- *Corresponding Author:
- S. Sivanandan
Department of Biotechnology and Bioinformatics
Saraswathy Thangavelu Extension Centre
A Research Centre of University of Kerala
KSCSTE-Jawaharlal Nehru Tropical Botanic Garden and Research Institute
Puthenthope, Thiruvananthapuram, 695 586, India
E-mail: drsreekumar@rediffmail.com
| Date of Received | 27 January 2020 |
| Date of Revision | 12 February 2021 |
| Date of Acceptance | 28 March 2021 |
| Indian J Pharm Sci 2021;83(2):307-321 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Discovery of novel drug against tuberculosis is inevitable since resistant bacterial strains are evolved against existing drugs and rapidly active single drug in short period is necessary for effective treatment.
https://betist.fun https://betlike.fun https://betmatik.fun https://betpark.fun https://bettilt.club https://elexbet.fun https://extrabet.fun https://hepsibahis.fun https://kingbetting.fun https://maksibet.fun https://marsbahis.xyz https://matadorbet.fun https://pulibet.fun https://restbet.fun https://milanobet.fun https://supertotobet.fun https://vevobahis.fun https://imajbet4.com https://maltcasinocu.fun https://sekabetgiris.fun
When compared to synthetic drugs, natural drugs particularly phytochemicals induce less side effects and long term stability. In Indian traditional medicine, several plant species have been used for treating tuberculosis and each plant species contains a plethora of phytochemicals. The efficacy of such treatment system and identification of the phytochemical with drug activity from plants has been seldom investigated scientifically. In this backdrop, the authors have validated the efficacy of anti-tuberculosis activity and identified lead molecule from a widely used plant species against several disease including tuberculosis, viz Punica granatum L. The four promising target proteins viz mycolyltransferase antigen protein 85C involved in cord factor synthesis, filamentous temperature sensitive protein Z involved in bacterial cell division, pantothenate kinase involved in co-enzyme A pathway and decaprenylphosphoryl β-D-ribofuranose-2 epimerase involved in the synthesis of virulent factor arabinan were docked with 243 phytochemicals derived from Punica granatum. The docked molecules having binding energy ≤-6 kcal/ mol were considered as active/hit molecules. The docked results showed that out of 243 phytochemicals, 126 have inhibitory activity on all selected target proteins. Further docking study using Glide and absorption, distribution, metabolism, excretion and toxicity studies revealed that the compound derived from the lowers, pomegranatate can be recommended as the best lead compound against tuberculosis.Keywords
Mycobacterium tuberculosis, multi drug resistance, extensively drug resistance, totally drug resistance, Punica granatum, molecular docking Tuberculosis (TB) remains as one of the top killer diseases worldwide from a single infectious agent, Mycobacterium tuberculosis. Globally 10 million people infected with TB and 1.5 million died in 2018[1]. Although several drugs are available to treat against TB, the emergence of resistance strains of M. tuberculosis such as MDR (Multi Drug Resistance) and TDR (Totally Drug Resistance Strains) is a major threat. Besides, TB patients with HIV are more vulnerable. The forgoing problems necessitate novel drug against TB. Generally, the drug molecules derived from natural sources especially from plants have long-term stability, effectiveness and find more potential therapeutic uses with no or less side effects to the human body since those molecules are derived within the living system through repeated interactions with other bio molecules and modifications through a long term evolutionary process. Moreover, the traditional herbal treatment systems serve as real indicator to make the drug discovery process easy and reduce the initial investment. In these backdrops, search on novel drugs from plants are considered as the best source.
The Greek physician Hippocrates said that “Let food is thy medicine and medicine is thy food”. In natural food materials, nutrients and medicines are enriched in a balanced form and consumption of such food materials can control or prevent ailments in the human body. Punica granatum (common name pomegranate), a widely used fruit, has been used as a polychrest against many ailments in traditional medicine in all over the world since time immemorial. It’s nutritional and medicinal values have been well reviewed[2-4]. The research reports demonstrated its anti-cancer, anti-inflammatory, anti-atherogenic, anti- diabetes, hepatoprotective and antioxidant activity[5-7]. It’s antimicrobial activity has also been reported[8,9]. The present investigation was aimed to validate the efficacy of anti-tuberculosis activity of P. granatum and identification of lead molecules through in silico method.
