, and then the GO enrichment and KEGG pathway analyses were conducted. A total of 14 KEGG pathways were enriched from CQ-target network as shown in fig. 3A (p<0.05), while 10 CQ pathways were retrieved from cluster 1 as shown in fig. 3B. Most of the retrieved pathways were involved in immunity and inflammation, virus infection, lipid metabolism and hematologic tumour development. On the other hand, there were 5 pathways enriched for HCQ as shown in fig. 3C, while 7 pathways were enriched from cluster 1 as shown in fig. 3D. The HCQ pathways were involved in immunity and inflammation, viral infection, synapse function and hematologic tumour development.
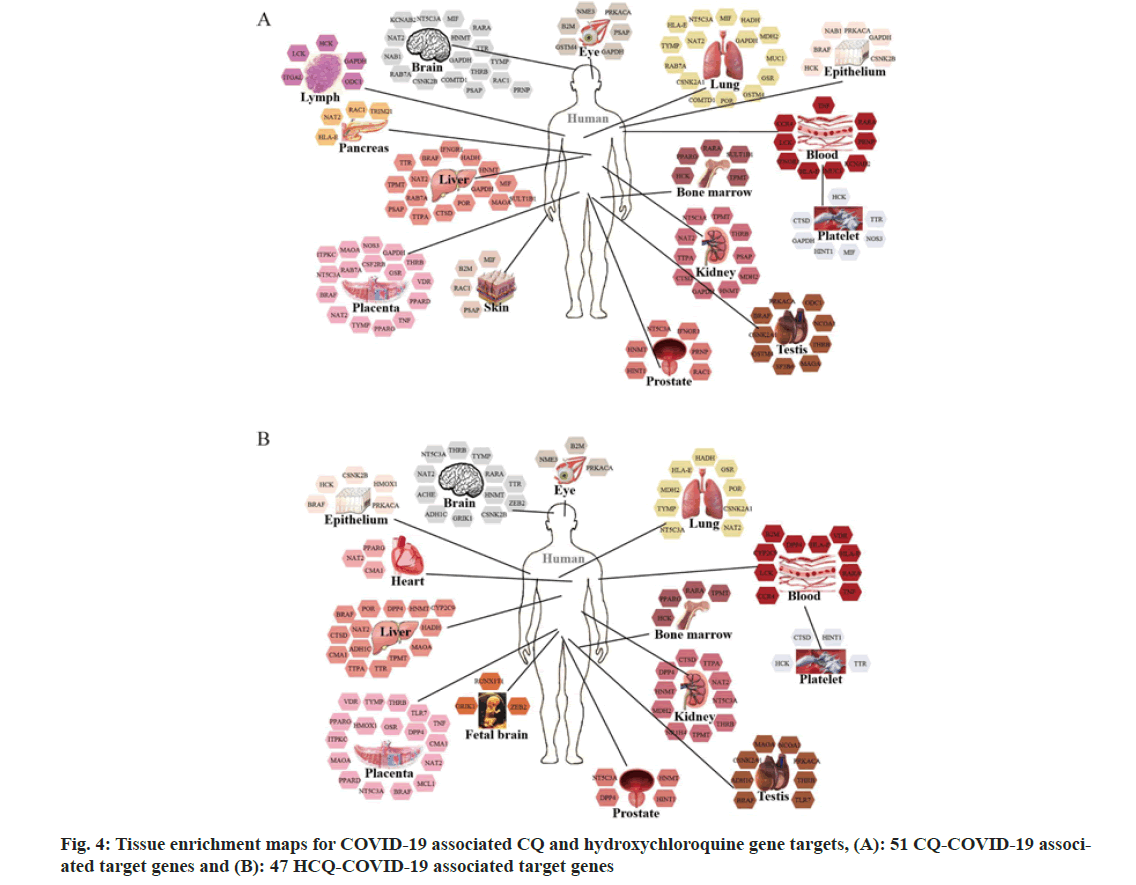
The 51 CQ-COVID-19 associated target genes were mainly enriched in the lung, liver, brain, placenta, kidney, blood, platelet, testis, prostate, eye, epithelium, bone marrow, lymph, pancreas and skin as shown in fig. 4A. On the other hand, the 47 HCQ-COVID-19 associated target genes were enriched in the lung, liver, brain, placenta, blood, platelet, kidney, testis, epithelium, bone marrow, prostate, eye, heart and fetal brain as shown in fig. 4B. These data may infer the potential tissue-specific regulation of CQ and HCQ against SARS-CoV-2 infection.
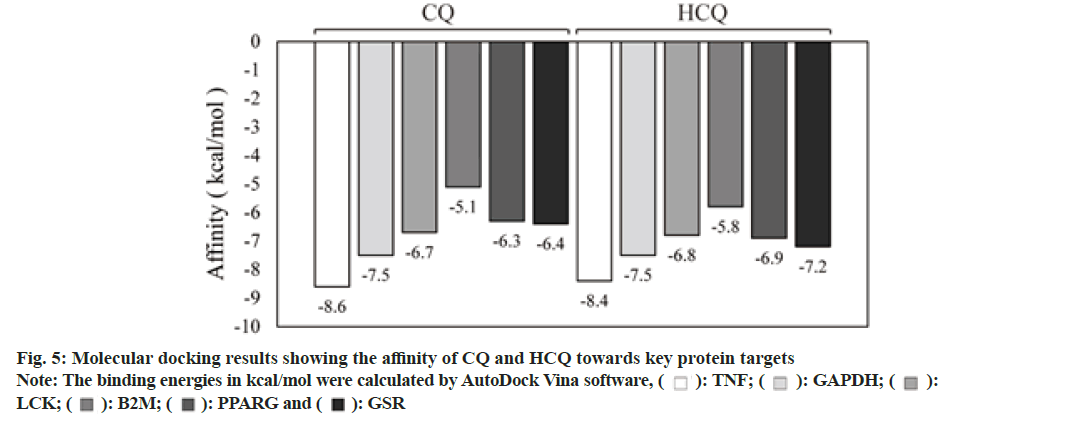
To establish and validate the binding affinity and modes between the drugs and the targets, we employed molecular docking experiments. The core target genes for CQ were Tumor Necrosis Factor (TNF), Glyceraldehyde 3-Phosphate Dehydrogenase (GAPDH), Lymphocyte-Specific Protein-Tyrosine Kinase (LCK), Beta-2 Microglobulin (B2M), Nuclear Receptor Coactivator 1 (NCOA1), Peroxisome Proliferator-Activated Receptor gamma (PPARγ) and Glutathione Disulfide Reductase (GSR). And the core target genes for HCQ included TNF, NCOA1, B2M, PPARG, LCK and GSR as shown in Table 1. Protein structures were obtained from PDB database: TNF (PDB:6OP0, X-ray diffraction, 2.55 Å resolution), NCOA1 (PDB:6GEV, X-ray diffraction, 1.54 Å resolution), LCK (PDB:1QPC, X-ray diffraction, 1.60 Å resolution), B2M (PDB:6TDQ, X-ray diffraction, 1.60 Å resolution), PPARG (PDB: 3ET3, X-ray diffraction, 1.95 Å resolution ), GAPDH (PDB: 6IQ6, X-ray diffraction, 2.29 Å resolution) and GSR (PDB: 3DK9, X-ray diffraction, 0.95 Å resolution).
