- *Corresponding Author:
- G. V. S. Kumar
Department of Pharmacology
A. U. College of Pharmaceutical Sciences
Andhra University Vishakhapatnam
Andhra Pradesh, India
E-mail: sampath.venu9@gmail.com
| Date of Received | 09 July 2020 |
| Date of Revision | 06 September 2020 |
| Date of Acceptance | 14 October 2020 |
| Indian J Pharm Sci 2020;82(5):826-832 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
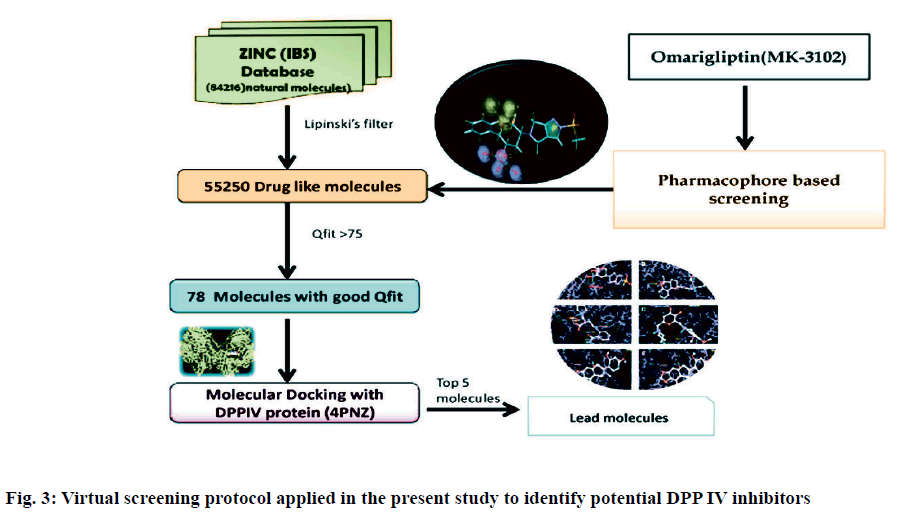
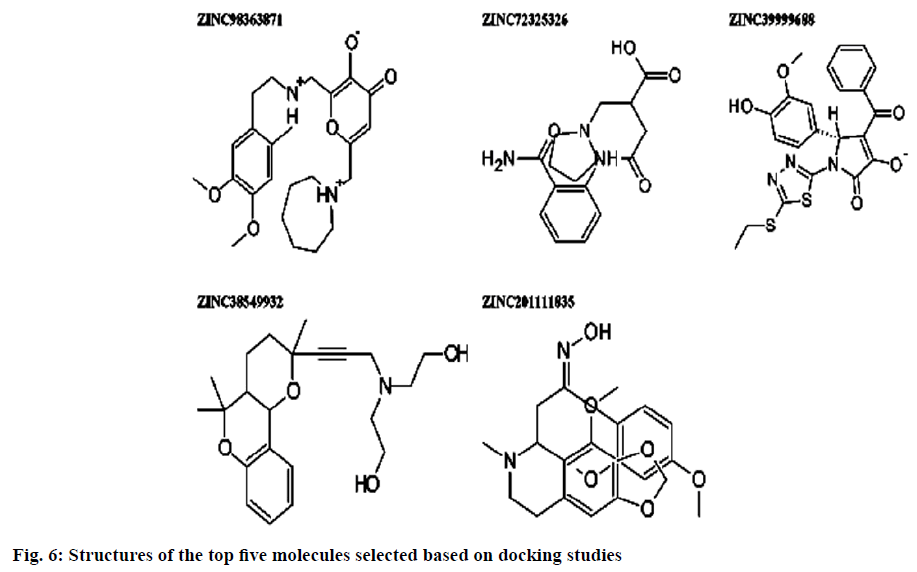
Dipeptidyl peptidase IV is a serine protease that rapidly cleaves glucagon-likepeptide-1 and glucosedependent insulinotropic polypeptide. Inhibition of Dipeptidyl peptidase IV would prolong the half-life of incretins which stimulates insulin secretion. Omarigliptin which is in phase 3 clinical trial has added benefits over currently available Dipeptidyl peptidase IV inhibitors in terms of Specificity towards Dipeptidyl peptidase IV and due to its pharmacokinetic profile. Hence, our present study aims at identifying potential Dipeptidyl peptidase IV inhibitors which are having similar structural features with that of omarigliptin. First, we have identified 3 D structural features of omarigliptin to build a pharmacophore model. The built pharmacophore model was used to screen 55 250 drug like molecules with diverse structures from natural origin. The 78 molecules with a good Qfit values were filtered for further docking study against Dipeptidyl peptidase IV receptor to validate the screening protocol. The docking scores were found to be in congruent with the virtual screening protocol. The candidate molecules which needs further attention was found to be the ZINC38549932, ZINC72325326, ZINC98363871, ZINC201111835 and ZINC39999688.The illustration regarding the virtual screening protocol followed for identifying the Dipeptidyl peptidase IV inhibitors is reported here.