Materials and Methods
Selection and preparation of target protein
The crystal structures of the target proteins viz. mycolyl transferase antigen 85C protein (Ag85C/FbpC, PDB ID: 1DQY), filamentous temperature sensitive protein Z (FtsZ, PDB ID: 2Q1X), pantothenate kinase (PanK, PDB ID: 4BFS) and decaprenyl phosphoryl β-D-ribose 2’-epimerase (DprE1, PDB ID: 4FDO) were retrieved from Protein Data Bank (PDB). The foregoing target proteins were analyzed using a web servers, VADAR (Volume, Area, Dihedral Angle Reporter) [10] and ProtParam[11]. Active sites of the proteins were determined using Computed Atlas of Surface Topography of proteins (CASTp) server and PDBSum server[12].
Selection and preparation of ligands
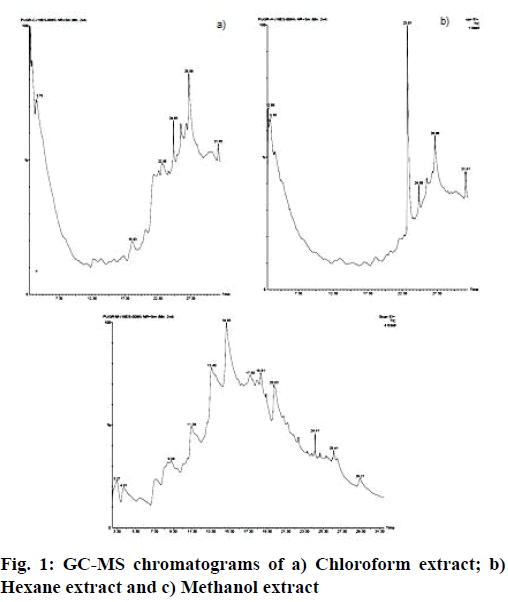
A total of 243 phytochemicals reported from Punica granatum were selected as ligand molecules. Of these, information on 219 phytochemicals was collected from literature and databases like PubChem, Foodb, ChEBI and ZINC. The remaining 24 phytochemicals were identified from fruit extract through Gas chromatography-mass spectrometry (GC-MS) analysis (fig. 1). The structure-data file (SDF) or MOL formats of the phytochemical structures were retrieved from open access databases and converted to PDB format using Open Babel version 2.4.1[13]. The Open Babel represents chemical toolbox which is designed to search, convert, analyze or store data from molecular modeling, chemistry or related areas.
Preparation of fruit extract and GC-MS analysis
Fresh pomegranate fruits (2 kg) collected from farmers’ field in Wayanad district of Kerala, India were cleaned well first in running tap water followed by distilled water thrice. Then water content was removed from the surface of the fruit using blotting paper. The fruits were cut open and removed the peels. The arils along with the seeds were weighed (1.300 kg) and oven dried at a temperature 55° for 3 d till the weight becomes constant. The dried sample was powdered and stored in an air tight glass container. 25 g samples were taken and extracted with 250 ml each of the solvents such as hexane, chloroform and methanol using soxhlet apparatus for 6 h according to their boiling point. Further, the extracts were concentrated in a rotary evaporator and the dried samples were stored in air tight bottles.
The GC-mass analysis was performed using Perkin Elmer-Clarus 680 which employs a fused silica column, packed with Elite-5MS (5 % biphenyl 95 % dimethylpolysiloxane, 30 m×0.25 mm ID×250 μm df) and the components were separated using helium as carrier gas at a constant flow of 1 ml/min. 1 μL of extract sample was injected into the instrument. The injector temperature was set at 260° during the chromatographic run. Initial temperature of the oven was set at 600° for 2 min followed by ramp to 300° at the rate of 10° min-1; and at 300°, where it was held for 6 min. The mass detector conditions include a transfer line temperature of 240°, ion source temperature 240° and ionization mode electron impact at 70 eV, a scan time 0.2 s and a scan interval of 0.1 s. The fragments from 40 to 600 Da were detected using this technique. The spectrums of the components were compared with the database, TurboMass ver 5.4.2. spectrum of known components stored in the GC-MS National Institute of Standards (NIST) (2008) library.