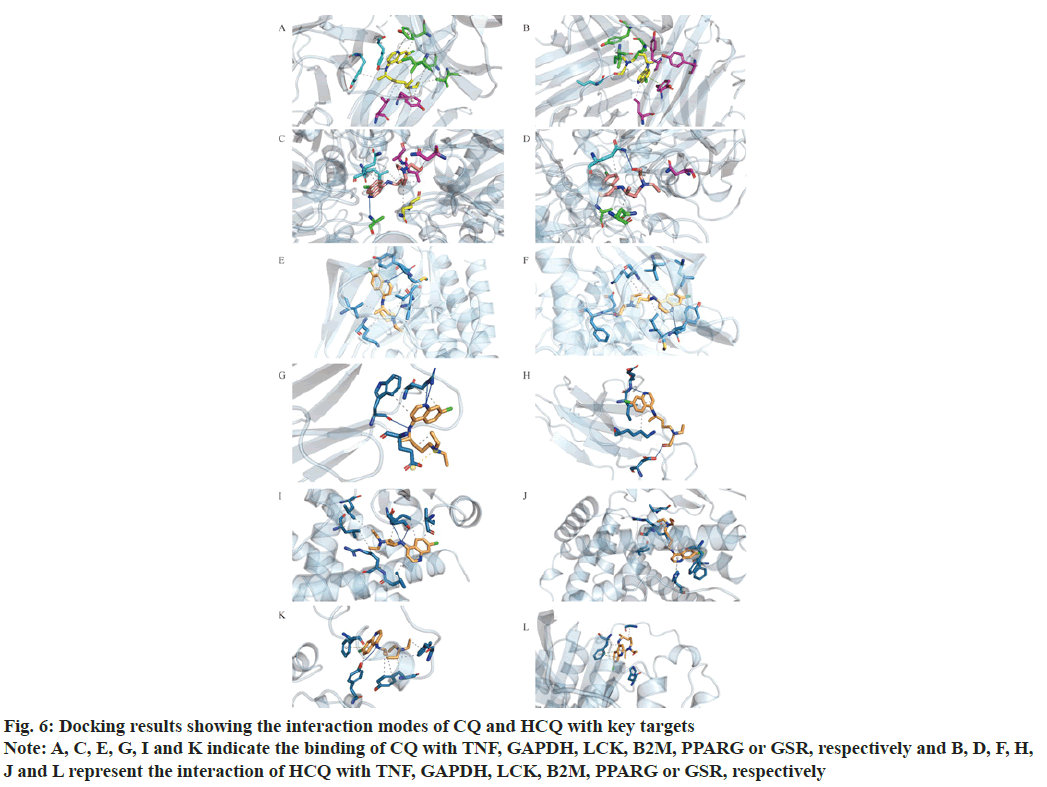
As shown in Table 2 and fig. 5-fig. 7, both CQ and HCQ had good binding affinity towards TNF (affinity=-8.6 and -8.4 kcal/mol, respectively) and GAPDH (affinity=-7.5 and -7.5 kcal/mol), but HCQ showed a better binding affinity towards GSR (affinity=-7.2 kcal/ mol). Both drugs showed marginal binding with LCK, B2M, PPARG, ACE2, 3CLpro and TMPRSS2 (affinity ranged from -7.0 to -5.0 kcal/mol). However, both drugs had a binding energy of more than -5 kcal/mol with NCOA1.
