- *Corresponding Author:
- H. Padh
Provost, Vanita Vishram Women's University
Athwa Gate, Surat, Gujarat 395007, India
E-mail: hpadh@yahoo.com
| Date of Submission | 05 December 2020 |
| Date of Revision | 03 July 2021 |
| Date of Acceptance | 04 November 2021 |
| Indian J Pharm Sci 2021;83(6):1107-1113 |
Abstract
Remarkable advancement in next generation sequencing technology has made personal genome analysis feasible and affordable. Here we present the whole genome sequence analysis of an individual from the Kashmir region of India who is from the pandit community (IHGP04). The Indian male’s genome was sequenced at 38.6 X coverage with a total of 110 Gb of sequence data generated using the Illumina HiSeq 2000 platform. The variant analysis revealed over 3.6 million single nucleotide variants and 5 46 681 small insertions and deletions including about 2.7 % of novel (unreported) variants. The known variants were analyzed for their health and disease relevance and drug pharmacogenetic profile. The mitochondrial haplogroup (L3e2b1a1) is indicative of maternal ancestors’ arrival to the Indian subcontinent about 70 000 y ago, while the Y-chromosome haplogroup (J) is suggestive of the arrival of paternal ancestors less than 25 000 y ago. The haplogroups also suggest different routes used by the ancestors to arrive on the subcontinent. The analysis and validation of the novel variants and relationship with health and diseases should be the next logical step.
Keywords
Genome analysis, genome and health, chromosomes, lymphocytes, blood
Indian population has two features that collectively make it a unique population to understand: 1.3 billion Indians make up 1/6th of the world population and is perhaps the most diverse in its anthropological as well as genetic makeup. It is a matter of concern that the Human reference genome originally developed in 2003[1] is derived from about 200 anonymous individuals from six countries: China, France, Germany, Great Britain, Japan and the United States, but did not have Deoxyribonucleic Acid (DNA) from any Indian. Subsequently, significant work has been done on Indian genomes but not much variant data from the Indian population has been added to the human genome variant database[2-7]. This leaves the reference human genome not an ideal representative of the human race. The reference Human Genome and related variant databases need to be enriched with data from underrepresented populations.
It has been an effort of our laboratory to sequence and analyze Indian genomes with the following broader objectives: To help develop the variant database of the Indian population; to enrich the reference human genome variants with new variants reported from the Indian population; to understand the human migration waves and admixing during past 70 000 y which now make up the Indian population.
With affordable technology and accessible reference genome database, analyzing individual genome has become feasible to understand one’s health and associated health related risk factors. Collectively the database has also revealed important aspects of the development of the human race. Particularly, Y-chromosome and mitochondrial haplogroups have helped to develop a high resolution human migration map during the past 100 000 y. With individual genome analysis, we are now able to ascertain the ethnic affiliation of the ancestors and develop a migratory pattern for that particular group and develop a health profile of the individual.
The contemporary Indian population is an admixture of several waves of human migration from various directions, which is believed to have been segregated into various communities over the past 2000 y or so[2]. In this context, we have recently reported a complete sequence analysis of a male from the Western part of The contemporary Indian population is an admixture of several waves of human migration from various directions, which is believed to have been segregated into various communities over the past 2000 y or so[2]. In this context, we have recently reported a complete sequence analysis of a male from the Western part of
The objectives of the present study were-DNA sequencing of a male Indian individual from Kashmir and analysis of the genome variants for known risk factors for health and disease; to identify unreported novel variants present in this individual; and through the analysis of mitochondrial and Y-chromosome haplogroups decipher the migratory routes as well as the time for the ancestors of this individual to reach India.
Materials and Methods
Sample selection and DNA isolation:
A healthy 61 y old male individual (IHGP04) who is a Hindu Brahmin resident of Kashmir in Northern India, with no chronic disease condition was selected for this study. The institutional ethics committee approved the study and the signed informed consent was obtained from the individual before initiation of the study. After the approval, the blood sample (10 ml of the peripheral blood) was withdrawn using a heparinized needle and was collected in anticoagulant (Dipotassium Ethylenediaminetetraacetic Acid (K2 EDTA) or sodium heparin) precoated collection tubes under aseptic conditions. Genomic DNA was extracted using the phenol-chloroform method[8-11]. Assessment of the isolated DNA sample was carried out in terms of quality, quantity and integrity of the DNA. The DNA quality and quantity were measured using NanoDrop 2000 (Thermo Fisher Scientific Inc., USA) and Qubit 2.0 DNA Broad Range Assay (Invitrogen, USA). The sample was also run on 1 % agarose gel to determine the quality of genomic DNA.