Keywords
Dipeptidyl peptidase IV, molecular docking, omarigliptin, pharmacophore
Type 2 diabetes mellitus (T2DM) is possibly the world’s fastest growing metabolic syndrome of multiple aetiologies causing hyperglycemia[1]. It accounts for 90-95 % of all diabetic cases[2]. DM may be genetic or acquired by inappropriate production of insulin from beta cells of pancreas[3]. While the current T2DM drugs that enhance insulin release have been therapeutically beneficial, they are associated with several side effects including unpredictable hypoglycemia and body weight gain. Thus, there is a need for more effective drugs, with fewer side effects, to treat T2DM[4].
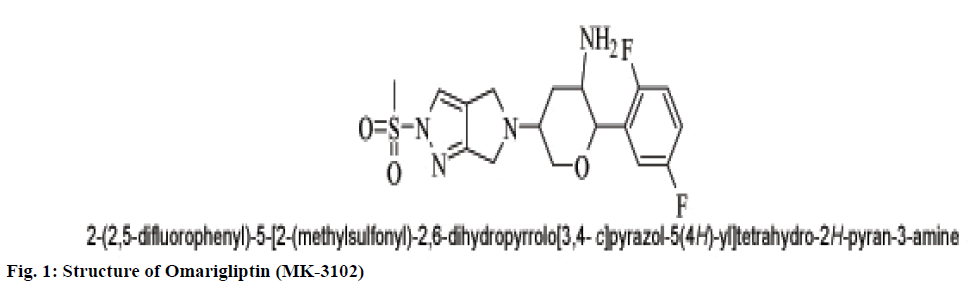
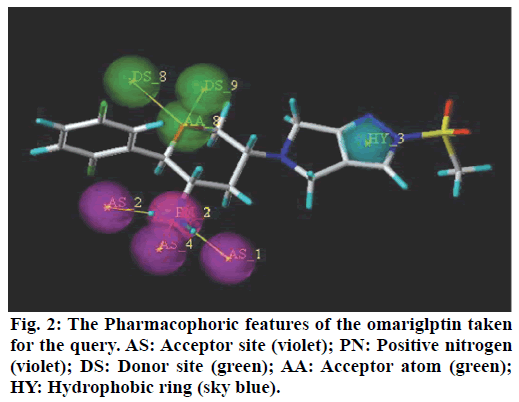
A popular theory on meal-induced insulin secretion, the ‘incretin effect’ states that glucose or any other drug is more effective on the pancreatic cells when administered orally than given through intravenous or subcutaneous injections[5]. Glucagon-likepeptide-1 (GLP-1) and glucose-dependent insulinotropic polypeptide (GIP), two major incretin hormones that are secreted into the circulation by ‘L’ and ‘K’ cells of the small intestine, respectively, are responsible for 50 % to 70 % of glucose dependent insulin release[6]. These compounds have emerged as a new class of drugs for the treatment of type 2 diabetes[7]. Dipeptidyl peptidase IV (DPP IV) is a serine protease responsible for in vivo degradation of endogenous peptides by cleaving the penultimate (N-terminal) proline or alanine[8]. Dipeptidyl peptidase IV (DPP IV, EC 3.4.14.5), a serine peptidase, is one of the newest pharmaceutical targets for type 2 diabetes treatment[9]. Sitagliptin, Merck’s lead DPP-4 inhibitor which was the first DPP-4 inhibitor approved by the United States Food and Drug Administration in October 2006.Subsequently, additional DPP-4 inhibitors have entered the market. These include vildagliptin, saxagliptin, linagliptin, and alogliptin. The sideeffects observed with the ‘Gliptins’ are specific to the molecules which inhibit other substrates rather than DPP IV per se. To avoid the side effects associated with the inhibition of other members of DPP family like DPP-II, DPP-VIII, and DPP-IX it becomes necessary to design selective inhibitors of DPP IV enzyme[10]. Omarigliptin (MK-3102) is a potent, selective, oral DPP-4 inhibitor with a half-life that supports once-weekly dosing[11,12] (fig. 1). The large scaffold diversity and properties of natural products (NPs), such as structural complexity and drug similarity, makes these molecules ideal starting points for drug design[13].Therefore the present study aims at identification of DPP IV inhibitors which are of natural origin based on the pharmacophoric features of omarigliptin and further molecular docking studies with DPP IV protein (4PNZ) (fig. 2).