Molecular Docking
Molecular docking procedure was carried out using an open access molecular docking program AutoDock Vina[14] and is compatible with Molecular Graphics Laboratory (MGL) Tools. Vina uses a hybrid scoring function and an iterated local optimization algorithm-Broyden-Fletcher-Goldfarb-Shanno algorithm. Prior to virtual screening, the PDB format of the macromolecules and phytochemicals were converted to PDBQT using MGL Tools wherein the Gasteiger partial charges and polar hydrogens were added. Like Autodock, Vina also require the specification of 3D search space centered on the protein’s active site in the form of grid points and grid box in different dimensions along with spacing. Vina allows the user to make the side chains of the target protein flexible as in Autodock. Hence both rigid and flexible part of the target proteins were saved separately. In Vina, spacing is always set to 1 Å. In order to execute the screening process, a configuration file in .txt format has to be setup wherein we should specify all the above mentioned parameters. Virtual screening of 243 phytochemicals was performed with the help of bash scripting.
Post virtual screening analysis
The docked complexes of the protein together with the top hit phytochemicals were analyzed using Hbind tool[15]. Hbind rigorously calculate and define intermolecular H-bonds by donor/acceptor chemistry and geometric constraint. The criteria for finding H-bonds is well described[16,17].
Analysis of molecular properties and ADMET properties
The hit molecules were subjected for molecular property calculation using OSIRIS Data Warrior. OSIRIS Data Warrior is a free cheminformatics program for data visualization and analysis[18]. Pharmacokinetics refers to the movements of drug into, through and out of the body, during the process of absorption, bioavailability, distribution, metabolism and excretion. The absorption, distribution, metabolism, excretion and toxicity (ADMET) property analysis was performed using the tool pkCSM, a platform for the analysis and optimization of pharmacokinetic and toxicity properties. Further, Extra Precision Glide[19] docking and QikProp analysis of the top prioritized hit molecules were also performed to finalize the best lead compound[20]. Qikprop analysis provides accurate results in predicting properties for molecules with novel scaffolds as for analogs of well- known drugs.
Results and Discussion
Most of the screening studies for anti-TB agents considered only small homogenous molecules against only one target[21], while in the present study a total of 243 phytochemicals derived from Punica granatum were screened against four target proteins namely, Ag85C, FtsZ, PanK and DprE1.
The antigen 85C represents protein C in Ag85 antigenic protein complex containing Ag85A, Ag85B and Ag85C which catalyze the synthesis of trehalose 6, 6’-dimycolate or cord factor which is considered to be one of the major toxic components of cell wall and is present only in virulent strains of Mycobacterium tuberculosis. The protein, Ag85C is also known as FbpC as it has high affinity with fibronectin, which facilitates the attachment of M. tuberculosis to murine alveolar macrophages[22] and also it contributes to the low permeability of the cell wall[23]. The protein consists of 39 % helices, 21 % beta sheets, 38 % coils and 19 % beta turns with mean hydrogen bond distance of 2.2 Å and mean hydrogen bond energy -1.8 kJ/mol and about 80 % of the residues form hydrogen bonds. The theoretical isoelectric point and average hydropathicity values were 4.99 and -0.417 respectively. The X-ray crystal structure of protein possess only a single chain with two unique ligands namely (4R)-2-methylpentane- 2,4-diol and diethyl phosphonate without H-bonds. The catalytic residues of the protein include Ser124, Glu228 and His260.
FtsZ, a bacterial tubulin homolog plays a critical role in cell division by the formation of Z ring and recruiting other proteins for septum formation leading to a new cell wall between dividing cells which is conserved in most prokaryotes and several organelles[24,25]. These proteins consists of 41 % helices, 29 % beta sheets, 28 % coils and 10 % with a mean H-bond distance of 2.2 Å and mean H-bond energy -1.8 kj/mol. The theoretical isoelectric point and average hydropathicity appears to be 4.50 and 0.080 respectively. The crystal structure holds citric acid as the ligand molecule having five H-bonds with four residues including Gly105, Thr106, Gly18 and Gly19.