Biochemical analysis and family disease profiling:
The basic biochemical analysis like haemogram profiling, lipid profiling [total cholesterol, triglycerides, High-Density Lipoprotein (HDL), Low-Density Lipoprotein (LDL)] and biochemical profiling (blood urea, serum creatinine, liver tests, blood and urine glucose) was performed using the manufacturer’s protocol. The demographic details and the disease history profiling of the individuals and their relatives were also recorded.
Cytogenetic analysis:
Chromosomal aberrations accounting for the identification of genetic diseases through the photographic representation was performed using karyotyping technique. Karyotyping was carried out using peripheral blood lymphocytes by applying standard techniques and Giemsa Banding (GTC banding) with 400-500 band resolution. No consistent structural and numerical chromosomal abnormalities were detected in the cytogenetic analysis through G-banded karyotyping chromosome imaging.
Library construction and sequencing:
The isolated genomic DNA was sent to Theragen BioInstitute, the Republic of Korea for sequence analysis. After passing the quality check measures the isolated sample was processed for DNA library preparation.
Library construction: Genomic DNA was fragmented using Covaris S220 (Covaris Inc, USA) to a targeted size of 350–500 bp. The fragmented DNA was then end-repaired, ligated to Illumina TruSeq adapters and Polymerase Chain Reaction (PCR)-enriched using TruSeq DNA sample preparation kit (Illumina, USA) according to the manufacturer’s protocol. The final sequencing library was quantified using KAPA kit (KAPA Biosystem, USA) on Agilent Stratagene Mx- 3005p quantitative PCR (Agilent, USA). The library size was confirmed using Agilent Bioanalyzer High Sensitivity DNA Chip (Agilent, USA). The parameters of the library constructed were similar to what was published earlier.
Whole-genome sequencing: The resulting library obtained was sequenced using an Illumina flow cell and 202 cycles on the Illumina HiSeq 2000 platform (Illumina, USA). A total of 98 GB of raw data was generated on the sequencer for IHGP04 sample, 98.9 % of mappable reads were mapped with a mean depth of 34.6 X.
Alignment and variant analysis:
Pre-processing: The raw data generated after the sequencing in the form of Binary Base Call (BCL) files were first converted plain-text QSeq format files, followed by FASTQ format using Illumina’s BCL converter. After conversion, the pre-processing was carried out. In pre-processing, the duplicated pair- end reads and reads with more than 2Ns (N bases representing uncalled nucleotides) were filtered. This ensured the high quality reads in the sample.
Mapping of reads: We first filtered low-quality reads by using Next Generation Sequencing Quality Control (NGS QC) toolkit v 2.3.3 with ‘-l 70 -s 20’ options cutoff read length for HQ=70 %, cutoff quality score=20). After filtering, the filtered reads were aligned onto the Human Genome version 19 (hg19) using Burrows-Wheeler Alignment-Maximal Exact Match (BWA-MEM) 0.7.8 with the default option. In addition, SnpEff with dbSNP138, dbSNP142, 1000 Genomes Phase 1 release v3, and ESP6500 were used to supplement hg19. SAM files were converted to BAM files using Samtools 0.1.19. PCR duplicated reads were removed by MarkDuplicate subroutine in Picard v1.9.2 (http://broadinstitute.github.io/picard/). To increase the accuracy of variants call, IndelRealigner and BaseRecalibration in GATK v2.3.9 were used. The variants were called by GATK UnifiedGenotyper with ‘--heterozygosity 0.0010 -dcov 200 -stand_call_conf 30.0 -stand_emit_conf 30.0’ options.