Sybyl X 2.1.1software from Tripos Inc., St. Louis, MO, USA, All in silico studies were performed on hp workstation with windows platform, xenon processor, 32 GB RAM and NVDIA graphics card.
Pharmacophore query search generation and Database screening
Pharmacophoric features of MK-3102 were used as a 3D search query. The pharmacophoric feature contains 3Acceptor site1Positive nitrogen2Donor sites, 1Acceptor atom and 1Hydrophobic ring. 84216 natural molecules from different vendors (Afrodb and Inter bioscreen) of the ZINC database were filtered by applying Lipinski’s rule-of-five that sets the criteria for drug-like properties. 55 250 drugs like molecules which passes lipinki’s filter were screened based on the pharmacophoric features of omarigliptin (Table 1). Those molecules which satisfied all the features of the pharmacophore model were retrieved as hit molecules. Then the 78 molecules which are having Qfit score above 75 % were chosen for molecular docking study to validate our Virtual screening protocol (fig. 3). Top five molecules based on molecular docking scores and binding interactions were chosen for further studies (Table 2).
| Name | Acceptor count | xlogP | Donor count | Molecular weight |
|---|---|---|---|---|
| ZINC98363871 | 8 | 2.05 | 1 | 429.537 |
| ZINC72325326 | 7 | 0.21 | 4 | 319.361 |
| ZINC39999688 | 8 | 3.23 | 1 | 468.536 |
| ZINC38549932 | 5 | 3.13 | 3 | 374.501 |
| ZINC20111835 | 8 | 3.29 | 1 | 414.458 |
| Omarigliptin | 8 | 0.3 | 1 | 398.42 |
Table 1: Lipinski’s Properties
| Name | Qfita | Total scoreb | Crashc | Polard | Number of H-bonds |
Binding interactions | Distance (A°) |
|---|---|---|---|---|---|---|---|
| ZINC38549932 | 76.84 | 8.3493 | -1.7524 | 5.0494 | 6 | SER630(OH)---Lig(O) TYR547(OH)---Lig(O) SER209(OH)---Lig(NH) VAL207(O)---Lig(OH) ARG669(O)---Lig(OH) GLU206(O)---Lig(OH) |
2.058 2.015 2.142 1.928 2.0761 and 2.007 1.916 |
| ZINC72325326 | 79.25 | 7.3768 | -1.9876 | 3.9818 | 6 | SER630(O)---Lig(NH) SER630(O)---Lig(OH) TYR547(OH)---Lig(O) ARG125(O)---Lig(OH) ARG125(OH)---Lig(O) GLU205(O)---Lig(NH) |
2.657 1.780 1.878 1.891 2.489 1.907 |
| ZINC98363871 | 82.89 | 7.2369 | -1.6883 | 2.6849 | 2 | SER209(OH)---Lig(N) ARG669(NH)---Lig(O) |
2.238 2.084 |
| ZINC201111835 | 75.51 | 7.1447 | -2.2501 | 2.9498 | 3 | TYR662(OH)---Lig(O) GLU206(O)---Lig(OH) ARG358(N)---Lig(OH) |
1.779 2.221 1.874 |
| ZINC39999688 | 77.51 | 5.7449 | -0.7888 | 2.6895 | 2 | ARG669(NH)---Lig(O) GLU206(O)---Lig(OH) |
2.025 and 2.022 1.934 |
| Omarigliptin | 7.3991 | -1.4197 | 4.0526 | 4 | GLU206(OH)---Lig(NH) GLU205(O)---Lig(NH) TYR662(OH)---Lig(N) ASN710(NH)---Lig(F) |
1.899 and 2.051 1.987 2.897 2.590 |
aThe Qfit score represents how close the ligand atoms match the pharmacophore query, which is generally in the range of 0 to 100. bTotal score is the total Surflex Dock score expressed as -log (Kd) to represent binding affinities which include hydrophobic, polar, repulsive, entropic and solvation terms. cCrash value is the degree of inappropriate penetration by the ligand into the protein and of interpenetration between ligand atoms (self-clash) that are separated by rotatable bonds. Crash scores close to 0 are favorable. dPolar value is contribution of the hydrogen bonding and salt bridge interactions to the total score. The polar score may be useful for excluding docking results that make no hydrogen bonds.