Figure 6: Docking results showing the interaction modes of CQ and HCQ with key targets
Note: A, C, E, G, I and K indicate the binding of CQ with TNF, GAPDH, LCK, B2M, PPARG or GSR, respectively and B, D, F, H, J and L represent the interaction of HCQ with TNF, GAPDH, LCK, B2M, PPARG or GSR, respectively
| Drugs | Affinity (kcal/mol) | ||
|---|---|---|---|
| ACE2 | 3CLpro | TMPRSS2 | |
| CQ | -6.5 | -5.4 | -5.2 |
| Hydroxychloroquine | -6.6 | -6.1 | -6.1 |
Table 2: Binding Affinity of CQ and HCQ with Proteins Associated with the SARS-COV-2 Infection
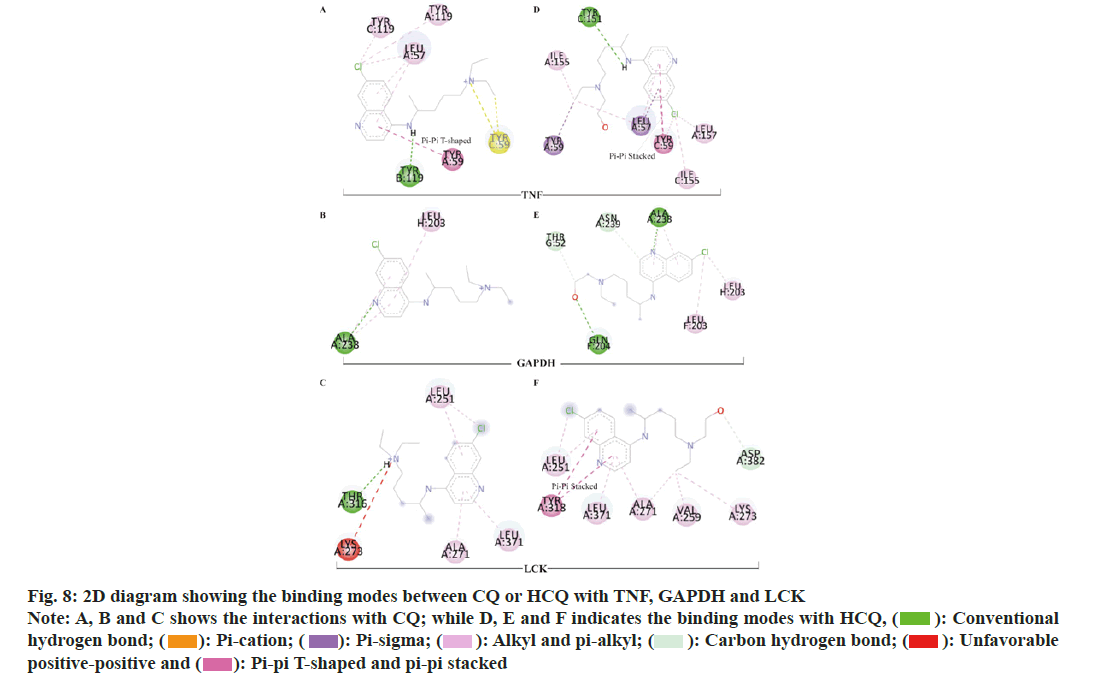
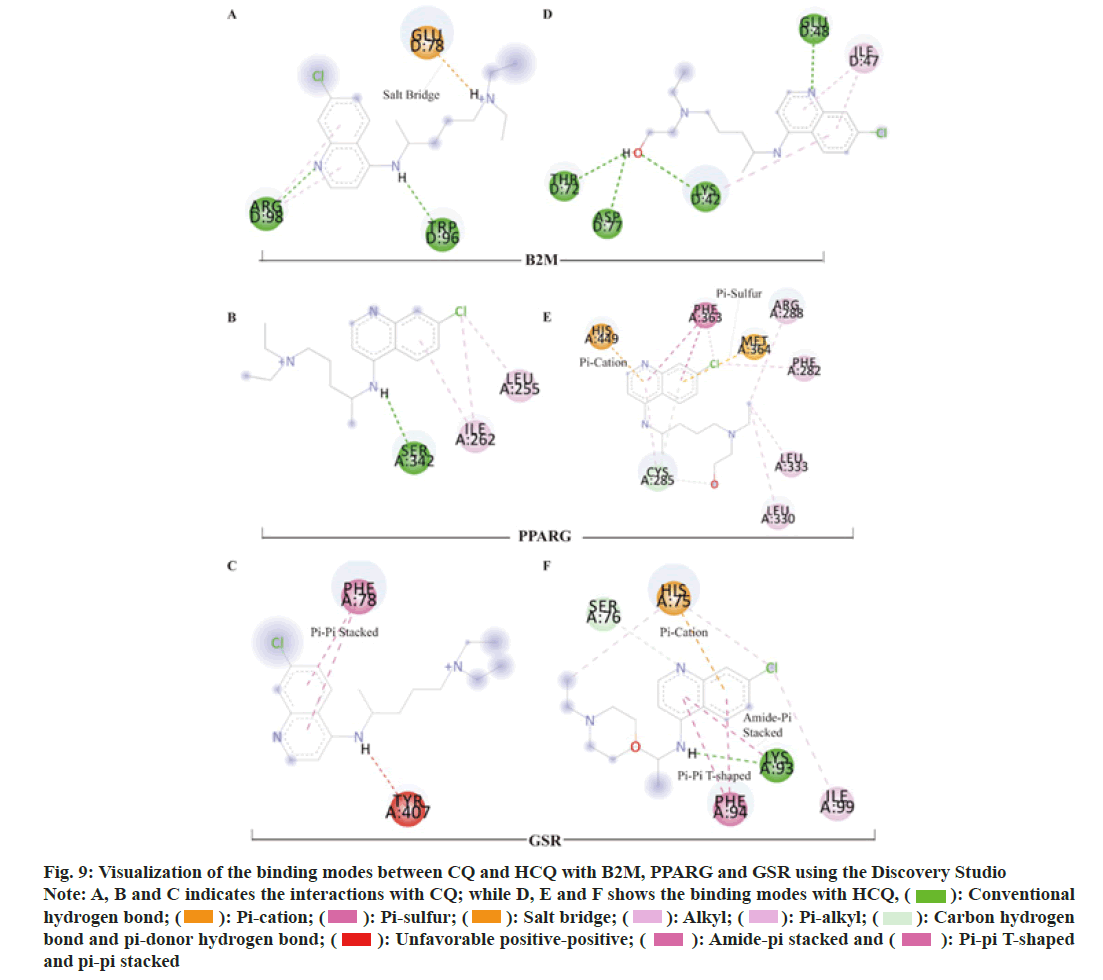
To elucidate the potential mechanisms underlying the binding differences between CQ and HCQ with their targets, the BIOVIA Discovery Studio v.16.1 was applied to assess the receptor-ligand interactions. Two- Dimensional (2D) diagram in the Discovery Studio was used to visualize and analyze the interaction modes of CQ and HCQ with TNF, GAPDH, LCK as shown in fig. 8, B2M, PPARG and GSR as shown in fig. 9. The detailed interaction types and sites are as listed in Table 3.