Identification & Classification of Variants: The identification of all the variants like Single Nucleotide Polymorphism (SNPs), Single Nucleotide Insertions and Deletions (InDel), structural variants, Copy Number Variations (CNV) and multiple nucleotide variants were carried out using the SnpEff software [(SnpEff version 3.4 (build 2013-11-23), by Pablo Cingolani]. The identified classes of variants like SNPs, InDels and multi-base Indels were subjected to classification in the form of known and Novel variations by aligning the data to dbSNP v137.
Variant analysis: A comprehensive list of 500 plus genes associated with various disease categories was screened and generated from the databases like Online Mendelian Inheritance in Man (OMIM), The National Center for Biotechnology Information (NCBI), SNPedia etc., The generated gene list consisted of genes associated with major diseases such as neurological disorders, cardiovascular diseases, cancer, diabetes, obesity, asthma and allergy. In addition, genes involved in drug metabolism and nutrigenomics were included in the data set.
The known SNPs in the coding region of the sample (IHGP04) were analyzed using the comprehensive gene list to assess the presence of the potential risk variants involved in the disease susceptibility.
The amino acid position of the novel variants in the coding region of disease-associated genes was determined using NCBI. The novel variants were then evaluated for their Polymorphism Phenotyping v2 (Polyphen score)[12] to predict the variants with significant damaging/deleterious effects.
Results and Discussion
The isolated human DNA sample (IHGP04) was subjected to Whole Genome Sequencing (WGS) using Illumina HiSeq 2000. A total of 98 Gb of sequence data (736 million high-quality pair-end reads of length 101 bp) was generated for the analysis. The sequence data thus generated was then aligned to the human genome reference v37.1 (hg19). Out of the total reads generated, 98.9% of the reads were successfully mapped with an average of 38.6 X coverage of the human reference genome (Table 1).
| Total Reads | 736 428 860 |
|---|---|
| Read Length (bp) | 150 |
| Total yield (Mbp) | 110 464 |
| Reference size (Mbp) | 2858 |
| Throughput mean depth (X) | 38.60 |
| De-duplicated reads | 695 64 382 |
| Mappable Reads | 659 280 879 |
Table 1: Preprocess Data Statistics Summary
The sample IHGP04 was aligned with the human genome reference v37.1 (hg19) for identification of variants (includes SNPs, Indels, CNVs and SV), as shown in Table 2A, Table 2B and Table 2C. In total 3 608 953 Single Nucleotide Variants (SNVs) and 546 681 small INDELs were identified in the IHGP04 genome. To get the estimate of novel variants, the SNVs were compared with those of the dbSNP 138 database. Out of the total 3 642 767, 96.3 % SNVs were reported in the dbSNP 137 database and subsequently, 97.6 % were found in dbSNP142. Total of 208 604 novel SNV and 68 354 novel INDELs were identified. Further classification of the SNVs showed 18 631 were homozygous and 32 961 were heterozygous in intragenic regions. Apart from SNVs, a total of 3545 short indels (up to ±20 bases) were identified in intragenic homozygous state and 6714 heterozygous INDELs were also identified, of which 789 were novel INDELs in the intragenic region. These and other characterizations of the variants are provided in Table 3. The genome also contained larger structural variants including inversions, translocations and duplication which are listed in Table 2C.
| SNPs | Small insertions | Small deletions | |
|---|---|---|---|
| Variants | 3 608 953 | 273,806 | 272,875 |
| Synonymous variants | 11 548 | ||
| Non-synonymous variants | 10 267 | ||
| Splicing variants | 256 | ||
| Stop gained | 66 | ||
| Stop loss | 46 | ||
| Frame shift | 346 | ||
| Found in dbSNP138, % | 96.3 | ||
| Found in dbSNP142, % | 97.3 | ||
| Het/Hom ratio | 1.67 | ||
| Ts/Tv ratio | 2.0771 |
Table 2A: Variant Analysis
| No. of Variants | |
|---|---|
| Copy number gains (>2) | 618 |
| Copy number losses (<2) | 380 |
Table 2B: Copy Number Variant
| SV type | No. of Variants |
|---|---|
| Duplications | 79 |
| Insertions | 2173 |
| Deletions | 5229 |
| Inversions | 88 |
| Translocations | 130 |
Table 2C: Structural Variants
Coding SNPs (cSNPs) may lead to amino-acid substitution in proteins encoded by the gene. Out of a total of 24 620 cSNPs in the IHGP04 genome, 12 091 were identified as non-synonymous substitutions (nsSNPs), of which about one-third were homozygous SNPs. In addition, 685 novel SNPs were of the nsSNPs category. Any possible significance of nsSNPs needs to be analyzed to determine the contribution to phenotype. Among the novel INDELs, 74 indels were found in the coding region (Table 3).