Table 2: Molecular Docking Scores and Binding Interactions of Top Five Molecules
Protein structure preparation and protomol generation
The protein was downloaded from Protein Data Bank (PDB) then Biopolymer preparation wizard is used for protein structure preparation. Co-crystalized ligand was removed from the structure then water molecules were removed, H atoms were added and side chains were fixed during protein preparation. Protein structure minimization was performed by applying Molecular modeling force field 94s (MMFF94s) force field and partial atomic charges were calculated by Gasteiger-Marshalli method. Protomol was generated by considering the active ligand’s binding pose. The ligands were taken from ZINC database, and then were prepared using ligand preparation wizard. The energy minimization was done using MMFF94s force field and atomic charges were added with Gasteiger-Huckel.
Surflex-Dock was used for molecular docking. Surflex module docks ligands automatically into ligand binding site of receptor using a protomol based method and an empirically derived scoring function. The protomol is a unique and important factor of the docking algorithm and is a computational representation of assumed ligands that interact with the binding site of receptor. Surflex-Dock’s scoring function contains hydrophobic, polar, repulsive, entropic, and solvation terms. Scoring function of Surflex can also be improved by matching algorithm which allows docking of a fragment of ligand to protein. The fragment is allowed to rotate from its original position during optimization of pose. Docking results contain three information: total score, crash value and polar value.
DPP IV is a serine protease responsible for the degradation of the insulinotropic incretin GLP-1 (fig. 4). Pharmaceutical inhibitors of this enzyme have successfully reduced hyperglycemia and hemoglobin A1C levels as a monotherapy and in combination with other anti-diabetic agents[14]. Natural extracts play an important role in traditional medicines for the treatment of diabetes mellitus and Natural products have proven historically to be a promising pool of structures for drug discovery[15,16]. Hence the present study was undertaken to identify potential DPP IV inhibitors from natural origin.
Recently, Bhadauriya suggested that a synergistic combination of pharmacophore-based virtual screening and docking-based virtual screening was an efficient approach to the design and development of novel drugs since this combination could potentially nullify limitations and augment mutual strengths. The ligandbased virtual screening method works on the principle of similarity, wherein similar compounds are assumed to produce similar effects[17]. Hence in this study we used the pharmacophoric features of MK-3102 to identify natural molecules taken from ZINC database as potential DPP IV inhibitors. The detailed virtual screening workflow was given in (fig. 1). 3 Acceptor site, 1 Positive nitrogen, 2 Donor sites, 1 Acceptor atom and 1 Hydrophobic ring are the given pharmacophoric features of omarigliptin, employed as a 3D search query for retrieving potential hits. Then the molecules with good Qfit were docked with 4PNZ protein.
The binding site of DPP IV consists of Ser630 (that together with Asp708 and His740 form the enzyme catalytic triad), the hydrophobic S1 pocket (formed by Tyr631, Val656, Trp659, Tyr662, Tyr666 and Val711), the hydrophobic S2 pocket (formed by Arg125, Phe357, Arg358, Tyr547, Pro550 and Asn710) and the N-terminal recognition region (formed by Glu205, Glu206 and Tyr662).Based on the analysis of the DPP IV crystal structures and interpretation of the structureactivity relationship (SAR) data, both the lipophilic S1 pocket and the Glu205/Glu206 dyad can be considered as crucial molecular anchors for DPP IV inhibition[12].