| Targets | Drug | Interaction Types | Sites |
|---|---|---|---|
| TNF | CQ | H-bond | Chain B: TYR 119 |
| Pi-pi T-shaped interaction | Chain A: TYR 59 | ||
| Pi-cation | Chain C: TYR 59 | ||
| Pi-alkyl and pi-sigma | Chain A: LEU 57, TYR119; Chain C: TYR119 | ||
| HCQ | H-bond | Chain C: TYR 151 | |
| Pi-pi stacked | Chain C: TYR 59 | ||
| Pi-sigma | Chain A: TYR 59, LEU 57 | ||
| GAPDH | CQ | H-bond | Chain A: ALA 238 |
| Pi-alkyl | Chain H: LEU 203 | ||
| HCQ | H-bonds | Chain A: ALA 238; Chain F: GLN 204 | |
| C−H bonds | Chain A: ASN 239; Chain G: THR 52 | ||
| Pi-alkyl | Chain A: ALA 238 | ||
| LCK | CQ | H-bond | Chain A: THR 316 |
| Pi-alkyl | Chain A: LEU 251, ALA 271, LEU 371 | ||
| Unfavorable positive-positive | Chain A: LYS 273 | ||
| HCQ | C−H bond | Chain A: ASP 382 | |
| Pi-pi stacked | Chain A: TYR 318 | ||
| Pi-alkyl interactions | Chain A: LEU 251, ALA 271, LEU 371 | ||
| B2M | CQ | H-bonds | Chain D: ARG 98 and TRP 96 |
| Salt-bridge | Chain D: GLU 78 | ||
| Pi-alkyl interactions | Chain D: ARG 98 | ||
| HCQ | H-bonds | Chain D: LYS 42, GLU 48, THR 72 and ASP 77 | |
| Pi-alkyl interactions | Chain D: ILE 47 and LYS 42 | ||
| PPARG | CQ | H-bond | Chain A: SER 342 |
| Pi-alkyl | Chain A: ILE 262 | ||
| HCQ | C−H bond | Chain A: CYS 285 | |
| Pi-donor H-bond | Chain A: CYS 285 | ||
| Pi-cation | Chain A: HIS 449 | ||
| Pi-sulfur | Chain A: MET 364 | ||
| Pi-pi stacked | Chain A: PHE 363 | ||
| Pi-pi T-shaped | Chain A: PHE 363 | ||
| Pi-alkyl | Chain A: CYS 285 | ||
| GSR | CQ | Pi-pi stacked interaction | Chain A: PHE 78 |
| Unfavorable donor-donor interaction | Chain A: TYR 407 | ||
| HCQ | H-bond | Chain A: LYS 93 | |
| C−H bond | Chain A: SER 76 | ||
| Pi-cation | Chain A: HIS 75 | ||
| Pi-pi T-shaped | Chain A: PHE 94 | ||
| Amide-pi stacked | Chain A: LYS 93 | ||
| Pi-alkyl | Chain A: PHE 94 |
Table 3: CQ and HCQ Interaction Types and Modes With TNF, GAPDH, LCK, B2M, PPARG OR GSR
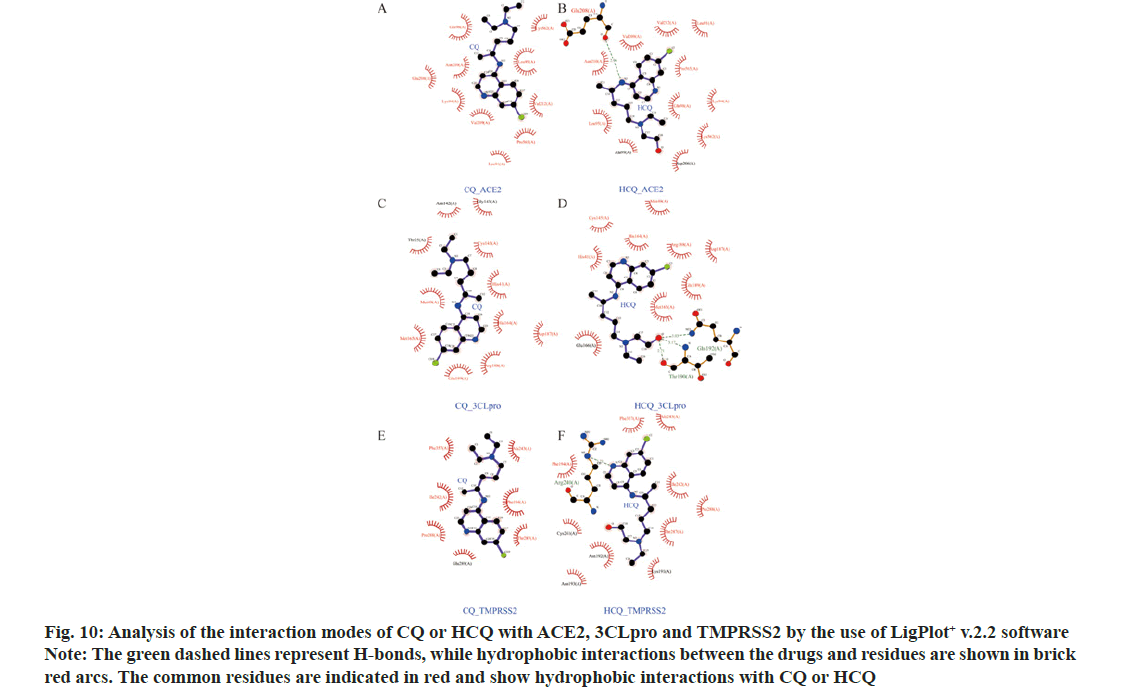
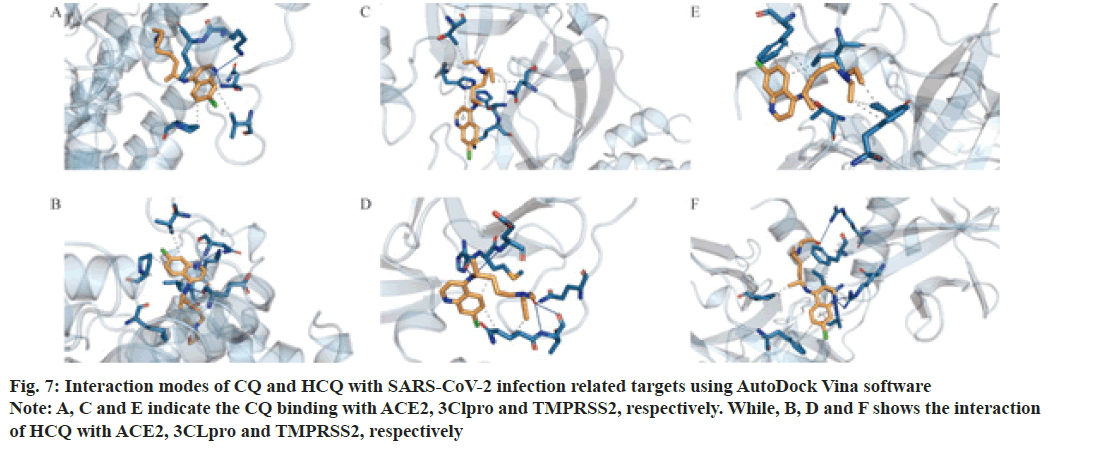
In order to understand the modes of binding between CQ or HCQ with ACE2, 3CLpro and TMPRSS2, we employed LigPlot+ v.2.2 software. HCQ had H-bond interactions with ACE2 (Chain A: Glu208), 3CLpro (Chain A: Thr 190 and Gln 192) and TMPRSS2 (Chain A: Arg 240) (Green dashed lines in fig. 10). Both CQ and HCQ shared most part of hydrophobic residues and the common residues are highlighted in red. Both CQ and HCQ were in the hydrophobic pockets which consisted of hydrophobic interactions between residues and drugs.