| IHGP04 | SNP | InDel | ||||
|---|---|---|---|---|---|---|
| Homozygous | Heterozygous | Novel SNP | Homozygous | Heterozygous | Novel INDEL | |
| Total | 1 495 832 | 25 83 170 | 2 08 796 | 2 80 604 | 4 93 097 | 68 354 |
| INTERGENIC | 830 954 | 14 27 671 | 1 25 346 | 1 46 733 | 2 48 154 | 35 749 |
| INTRAGENIC | 18 631 | 32 961 | 1777 | 3545 | 6714 | 789 |
| UPSTREAM | 36 878 | 67 479 | 6718 | 8305 | 14 357 | 2388 |
| DOWNSTREAM | 41 323 | 79 606 | 7571 | 9147 | 17 158 | 2517 |
| UTR_5_PRIME | 1552 | 2807 | 276 | 303 | 317 | 89 |
| UTR_3_PRIME | 8940 | 15 692 | 1294 | 2199 | 3 805 | 398 |
| INTRON | 545 130 | 9 32 742 | 63 764 | 1 09 631 | 2 01 394 | 26 222 |
| Noncoding exon variant | 3761 | 8255 | 892 | 493 | 883 | 128 |
| Synonymous mutation | 4488 | 8041 | 473 | 0 | 0 | 0 |
| Non-synonymous mutation | 4175 | 7916 | 685 | 1 | 1 | 0 |
| Indels in coding region | 0 | 0 | 0 | 247 | 314 | 74 |
Table 3: Variant Mapping Statistics
Based on analyses several variants in the non-coding intragenic regions can also be ‘probably damaging’[12-14]. Novel variants need to be thoroughly analyzed for their locations in the coding region, possible effect on the structure-function of the putative proteins and overall impact on the phenotype. The frequency of the occurrence of the novel variants in the target population should be ascertained.
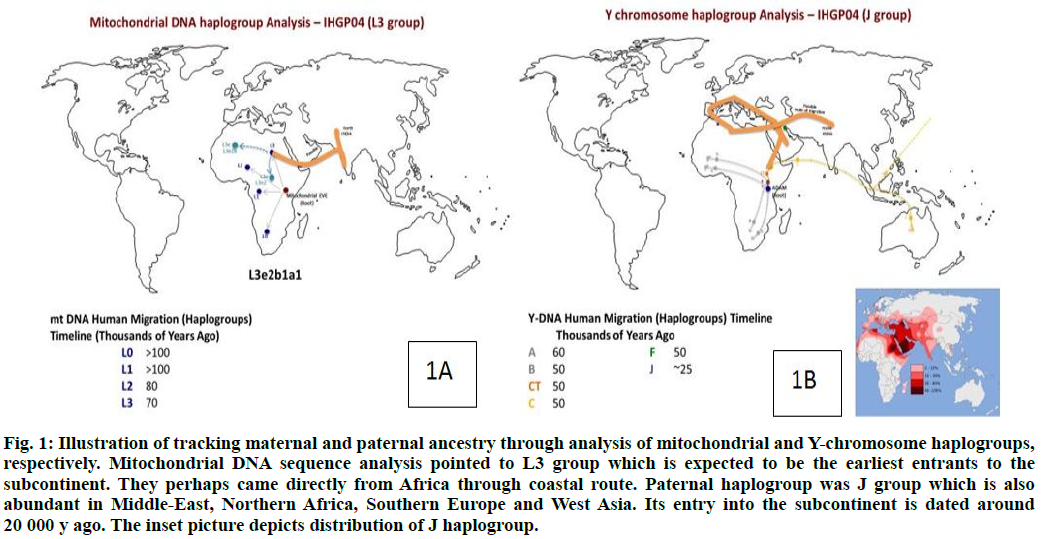
Mitochondrial DNA (mtDNA) sequence is widely used to understand the maternal genetic and migration history of human populations[15]. Variation in the mtDNA sequence has been analyzed in the human population, both in terms of evolution and population dispersals and in terms of the role that mtDNA mutations play in human disease[16-19]. The analysis of the IHGP04 mitochondrial genome (16 569 bp) showed that it had 41 SNPs as compared to the Cambridge Reference Sequence (rCRS)[17]. The haplogroup analysis of the mitochondrial sequence was carried out using Mito Tool (http://www.mitotool.org/)[20-22] that revealed L3e2b1a1 haplogroup to be the most related haplogroup for IHGP04. L3 haplogroup is found in the Indian subcontinent, particularly in the southern and eastern parts of India. The members of this group are believed to be the earliest arrival on the subcontinent directly from Africa probably through the coastal route and spread further to the Australian Aboriginal population.