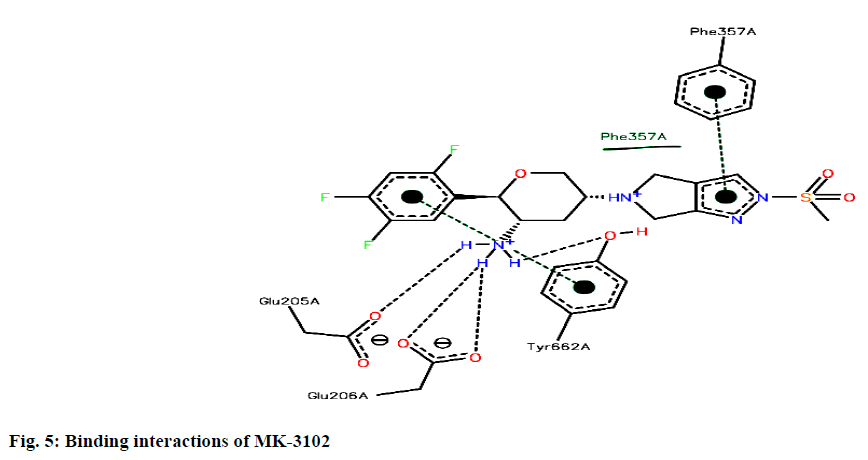
Binding interactions of Omarigliptin there are four hydrogen bonding interactions found at the active site. GLU205 is making an hydrogen bonding interaction with amino portion of the omarigliptin at a distance of 1.987Ao, with the same amine portion TYR 662 is forming an hydrogen bond at a distance of 2.897 Ao with the same amine portion GLU 206 is forming two hydrogen bonds at a distance of 1.899 Ao and 2.051 Ao respectively. Finally there is hydrogen bond is found between ortho-Fluorine along with ASN710 at a distance of 2.590 Ao (fig. 5).
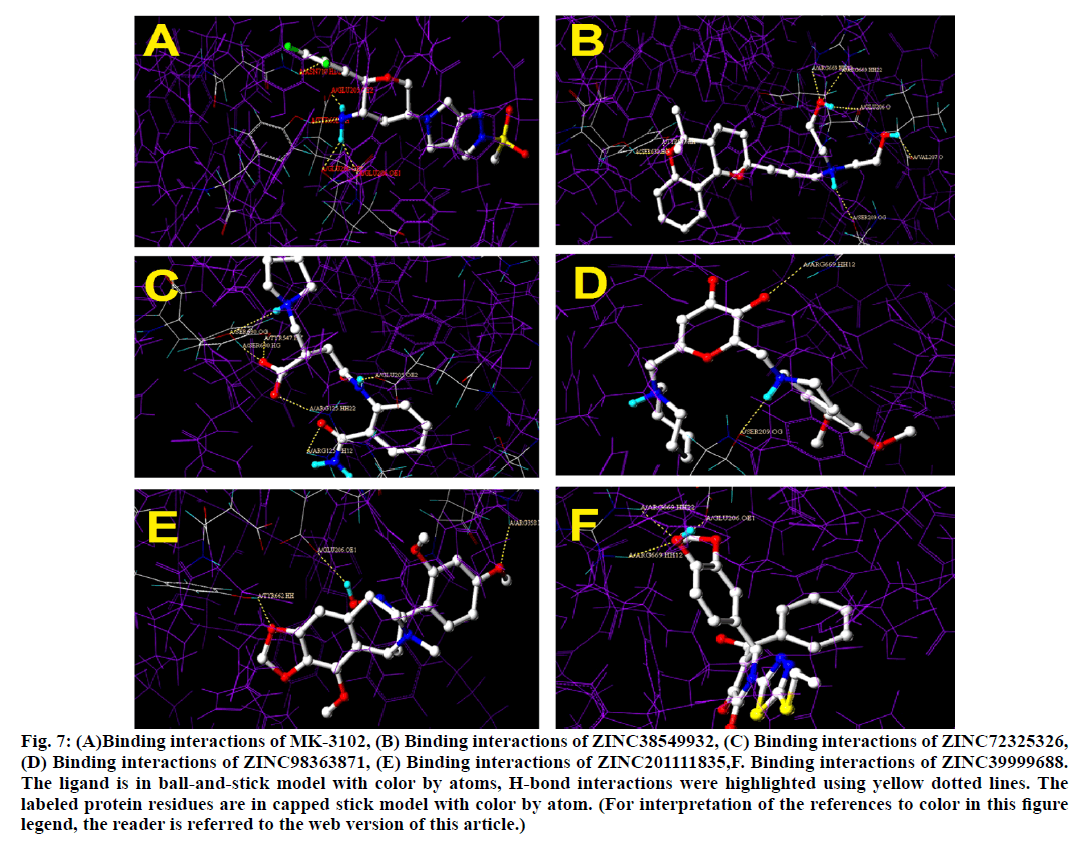
Binding interactions of ZINC38549932: There are six hydrogen bond interactions have found.SER630 formed hydrogen bond with the tricyclic ethoxy portion at a distance of 2.058 Ao, then to the same portion TYR547 also formed a hydrogen bond at a distance of 2.015 Ao.SER209 has formed hydrogen bond with 30 nitrogen at a distance of 2.142 Ao, then GLU206 has formed hydrogen bond at a distance of 1.916 Ao and to the same hydroxyl group,ARG669 formed two simultaneous hydrogen bonds at a distance of 2.076 Ao and 2.007 Ao respectively. Finally to the other hydroxyl portion VAL207 formed a hydrogen bond at a distance of 1.928 Ao.
Binding interactions of ZINC72325326.There are six hydrogen bonding interactions are present out of which ARG125 formed two simultaneous hydrogen bonds one with hydroxyl moiety and other with keto moiety of amide linkage at a distance of 1.891 Ao and 2.489 Ao respectively. Then other interactions like SER630 also formed two interactions with one with amine and other with hydroxyl moiety at a distance of 2.657 Ao and 1.780 Ao respectively. Then finally TYR 547 formed hydrogen bond interactions with hydroxyl moiety at a distance of 1.878 Ao and GLU205 formed interactions with amine moiety at a distance of 1.907 Ao.