PanK, pantothenase kinase catalysesthe phosphorylation of pantothenate, the first committed and rate limiting step in the biosynthesis of coenzyme A, which is an essential acyl group carrier indispensable for respiration and lipid metabolism. Based on the differences in biochemical and structural characteristics, three types- PanK type I, II and III were present, among which PanK type I is essential for the growth of bacteria in vitro and in vivo[26]. The structure contains 43 % helices, 23 % beta sheets, 33 % coils and 25 % turns with a mean H-bond distance and mean H-bond energy 2.2Å and -1.7 kj/mol respectively. The theoretical isoelectric point of the protein was 7.47 and average hydropathicity -0.249. The crystal structure consist of N-[1-(5-{[2-(4- fluorophenoxy) ethyl]sulfanyl}-4-methyl-4h-1,2,4- triazol-3-yl) ethyl]-2-(trigluoro-methyl) benzamide that forms H-bonds with Tyr235 and Asn277 respectively.
DprE1, Decaprenylphosphoryl-β-D-ribose oxidase belongs to DprE1-DprE2 complex, catalyses the formation of decaprenyl-phospho-arabinose (DPA), which is the sole arabinosyl donor of mycobacterium cell wall through epimerization reaction[27]. DprE1 is a flavin adenine dinucleotide (FAD) dependent enzyme. The enzyme DprE1 first epimerizes decaprenylphosphoryl-β-D-ribofuranose (DPR) to a keto intermediate decaprenylphosphoryl-D-2- keto-erythro-pentofuranose (DPX) which is then catalysed by DprE2 to decaprenylphosphoryl-β-D- arabinofuranose (DPA). The structures consists of 28 % helices, 33 % beta sheets, 38 % coil, 20 % turns with mean H-bond distance and mean H-bond energy of 2.2 Å and -1.7 kj/mol respectively. The protein possesses a theoretical isoelectric point of 7.86 and average hydropathicity of -0.147. The 3-nitro-N- [(1R)-1-phenylethyl]-5-(trifluoromethyl) benzamide (His132 and Asn385-hydrogen bonding residues) and flavin-adenine dinucleotide (Gly117, His132, Tyr415, Gly125, Gly55, Gly57, Asn63, Ala53, Ile184, Thr122, Ser59, Leu56-hydrogen bonding residues) were the unique ligands present in the crystal structure. PDB format of the target proteins were converted to PDBQT using AutoDock tools.
Considering the ligands molecules, out of 243 phytochemicals selected for the study, the details of 219 molecules were procured from various literature and open access databases and remaining 24 phytochemicals were identified from the fruit of P. granatum through GC-MS analysis, of which oleic acid, beta carotene and methyl 3-bromo-1- adamentacetate were already reported. Lists of compounds identified through GC-MS analysis were depicted in Table 1.
| Name of the extract | RT | Name of the compound | Peak area % |
|---|---|---|---|
| Chloroform | 21.026 | Dodecane, 1-chloro- | 6.004 |
| 21.446 | 14-heptadecenal | 9.333 | |
| 21.551 | Tetradecane, 1-chloro- | 10.239 | |
| 21.736 | Oleic acid | 2.795 | |
| 22.526 | 10,12-tricosadiynoic acid | 12.093 | |
| 22.916 | Beta Carotene | 2.864 | |
| 23.587 | Pentanoic acid, 3-[(adamantan-1-ylmethyl)carbamoyl]-4-phenyl | 2.132 | |
| 24.637 | 1-methylene-2b-hydroxymethyl-3,3-dimethyl-4b-(3-methylbut-2-enyl)-cyclohexane | 8.594 | |