The timeline of arrival in India is about 70 000 y back (http://www.eupedia.com/europe/Haplogroup_J_ mtDNA.shtml) (fig. 1A).
Fig. 1: Illustration of tracking maternal and paternal ancestry through analysis of mitochondrial and Y-chromosome haplogroups, respectively. Mitochondrial DNA sequence analysis pointed to L3 group which is expected to be the earliest entrants to the subcontinent. They perhaps came directly from Africa through coastal route. Paternal haplogroup was J group which is also abundant in Middle-East, Northern Africa, Southern Europe and West Asia. Its entry into the subcontinent is dated around 20 000 y ago. The inset picture depicts distribution of J haplogroup.
Similarly, the Y haplogroup was calculated using the Y-chromosome DNA sequence SNP index (http://www.isogg.org/tree/ISOGG_YDNA_SNP_Index.html) and the variants for IHGP04 individual. J haplogroup was determined for the IHGP04 sample (fig. 1B), which is found in southern Europe, the Northern Sub-Saharan region around the Mediterranean Sea, along with West Asia and the Indo-European belt[22-24]. The arrival of this haplogroup members in India is estimated to be later than 20 000 y back through land routes from the North-West region. Drastically different ancestors of the maternal and paternal lineages, in terms of the time of arrival and the haplogroups, indicate extensive admixing of the populations on this continent. Earlier, we have analyzed the genome of the Gujarati male from the western region of India[8]. In that particular case, the haplogroup migration pattern for both mtDNA and Y chromosome followed a similar timeline and route of migration[8,22-24].
The analysis of the Copy Number Variation (CNV) was carried out using the readDepth program. This program detects CNVs by measuring the depth of coverage. The CNV in terms of gene loss and gene gain was reported for the IHGP04 sample. Out of the total 998 CNVs, the genome contained a total of 380 deleted genes (copy number <2) and 618 duplicated genes (copy number >2), shown in Table 2B.
To have a health profile of an individual, the cSNPs identified in the IHGP04 genome were annotated using several databases like OMIM, SNPedia, Human Gene Mutation Database (HGMD) and others[25-31]. A comprehensive list of 500 plus genes and their variants that have been associated with disease susceptibility or risk was developed. Further, all the variants of the IHGP04 genome were analyzed, which revealed susceptibility to hypertension, lung cancer, heart disease and others. The variant genes and their correlation to health and disease are listed in Table 4. The individual has a lower risk of Alzheimer's and dementia due to variants in the Cholesteryl Ester Transfer Protein (CETP) gene.