Binding interactions of ZINC98363871 with DPP IV at the active site. There are only two hydrogen bonding interactions are present in the active site. One is with SER209 to the amine portion at a distance of 2.238 Ao and other one is ARG669 to the oxo portion at distance of 2.084. Binding interactions of ZINC201111835. There are three hydrogen bond interactions are present one with TYR662 to the oxygen of the heterocyclic ring portion at a distance of 1.779 Ao then GLU206 has formed the hydrogen bond with oxime portion at a distance of 2.221 Ao. Finally ARG358 formed the hydrogen bond with hydroxyl moiety at a distance of 1.874 Ao (fig. 6).
Binding interactions of ZINC39999688.There are two hydrogen bond interactions are found. Two simultaneous interactions of ARG669 with hydroxyl moiety at a distance of 2.025 Ao and 2.022 Ao respectively. Finally GLU206 formed the hydrogen bond interaction with the same moiety at a distance of 1.934 Ao (fig. 7).
Figure 7: (A)Binding interactions of MK-3102, (B) Binding interactions of ZINC38549932, (C) Binding interactions of ZINC72325326, (D) Binding interactions of ZINC98363871, (E) Binding interactions of ZINC201111835,F. Binding interactions of ZINC39999688. The ligand is in ball-and-stick model with color by atoms, H-bond interactions were highlighted using yellow dotted lines. The labeled protein residues are in capped stick model with color by atom. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
In the present study the virtual screening was performed based on pharmacophoric features of omarigliptin which contains 3 Acceptor site 1 Positive nitrogen 2 Donor sites, 1 Acceptor atom and 1 Hydrophobic ring. The molecules with good Qfit score were docked with DPP IV protein (4PNZ) based on the docking scores and binding interactions the lead molecules were identified. ZINC38549932, ZINC72325326, ZINC98363871, ZINC201111835 and ZINC39999688 are the molecules which possess good docking score and possess potential Anti-diabetic properties.
Conflict of interests:
The authors declared no conflict of interest.
References
- Bharti SK, Krishnan S, Kumar A, Rajak KK, Murari K, Bharti BK, et al. Antihyperglycemic activity with DPP-IV inhibition of alkaloids from seed extract of Castanospermum australe: Investigation by experimental validation and molecular docking. Phytomedicine 2012;20:24-31.