Figure 10: Analysis of the interaction modes of CQ or HCQ with ACE2, 3CLpro and TMPRSS2 by the use of LigPlot+ v.2.2 software
Note: The green dashed lines represent H-bonds, while hydrophobic interactions between the drugs and residues are shown in brick red arcs. The common residues are indicated in red and show hydrophobic interactions with CQ or HCQ
This study aimed to elucidate the common and diverging mechanisms of CQ and HCQ against COVID-19. We employed network pharmacology and in silico molecular docking tools to dissect the common and different pathways affected by the drugs. This study could provide more insights into the modes of action and may facilitate a better understanding of CQ and HCQ with their potential regulation effects against SARS-CoV-2 infection.
The study analyzed the top 6 target genes as shown in Table 1 and gene functions were retrieved from GeneCards. The main targets for CQ are TNF, GAPDH, LCK, B2M, NCOA1 and PPARG. On the other hand, TNF, NCOA1, B2M, PPARG, LCK and GSR are the key targets for HCQ. Briefly, TNF is a pro-inflammatory cytokine secreted by macrophages, which participates in the immune and inflammatory responses. GAPDH is involved in glycolysis, transcription, RNA transport, DNA replication and apoptosis. LCK plays an important role in the development and maturation of T cells and participates in signal transduction related to T-cell antigen receptors. B2M is a component of histocompatibility complex and is involved in the expression of peptide antigen to the immune system. NCOA1 is a nuclear receptor-assisted activator and participates in transcription activity in a hormone- dependent manner. PPARG is an important regulator of adipocyte differentiation and glucose status, and is involved in the regulation of energy metabolism. GSR is a key enzyme in the cellular antioxidant defense system.
Through network and functional enrichment analysis, our data showed that the CQ targets participate in the regulation of signalling pathways associated with viral infection, immune inflammation, bacterial infection, steroid metabolism, amino acid metabolism or drug metabolism enzymes. On the other hand, HCQ mainly involved pathways such as immune inflammation, drug metabolism and serotonergic nerve synaptic function. The regulation of immune inflammation and drug metabolism enzymes was common with CQ and HCQ. However, the drugs showed some divergence which may be related to their structures. CQ is more liposoluble, while HCQ shows regulation effect on nerve synapse.
Both CQ and HCQ had good binding affinity towards TNF (affinity=-8.6 and -8.4 kcal/mol, respectively) and GAPDH (affinity=-7.5 and -7.5 kcal/mol). However, HCQ showed a better binding affinity towards GSR (affinity=-7.2 kcal/mol), because HCQ had more H-bonds and binding interactions than CQ as shown in Table 3.
SARS-CoV-2 infection related target proteins, ACE2, 3CLpro and TMPRSS2 were also used to carry out molecular docking with CQ and HCQ. Our results showed that CQ and HCQ both had good affinity with ACE2, 3CLpro and TMPRSS2. However, HCQ manifested better binding affinity with the three proteins comparing with that of CQ. The common residues of CQ and HCQ constitute hydrophobic pocket, in which the target proteins showed interactions with CQ or HCQ. However, HCQ had H-bond interactions with ACE2 (Chain A: Glu208), 3CLpro (Chain A: Thr 190 and Gln 192) and TMPRSS2 (Chain A: Arg 240) (Green dashed lines in fig. 10), while CQ had no H-bond in the hydrophobic pocket. Other studies also showed a better binding affinity of HCQ with TMPRSS2 than that of CQ[9,10].
Previous studies on SARS-COV-2 in Vero E6 cell model showed that CQ had potential activity in the prevention and treatment of novel coronavirus[11]. HCQ could block the entry of SARS-CoV virus into human cells by changing the glycosylation of ACE2 receptor protein and played an inhibitory effect on the invasion of the virus[12]. In addition, COVID-19 patients exhibit an overactive immune inflammatory response, which is also known as "cytokine storm", which causes detrimental effects to the patients[13]. Autopsy reports of patients who died from COVID-19 revealed substantial lung lesions, liver enlargement, hepatocellular degeneration and cardiovascular system damage[3]. Due to the fact that ACE2 is expressed in the myocardium, the heart has become an important target organ for SARS-COV-2. Studies have summarized the mechanisms of heart damage associated with SARS- COV-2 infection, providing a basis for CQ and HCQ to play roles in combating cardiac detriments[14].The role of CQ and HCQ in regulating immune inflammation may be the main mechanism for their potential anti- COVID-19 effects. Meanwhile, both drugs may be involved in the regulation of drug metabolic enzymes. Besides, CQ affects the metabolism of small lipid molecules and amino acids, and may also be involved in the protection of impaired cardiac and liver functions in COVID-19 patients. HCQ is involved in the regulation of synapses, and may play a role in arachidonic acid metabolism and neuroprotection.