| Disease profile | |
|---|---|
| Increased risk | |
| AGT, CYP4A11 | Higher hypertension risk |
| HNF1A, MMP9, XPC | ~2X increased lung cancer risk |
| LRP8, SNX19, GIPR & ROS1 | Increased risk for heart disease; better response to statins for SNX19 |
| HCRTR2 | 2-6X increased risk for cluster headaches |
| TAS2R38 | possible unable to taste bitter |
| ATF4 | 1.78X increased risk for schizophrenia in males |
| NAT2 | Increased risk of hearing loss |
| EDA2R | Increased risk of baldness |
| ERAP1 | Higher risk for spondylitis |
| ERAP2 | 1.3X increased risk for preeclampsia in most population |
| FAM71F1 | >2X increased risk of familial obesity |
| Protection/reduced risk | |
| CEPT | Lower risk of dementia and Alzheimer’s |
| Pharmacogenomics profile | |
| KCNJ11 | Better response to Repaglinide in case of T2D |
| SLC22A2 | may have reduced risk to nephrotoxicity in response to cisplatin treatment |
| HSPA1L | May be protective against adverse reaction to carbamazepine |
| KIF6 | May have increased risk for adverse cardiovascular events when treated with statins |
Table 4: Disease Association And Drug Response Prediction In Genome Ihgp04
The genetic variants (SNPs) present in an individual can be used for the prediction of adverse drug reactions and also manage the effective dose of drugs prescribed. To understand the pharmacogenomic profile of IHGP04 individual, the variants (SNPs) present in the genome were annotated based on the published literature relevant to pharmacokinetic and pharmacodynamics relevance[32-34]. Upon analysis, we found the genome to have several SNPs associated with the metabolism of or response to various drugs. The variant analysis suggests that the person may have a better response to the drug Repagalinide in case of type 2 diabetes and may have a reduced risk of nephrotoxicity to cisplatin therapy. Some of the relevant profile parameters are listed in Table 4.
With technology becoming accessible and affordable, individual genome analysis is becoming a reality. The outcome from such exercise is important to the individual to glimpse at one’s genetic background and impact on one’s health. A large number of unreported new and novel variants were present in the individual (208 796 SNPs and 68 354 Indels). The novel variants add to the database for human genetic variants. This study is particularly important because the human genetic variant database does not contain adequate representation from people of the Indian subcontinent, which makes up approximately 1/6th of the world population. More analysis of individual genomes from the underrepresented populations will eventually make the human genome database “complete” representing the entire human population. Analysis of mitochondrial DNA and Y-chromosome indicate timeline and route of human migration as revealed from the present analysis. Indian population is an admixture of several waves of migration for 80 000 y; it is therefore not possible to have a representative Indian genome. The current analysis has picked one individual from the Kashmir region of the North-Northwest part of India. Many more genome analyses across the country would help us to develop a better pic of the genetic background of Indians, which may help in the development of a better health policy of the nation.
Acknowledgments:
The author is indebted to Suhani Almal for analysis of some results; Milee Agarwal, Sweta Patel for their advice during the initial phase of the project; Je Hoon Jun and Jong Bhak of Personal Genomics Institute, Genome Research Foundation, Suwon, Republic of Korea. Financial support from GSBTM is gratefully acknowledged.
Conflict of interests:
The authors declared no conflicts of interest.
References
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature 2001;409:860-921.
- Basu A, Sarkar-Roy N, Majumder PP. Genomic reconstruction of the history of extant populations of India reveals five distinct ancestral components and a complex structure. Proc Natl Acad Sci 2016;113(6):1594-9.
- Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, et al. 1000 Genomes Project Consortium. A map of human genome variation from population scale sequencing. Nature 2010;467(7319):1061-73.
- 1000 Genomes Project Consortium. An integrated map of genetic variation from 1092 human genomes. Nature 2012;491(7422):56-65.
- The Indian genome variation database (IGVdb): a project overview. Hum Genet 2005;118(1):1-11.
- Indian Genome Variation Consortium. Genetic landscape of the people of India: a canvas for disease gene exploration. J Genet 2008;87:3-20.
- Narang A, Roy RD, Chaurasia A, Mukhopadhyay A, Mukerji M, Dash D. Indian Genome Variation Consortium. IGVBrowser–a genomic variation resource from diverse Indian populations. Database 2010;2010.
- Almal S, Jeon S, Agarwal M, Patel S, Patel S, Bhak Y, et al. Sequencing and analysis of the whole genome of Indian Gujarati male. Genomics 2019;111(2):196-204.
- Padh H. Sequencing and comparative genome analysis of three Indians. Mamm Genome 2021:1-2.
- Almal SH, Padh H. Frequency distribution of autoimmunity associated FCGR 3B gene copy number in Indian population. Int J Immunogenet 2015;42(1):26-30.