| 25.177 | Methyl 3-bromo-1-adamantaneacetate | 2.704 | |
| 26.973 | Pregn-5-en-20-one, 3-(acetyloxy)-16-bromo-, (3.beta.,16.alpha.)- | 30.044 | |
| Hexane | 3.209 | Hexane, 2-chloro | 3.110 |
| 3.259 | Pentafluoropropionic acid, hexyl ester | 3.526 | |
| 3.379 | 1,1-dimethylethylamine, n-methoxycarbonyloxy | 4.364 | |
| 22.997 | 1,2-benzenedicarboxylic acid, diisooctyl ester | 71.247 | |
| 24.637 | Psi-psi-carotene, 7,7',8,8',11,11',12,12',15,15'-decahydro- | 2.945 | |
| Methanol | 9.111 | 3-amino-2-oxazolidinone | 4.057 |
| 11.162 | Cyclohexanone, 3-hydroxy | 8.407 | |
| 13.408 | 4h-pyran-4-one, 2,3-dihydro-3,5-dihydroxy-6-methyl | 19.957 | |
| 14.948 | 4-hepten-3-one, 5-methyl | 26.949 | |
| 14.948 | T-butyl cyclopentaneperoxycarboxylate | 12.312 | |
| 18.605 | Alpha.-d-glucopyrnoside, o-.alpha.-d-glucopyranosyl-(1.fwdarw.3)-.beta.-d-fructofuranosyl | 10.438 | |
| 19.190 | Pentanoic acid, 2-(amino_oxy)- | 4.492 | |
| 20.015 | 3-deoxy-d-mannoic lactone | 12.442 | |
| 22.631 | 1,10-hexadecanediol | 0.946 |
Table 1: Compounds Identified from the Fruit of Punica granatum through GC-MS Analysis
The analysis of docked results between the targets viz Ag85C/FbpC, FtsZ, PanK and DprE1 and phytochemicals from Punica granatum through virtual screening revealed that out of 243 phytochemicals screened 126 have inhibitory activity (ΔGbind≤-6 kcal/ mol ) on all the above targets. In many reports[28,29] minimum free energy level of the active/hit molecules’ was considered as ≤-5 kcal/mol, however, as suggested by Shityakov[30] minimum free energy of binding of active/hit molecules was considered here as ≤-6 kcal/ mol. The number of active/hit molecules (ΔGbind≤-6 kcal/mol) obtained against each target protein such as Ag85C/FbpC, FtsZ, PanK and DprE1 was 155, 146, 166 and 176 respectively. The top five hit molecules obtained against Ag85C/FbpC based on least binding energy in the order of merit was pomegranatate (-9.5 kcal/mol), icariside D1 (-9.3 kcal/mol), luteolin- 3’-o-β-D-glucoside (-9.3 kcal/mol), 1-2-4-tri-o-galloyl- β-gluco-pyranose (-9.1 kcal/mol) and epigallocatechin-3-o-rhamnoside (-9.1 kcal/mol). Similarly, the compounds tercatain (-10.2 kcal/mol), granatin A (-9.4 kcal/mol), 1-2-4-tri-o-galloyl-β-gluco-pyranose (-9.7 kcal/mol), granatin B (-8.8 kcal/mol) and luteolin- 3’-o-β-D-glucoside (-8.8 kcal/mol) were the top ranked hit molecules against FtsZ and the compounds lupenone (-11.2 kcal/mol), terminalin (-10.9 kcal/ mol), gallocatechin-(4β→8)-catechin (-10.8 kcal/ mol), kaempferol-3-o-rhamnoglucoside (-10.8 kcal/ mol) and pomegranatate (-10.3 kcal/mol) were the top ranked hits against PanK. L-malic acid (-13.0 kcal/mol), (+)-gallocatechin-(4α→8)-(+)-catechin (-12.5 kcal/mol), serraten (-12.0 kcal/mol), 1,2,4,6-tetra- o-galloyl-β-D-glucose(-11.8 kcal/mol), procyanidin B1 (-11.8 kcal/mol) and pomegranatate (-11.6 kcal/mol) were identified as top ranked hits against DprE1. The docked results of phytochemicals that can inhibit all the four target proteins were depicted in Table 2.