What’s more, tissue enrichment analysis of the CQ and HCQ targets showed the potential regulation of susceptible tissue targets as shown in fig. 4. Both the 51 or 47 COVID-19-associated target genes for CQ or HCQ were enriched in the lung, liver, brain, placenta, kidney, blood, platelet, testis, prostate, eye, epithelium and bone marrow. Besides, CQ targets were exclusively enriched in the lymph, pancreas and skin, while HCQ targets were uniquely enrichment in the heart and fetal brain. ACE2 and its co-factor TMPRSS2 are primarily expressed in bronchial transient secretory cells[15], while the lung is the main organ that suffers the SARS-CoV-2 infection[16]. ACE2 receptors were also identified in gastrointestinal tract and the cholangiocytes of the liver. Hepatic dysfunction occurs in 14 %-53 % of COVID-19 patients[17]. On the other hand, study reported the first case of SARS-CoV- 2-associated meningitis/encephalitis[18]. So far, there are several studies discussing the potential invasion mechanisms of SARS-CoV-2 into the brain[19-21]. There are conflicting data on the effect of SARS- CoV-2 in female reproductive system. Some studies have reported the mechanisms of placenta invasion by the virus[22-24]. In addition, maternal COVID-19 may affect fetal development and phosphatidylcholine or choline supplements have been shown to help mitigate the detrimental fetal brain effects[25]. For hospitalized COVID-19 patients, 36.6 % developed acute kidney injury[26]. It has been shown that SARS-CoV-2 can infect podocytes and tubular epithelial cells which attributed to the renal abnormalities[27]. Hypoxia and hypercoagulability may also cause renal injury[28]. In the circulation system, morphological anomalies in the blood cells and coagulation dysfunction have been observed[29-31]. SARS-CoV-2 infection was associated with platelet hyperactivity and could contribute to blood coagulation[32]. In fact, COVID-19 often causes thrombosis during SARS-CoV-2 infection[33]. There is conflicting data on the effect of the SARS-CoV-2 on male reproductive. It has been reported that the human testis is a potential target for SARS-CoV-2[34] and ACE2 may be mediating the effect[35]. COVID-19 may cause significant seminiferous tubular injury, Leydig cells decrease and mild lymphocytic inflammation of the testis[36]. TMPRSS2 together with ACE2 are essential for the cell invasion of the SARS-CoV-2. TMPRSS2 is expressed in normal prostatic epithelium and is increased in malignant prostatic tissue. Report showed that expressed prostatic secretion of COVID-19 patients was absent of coronavirus[37]. Research indicated that nasolacrimal system is a conduit between the eye and the respiratory tract, so more attention should be paid to SARS-CoV-2 transmission through the eye[38]. SARS- CoV-2 appears to preferentially target respiratory epithelium. While ACE2 is mainly expressed in the respiratory epithelium, the different tissue epithelium, such as gastrointestinal epithelium, nasal cavity olfactory epithelium and conjunctival, limbal or corneal epithelium, are all susceptible to the SARS- CoV-2 infection[39-41]. There are few reports about SARS-CoV-2 invasion in the bone marrow. The first case reported the persistent pancytopenia attributed to the SARS-CoV-2 infection of bone marrow[42,43].
Our data showed that the CQ targets were exclusively enriched in the lymph, pancreas and skin, while HCQ targets were uniquely enrichment in heart and fetal brain. It has been demonstrated that severe and critical COVID-19 patients showed higher incidences of lymph node enlargement, compared with the ordinary patients[44]. Another study showed the first report of SARS-CoV-2 infection-associated hemophagocytic lymphohistiocytosis in autopsy[45]. Based on evidences concerning the multi-organ abnormalities attributed to the SARS-CoV-2, COVID-19 should be recognized as a multi-systemic and polyhedral disease. ACE2 receptors are expressed in various tissues, including respiratory tract, heart, kidney, gastrointestinal tract, liver, pancreas, nervous systems and skin[46]. ACE2 imbalance in the pancreas may cause acute beta-cell dysfunction[47]. Besides, the SARS-CoV-2 infection in the endocrine pancreas may activate Na+/H+ Exchanger 2 (NHE2) and increase lactate levels[48]. In addition, plenty studies have also reported various skin lesions and cutaneous manifestations of COVID-19, but the associated mechanisms need furth51er interrogation[49,50]. Single nucleus RNA sequencing study showed that pericytes with high expression of ACE2 might act as cardiac cell targets for SARS-CoV-2[51]. SARS-CoV-2 infection may cause direct and indirect damages to the heart, with abnormal cardiac injury biomarkers, structural and functional detriments. Myocarditis and heart failure are the common cardiac manifestations in COVID-19 patients[14,52].
Taken together, our data robustly demonstrate CQ and HCQ may have potential regulatory effects on COVID-19 disease network, which may affect multiple organs, protein targets and pathways. Despite the common and diverging aspects in the modes of multi- actions, CQ and HCQ both affect common pathways which are associated with SARS-CoV-2 entry and infection. The results indicate that both CQ and HCQ could have potential to inhibit over-activated immunity and inflammation. Routine measurements of the CQ and HCQ blood concentrations and tailored therapy regimen may be essential for individual COVID-19 patient. However, further rigorous and high quality randomized controlled clinical trials are warranted to validate the antiviral effects of CQ and HCQ against SARS-CoV-2. Our proposed strategy could facilitate the drug repurposing efforts for COVID-19 treatment.
Acknowledgements:
This research was funded by the Natural Science Foundation of Zhejiang Province (grant number LYY19H280005 and number LQ17H280001); the Scientific Foundation in Traditional Chinese Medicine of Zhejiang Province (grant number 2017ZQ008); the Natural Science Foundation of Hangzhou Medical College (grant number 2015B01); the Zhejiang Pharmaceutical Association (grant number 2018ZYY11); the “13th Five-Year” Chinese Medicine Key Discipline in Zhejiang Province-Chinese Medicine Quality and functional evaluation (grant number 2017- XK-A43 to J. Shi). We thank HOME for Researchers for its linguistic assistance during the preparation of this manuscript.
Conflict of interests:
The authors declared no conflicts of interest.
References
- Mehra MR, Desai SS, Ruschitzka F, Patel AN. Retracted: Hydroxychloroquine or chloroquine with or without a macrolide for treatment of COVID-19: A multinational registry analysis. Lancet 2020;6736(20):31180-6.
[Crossref] [Google Scholar] [Pub Med]
- Daily C. White paper–Fighting COVID-19: China in action. Recuperado desde; 2020.
- Wang GQ, Zhao L, Wang X, Jiao YM, Wang FS. Diagnosis and treatment protocol for COVID-19 patients (Tentative 8th Edition): Interpretation of updated key points. Infect Immun Dis 2021;1(1):17–9.
[Crossref]
- Wei ZX, Tang TT, Jiang SP. The antiviral mechanisms, effects, safety and adverse effects of chloroquine. Eur Rev Med Pharmacol Sci 2020;24(12):7164-72.