- Safavi M, Foroumadi A, Abdollahi M. The importance of synthetic drugs for type 2 diabetes drug discovery. Expert Opin Drug Discov 2013;8:1339-63.
- Sharma A, Paliwal G, Upadhyay N, Tiwari A. Therapeutic stimulation of GLP-1 and GIP protein with DPP-4 inhibitors for type-2 diabetes treatment. J Diabetes Metab Disord 2015;14:15.
- Lotfy M, Singh J, Kalász H, Tekes K, Adeghate E. Medicinal Chemistry and Applications of Incretins and DPP-4 Inhibitors in the Treatment of Type 2 Diabetes Mellitus. Open Med Chem J 2011;5:82-92.
- Bharti SK, Sharma NK, Kumar A, Jaiswal SK, Krishnan S, Gupta AK, et al. Dipeptidyl Peptidase IV inhibitory activity of seed extract of Castanospermum australe and molecular docking of their Alkaloids. Topclass J Herb Med 2012;1:1-7.
- Nisal K, Kela R, Khunti K, Davies MJ. Comparison of efficacy between incretin-based therapies for type 2 diabetes mellitus. BMC Med 2012;10:152.
- Bianchi R, Cervellini I, Porretta-Serapiglia C, Oggioni N, Burkey B, Ghezzi P, et al. Beneficial effects of PKF275-055, a novel, selective, orally bioavailable, long-acting dipeptidyl peptidase IV inhibitor in streptozotocin-induced diabetic peripheral neuropathy J Pharmacol Exp Ther 2012;340:64-72.
- Almasri IM, Taha MO, Mohammad MK. New leads for DPP IV inhibition: Structure-based pharmacophore mapping and virtual screening study. Arch Pharm Res 2013;36(11):1326-37.
- Fan J, Johnson MH, Lila MA, Yousef G, De Mejia EG. Berry and citrus phenolic compounds inhibit dipeptidyl peptidase IV: Implications in diabetes management. Evidence-based Complement Altern Med 2013;2013.
- Sagar SR, Agarwal JK, Pandya DH, Dash RP, Nivsarkar M, Vasu KK, et al. Design, synthesis and biological evaluation of novel pyrazolo-pyrimidinones as DPP-IV inhibitors in diabetes. Bioorg Med Chem Lett 2015;25(20):4428-33
- Sheu WH, Gantz I, Chen M, Suryawanshi S, Mirza A, Goldstein BJ, et al. Safety and Efficacy of Omarigliptin (MK-3102), a Novel Once-Weekly DPP-4 Inhibitor for the Treatment of Patients With Type 2 Diabetes. Diabetes Care 2015;38(11):2106-14.
- Coe D, Fox D, Road GW. Recent disclosures of clinical candidates. Drug Future 35:349-58.
- Guasch L, Ojeda MJ, González-Abuín N, Sala E, Cereto-Massagué A, Mulero M, et al. Identification of Novel Human Dipeptidyl Peptidase-IV Inhibitors of Natural Origin (Part I): Virtual Screening and Activity Assays. PLoS One 2012; 12;7(9):e44971.
- Bower AM, Real Hernandez LM, Berhow MA, De Mejia EG. Bioactive compounds from culinary herbs inhibit a molecular target for type 2 diabetes management, dipeptidyl peptidase IV. J Agric Food Chem 2014;62:6147-58.
- Guasch L, Ojeda MJ, González-Abuín N, Sala E, Cereto-Massagué A, Mulero M, et al. Identification of Novel Human Dipeptidyl Peptidase-IV Inhibitors of Natural Origin (Part II): In Silico Prediction in Antidiabetic Extracts. PLoS One 2012;7(9): e44971.
- Wang L, Waltenberger B, Pferschy-Wenzig EM, Blunder M, Liu X, Malainer C, et al. Natural product agonists of peroxisome proliferator-activated receptor gamma (PPARγ): a review. Biochem Pharmacol 2014;92:73-89.
- Muralidharan AR, Selvaraj C, Singh SK, Jesudasan CN, Geraldine P, Thomas PA. Virtual screening based on pharmacophoric features of known calpain inhibitors to identify potent inhibitors of calpain. Med Chem Res. 2014;23(5):2445-55.