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual (New York, Cold Spring Harbor Laboratory); 1982.
- Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res 2003;31(13):3812-4.
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods 2010;7(4):248-9.
- Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res 2012;40(W1):W452-7.
- Cann RL, Stoneking M, Wilson AC. Mitochondrial DNA and human evolution. Nature 1987;325(6099):31-6.
- Torroni A, Huoponen K, Francalacci P, Petrozzi M, Morelli L, Scozzari R, et al. Classification of European mtDNAs from an analysis of three European populations. Genetics 1996;144(4):1835-50.
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 1999;23(2):147.
- Wallace DC. Mitochondrial diseases in man and mouse. Science 1999;283(5407):1482-8.
- Ingman M, Kaessmann H, Pääbo S, Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature 2000;408(6813):708-13.
- Fan L, Yao YG. MitoTool: a web server for the analysis and retrieval of human mitochondrial DNA sequence variations. Mitochondrion 2011;11(2):351-6.
- Olivieri A, Pala M, Gandini F, Kashani BH, Perego UA, Woodward SR, et al. Mitogenomes from two uncommon haplogroups mark late glacial/postglacial expansions from the near east and neolithic dispersals within Europe. PloS one 2013;8(7):e70492.
- gounder Palanichamy M, Sun C, Agrawal S, Bandelt HJ, Kong QP, Khan F, et al. Phylogeny of mitochondrial DNA macrohaplogroup N in India based on complete sequencing: implications for the peopling of South Asia. Am J Hum Genet 2004;75(6):966-78.
- Sharma S, Rai E, Sharma P, Jena M, Singh S, Darvishi K, et al. The Indian origin of paternal haplogroup R1a1* substantiates the autochthonous origin of Brahmins and the caste system. J Hum Genet 2009;54(1):47-55.
- Sahoo S, Singh A, Himabindu G, Banerjee J, Sitalaximi T, Gaikwad S, et al. A prehistory of Indian Y chromosomes: evaluating demic diffusion scenarios. Proc Natl Acad Sci 2006;103(4):843-8.
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, et al. Detection of large-scale variation in the human genome. Nat Genet 2004;36(9):949-51.
- Bare LA, Morrison AC, Rowland CM, Shiffman D, Luke MM, Iakoubova OA, et al. Five common gene variants identify elevated genetic risk for coronary heart disease. Genet Med 2007;9(10):682-9.
- Garin MC, James RW, Dussoix P, Blanché H, Passa P, Froguel P, et al. Paraoxonase polymorphism Met-Leu54 is associated with modified serum concentrations of the enzyme. A possible link between the paraoxonase gene and increased risk of cardiovascular disease in diabetes. J Clin Invest 1997;99(1):62-6.
- Serrato M, Marian AJ. A variant of human paraoxonase/arylesterase (HUMPONA) gene is a risk factor for coronary artery disease. J Clin Invest 1995;96(6):3005-8.
- Odawara M, Tachi Y, Yamashita K. Paraoxonase polymorphism (Gln192-Arg) is associated with coronary heart disease in Japanese noninsulin-dependent diabetes mellitus. J Clin Endocrinol Metab 1997;82(7):2257-60.
- Sinha S, Qidwai T, Kanchan K, Anand P, Jha GN, Pati SS, et al. Variations in host genes encoding adhesion molecules and susceptibility to falciparum malaria in India. Malar J 2008;7(1):1-9.
- Hofmann S, Franke A, Fischer A, Jacobs G, Nothnagel M, Gaede KI, et al. Genome-wide association study identifies ANXA11 as a new susceptibility locus for sarcoidosis. Nat Genet 2008;40(9):1103-6.
- Altman RB. PharmGKB: a logical home for knowledge relating genotype to drug response phenotype. Nat Genet 2007;39(4):426.
- Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res 2006;34:D668-72.
- Collet JP, Hulot JS, Pena A, Villard E, Esteve JB, Silvain J, et al. Cytochrome P450 2C19 polymorphism in young patients treated with clopidogrel after myocardial infarction: a cohort study. Lancet 2009;373(9660):309-17.