[Crossref] [Google Scholar] [Pub Med]
- “Notice on adjusting the usage and dosage of chloroquine phosphate for the COVID-19 Infection”. National Administration of Traditional Chinese Medicine Office Medical Letter; 2020.
- Costedoat Chalumeau N, Le Guern V, Piette JC. Routine hydroxychloroquine blood concentration measurement in systemic lupus erythematosus reaches adulthood. J Rheumatol 2015;42(11):1997-9.
[Crossref] [Google Scholar] [Pub Med]
- Hopkins AL. Network pharmacology: The next paradigm in drug discovery. Nat Chem Biol 2008;4(11):682-90.
[Crossref] [Google Scholar] [Pub Med]
- Wang J, Zhao S, Liu M, Zhao Z, Xu Y, Wang P, et al. ACE2 expression by colonic epithelial cells is associated with viral infection, immunity and energy metabolism. MedRxiv 2020.
- Dave GS, Rakholiya KD, Kaneria MJ, Galvadiya BP, Vyas SR, Kanbi VH, et al. High affinity interaction of Solanum tuberosum and Brassica juncea residue smoke water compounds with proteins involved in coronavirus infection. Phytother Res 2020;34(12):3400-10.
[Crossref] [Google Scholar] [Pub Med]
- Marciniec K, Beberok A, Boryczka S, Wrze?niok D. The application of in silico experimental model in the assessment of ciprofloxacin and levofloxacin interaction with main SARS-CoV-2 targets: S-, E-and TMPRSS2 proteins, RNA-dependent RNA polymerase and papain-like protease (PLpro)-preliminary molecular docking analysis. Pharmacol Rep 2021;73(6):1765-80.
[Crossref] [Google Scholar] [Pub Med]
- Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 2020;30(3):269-71.
[Crossref] [Google Scholar] [Pub Med]
- Savarino A, di Trani L, Donatelli I, Cauda R, Cassone A. New insights into the antiviral effects of chloroquine. Lancet Infect Dis 2006;6(2):67-9.
[Crossref] [Google Scholar] [Pub Med]
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395(10223):497-506.
- Ammirati E, Wang DW. SARS-CoV-2 inflames the heart. The importance of awareness of myocardial injury in COVID-19 patients. Int J Cardiol 2020;311:122-23.
[Crossref] [Google Scholar] [Pub Med]
- Lukassen S, Chua RL, Trefzer T, Kahn NC, Schneider MA, Muley T, et al. SARS?CoV?2 receptor ACE 2 and TMPRSS 2 are primarily expressed in bronchial transient secretory cells. EMBO J 2020;39(10):e105114.
[Crossref] [Google Scholar] [Pub Med]
- Barton LM, Duval EJ, Stroberg E, Ghosh S, Mukhopadhyay S. Covid-19 autopsies, oklahoma, USA. Am J Clin Pathol 2020;153(6):725-33.
[Crossref] [Google Scholar] [Pub Med]
- Jothimani D, Venugopal R, Abedin MF, Kaliamoorthy I, Rela M. COVID-19 and the liver. J Hepatol 2020;73(5):1231-40.
[Crossref] [Google Scholar] [Pub Med]
- Moriguchi T, Harii N, Goto J, Harada D, Sugawara H, Takamino J, et al. A first case of meningitis/encephalitis associated with SARS-Coronavirus-2. Int J Infect Dis 2020;94:55-8.
[Crossref] [Google Scholar] [Pub Med]
- Saavedra JM. COVID-19, angiotensin receptor blockers and the brain. Cell Mol Neurobiol 2020;40(5):667-74.
[Crossref] [Google Scholar] [Pub Med]
- Wu Y, Xu X, Chen Z, Duan J, Hashimoto K, Yang L, et al. Nervous system involvement after infection with COVID-19 and other coronaviruses. Brain Behav Immun 2020;87:18-22.
[Crossref] [Google Scholar] [Pub Med]
- Li Z, Liu T, Yang N, Han D, Mi X, Li Y, et al. Neurological manifestations of patients with COVID-19: Potential routes of SARS-CoV-2 neuroinvasion from the periphery to the brain. Front Med 2020;14(5):533-41.
[Crossref] [Google Scholar] [Pub Med]
- Algarroba GN, Rekawek P, Vahanian SA, Khullar P, Palaia T, Peltier MR, et al. Visualization of severe acute respiratory syndrome coronavirus 2 invading the human placenta using electron microscopy. Am J Obstet Gynecol 2020;223(2):275-8.
[Crossref] [Google Scholar] [Pub Med]
- Li M, Chen L, Zhang J, Xiong C, Li X. The SARS-CoV-2 receptor ACE2 expression of maternal-fetal interface and fetal organs by single-cell transcriptome study. PloS One 2020;15(4):e0230295.
- Hosier H, Farhadian SF, Morotti RA, Deshmukh U, Lu-Culligan A, Campbell KH, et al. SARS–CoV-2 infection of the placenta. J Clin Invest 2020;130(9):4947-53.
[Crossref] [Google Scholar] [Pub Med]
- Freedman R, Hunter SK, Law AJ, D'Alessandro A, Noonan K, Wyrwa A, et al. Maternal choline and respiratory coronavirus effects on fetal brain development. J Psychiatr Res 2020;128:1-4.
[Crossref] [Google Scholar] [Pub Med]
- Hirsch JS, Ng JH, Ross DW, Sharma P, Shah HH, Barnett RL, et al. Acute kidney injury in patients hospitalized with COVID-19. Kidney Int 2020;98(1):209-18.
[Crossref] [Google Scholar] [Pub Med]
- Martinez Rojas MA, Vega Vega O, Bobadilla NA. Is the kidney a target of SARS-CoV-2? Am J Physiol Renal Physiol 2020;318(6):F1454-62.
[Crossref] [Google Scholar] [Pub Med]
- Yang X, Jin Y, Li R, Zhang Z, Sun R, Chen D. Prevalence and impact of acute renal impairment on COVID-19: A systematic review and meta-analysis. Crit Care 2020;24(1):356.
[Crossref] [Google Scholar] [Pub Med]
- Connors JM, Levy JH. COVID-19 and its implications for thrombosis and anticoagulation. Blood 2020;135(23):2033-40.
[Crossref] [Google Scholar] [Pub Med]
- Zini G, Bellesi S, Ramundo F, d'Onofrio G. Morphological anomalies of circulating blood cells in COVID?19. Am J Hematol 2020;95(7):870-2.
[Crossref] [Google Scholar] [Pub Med]
- Lemke G, Silverman GJ. Blood clots and TAM receptor signalling in COVID-19 pathogenesis. Nat Rev Immunol 2020;20(7):395-6.
- Denorme F, Middleton EA, Portier I, Rowley JW, Stubben CJ. Platelet gene expression and function in COVID-19 patients. Blood 2020;136(11):1317-29.
[Crossref] [Google Scholar] [Pub Med]
- Bikdeli B, Madhavan MV, Jimenez D, Chuich T, Dreyfus I, Driggin E, et al. COVID-19 and thrombotic or thromboembolic disease: Implications for prevention, antithrombotic therapy and follow-up: JACC state-of-the-art review. J Am Coll Cardiol 2020;75(23):2950-73.
[Crossref] [Google Scholar] [Pub Med]
- Wang Z, Xu X. scRNA-seq profiling of human testes reveals the presence of the ACE2 receptor, a target for SARS-CoV-2 infection in spermatogonia, Leydig and Sertoli cells. Cells 2020;9(4):920.
[Crossref] [Google Scholar] [Pub Med]
- Younis JS, Abassi Z, Skorecki K. Is there an impact of the COVID-19 pandemic on male fertility? The ACE2 connection. Am J Physiol Endocrinol Metabol 2020;318(6):E878-80.
[Crossref] [Google Scholar] [Pub Med]
- Yang M, Chen S, Huang BO, Zhong JM, Su H, Chen YJ, et al. Pathological findings in the testes of COVID-19 patients: Clinical implications. Eur Urol Focus 2020;6(5):1124-9.
[Crossref] [Google Scholar] [Pub Med]
- Zhang S, Wang X, Zhang H, Xu A, Fei G, Jiang X, et al. The absence of coronavirus in expressed prostatic secretion in COVID-19 patients in Wuhan city. Reprod Toxicol 2020;96:90-4.
[Crossref] [Google Scholar] [Pub Med]
- Ho D, Low R, Tong L, Gupta V, Veeraraghavan A, Agrawal R. COVID-19 and the ocular surface: A review of transmission and manifestations. Ocul Immunol Inflamm 2020;28(5):726-34.
[Crossref] [Google Scholar] [Pub Med]
- Zhang H, Rostami MR, Leopold PL, Mezey JG, O’Beirne SL, Strulovici Barel Y, et al. Expression of the SARS-CoV-2 ACE2 receptor in the human airway epithelium. Am J Respir Crit Care Med 2020;202(2):219-29.
[Crossref] [Google Scholar] [Pub Med]
- Collin J, Queen R, Zerti D, Dorgau B, Georgiou M, Djidrovski I, et al. Co-expression of SARS-CoV-2 entry genes in the superficial adult human conjunctival, limbal and corneal epithelium suggests an additional route of entry via the ocular surface. Ocul Surf 2021;19:190-200.
[Crossref] [Google Scholar] [Pub Med]
- Uno Y. Why does SARS-CoV-2 invade the gastrointestinal epithelium? Gastroenterology 2020;159(4):1622-3.
[Crossref] [Google Scholar] [Pub Med]
- Issa N, Lacassin F, Camou F. First case of persistent pancytopenia associated with SARS-CoV-2 bone marrow infiltration in an immunocompromised patient. Ann Oncol 2020;31(10):1418-9.
[Crossref] [Google Scholar] [Pub Med]
- Prieto-Pérez L, Fortes J, Soto C, Vidal-González Á, Alonso-Riaño M, Lafarga M, et al. Histiocytic hyperplasia with hemophagocytosis and acute alveolar damage in COVID-19 infection. Mod Pathol 2020;33(11):2139-46.
[Crossref] [Google Scholar] [Pub Med]
- Li K, Wu J, Wu F, Guo D, Chen L, Fang Z, et al. The clinical and chest CT features associated with severe and critical COVID-19 pneumonia. Investigat Radiol 2020;55(6):327-31.
[Crossref] [Google Scholar] [Pub Med]
- Prilutskiy A, Kritselis M, Shevtsov A, Yambayev I, Vadlamudi C, Zhao Q, et al. SARS-CoV-2 infection-associated hemophagocytic lymphohistiocytosis: An autopsy series with clinical and laboratory correlation. Am J Clin Pathol 2020;154(4):466-74.
[Crossref] [Google Scholar] [Pub Med]
- Tuttolomondo D, Frizzelli A, Aiello M, Bertorelli G, Majori M, Chetta A. Beyond the lung involvement in COVID-19 patients. A review. Minerva Med 2020.
[Crossref] [Google Scholar] [Pub Med]
- Cuschieri S, Grech S. COVID-19 and diabetes: The why, the what and the how. J Diabetes Complication 2020;34(9):107637.
- Cure E, Cumhur Cure M. COVID-19 may affect the endocrine pancreas by activating Na+/H+ exchanger 2 and increasing lactate levels. J Endocrinol Invest 2020;43(8):1167-8.
[Crossref] [Google Scholar] [Pub Med]
- Vesely MD, Perkins SH. Caution in the time of rashes and COVID-19. J Am Acad Dermatol 2020;83(4):e321-2.
[Crossref] [Google Scholar] [Pub Med]
- Novak N, Peng W, Naegeli MC, Galvan C, Kolm?Djamei I, Brüggen C, et al. SARS?CoV?2, COVID?19, skin and immunology-What do we know so far? Allergy 2021;76(3):698-713.
- Chen L, Li X, Chen M, Feng Y, Xiong C. The ACE2 expression in human heart indicates new potential mechanism of heart injury among patients infected with SARS-CoV-2. Cardiovasc Res 2020;116(6):1097-100.
[Crossref] [Google Scholar] [Pub Med]
- Zeng JH, Liu YX, Yuan J, Wang FX, Wu WB, Li JX, et al. First case of COVID-19 complicated with fulminant myocarditis: A case report and insights. Infection 2020;48(5):773-7.
[Crossref] [Google Scholar] [Pub Med]
 and pi-pi stacked
and pi-pi stacked