| Punica granatum | |||||
|---|---|---|---|---|---|
| Targets | |||||
| Phytomolecule name | Ag85C (kcal/mol) |
DprE1 (kcal/mol) |
FtsZ (kcal/mol) |
PanK (kcal/mol) |
|
| (2E,6E)-9-(3,3’-Dimethyl-2-oxiranyl)-3-7-dimethyl-2-6-nonadienyl phenyl sulfide | -7.7 | -8.6 | -6.3 | -7.2 | |
| 1,2,3,4,6-penta-o-galloyl- β d glucose | -7.1 | -10.9 | -7.7 | -7.8 | |
| 1,2,3-tri-o-galloyl- β -4c-1-glucopyranose | -8.2 | -11.1 | -7.9 | -9.5 | |
| 1,2,3-tri-o-galloyl- β -4c1-glucose | -7.0 | -11.2 | -7.5 | -9.1 | |
| 1,2,3-tri-o-galloyl- β -d-glucose | -7.9 | -11.0 | -8.1 | -9.0 | |
| 1,2,4,6-tetra-o-galloyl-β-D-glucose | -7.1 | -11.8 | -7.9 | -7.6 | |
| 1,2,4 tri-o-galloyl-β-glucopyranose | -9.1 | -11.3 | -8.8 | -9.7 | |
| 2,3-(S)-Hexahydroxydiphenoyl-D-glucose | -7.7 | -10.5 | -7.3 | -8.8 | |
| 2-hydroxycyclopentadecanone | -6.9 | -8.2 | -6.5 | -7.9 | |
| 3,3,4-tri-O-Methylellagicacid | -7.4 | -8.7 | -7.1 | -8.0 | |
| 3-3'-di-O-methylellagic acid | -7.8 | -9.2 | -7.5 | -8.4 | |
| 3,4,8,9,10-pentahydroxydibenzo( b-d)-pyran-6-one | -7.6 | -8.8 | -7.4 | -7.5 | |
| 3-o-methylellagic acid | -7.6 | -9.4 | -7.3 | -8.0 | |
| 17alpha estradiol | -7.2 | -8.7 | -7.0 | -8.4 | |
| 17- β estradiol | -7.2 | -8.7 | -7.0 | -8.4 | |
| 17- β estriol | -7.1 | -9.0 | -7.2 | -8.7 | |
| Alpha tocopherol | -8.1 | -8.8 | -6.4 | -8.5 | |
| Amurensin | -7.7 | -10.7 | -8.3 | -8.4 | |
| Antirrhinin | -8.5 | -10.8 | -8.5 | -9.0 | |
| Apigenin | -8.7 | -9.5 | -7.1 | -7.7 | |
| Apigenin 4'-o-beta-glucopyranoside | -8.1 | -10.0 | -7.1 | -8.3 | |
| Apigenin 7-O-glucoside | -8.7 | -10.1 | -7.5 | -8.6 | |
| Astragalin | -8.1 | -10.5 | -7.6 | -7.9 | |
| β -Eleostearic acid | -6.6 | -6.8 | -6.1 | -6.7 | |
| β -sitosterol | -6.4 | -9.7 | -7.0 | -8.4 | |
| Betulic acid | -6.6 | -9.2 | -7.2 | -9.9 | |
| Betulinic acid | -6.4 | -9.3 | -7.2 | -9.9 | |
| Brevifolin-carboxylic-acid-10-monosulphate | -7.1 | -8.6 | -7.9 | -7.1 | |
| Brevifolin-carboxylicacid | -7.1 | -8.5 | -7.9 | -7.6 | |
| Caffeic acid | -6.4 | -6.7 | -6.1 | -6.0 | |
| Callistephin | -8.1 | -9.8 | -7.7 | -8.4 | |
| Campesterol | -6.0 | -10.0 | -6.7 | -8.9 | |
| Catechin | -8.6 | -9.0 | -7.2 | -7.8 | |
| catechin-(4β→8)-gallo-catechin | -7.6 | -12.5 | -7.7 | -7.9 | |
| Chlorogenic acid | -7.7 | -9.2 | -6.7 | -7.9 | |
| Chrysanthemin | -8.5 | -10.1 | -8.1 | -8.9 | |
| Conidendrin | -7.7 | -9.3 | -8.3 | -8.7 | |
| Corilagin | -8.5 | -9.9 | -7.9 | -9.8 | |
| Coumestrol | -8.0 | -9.2 | -6.6 | -8.2 | |
| Coutaric acid | -7.4 | -8.1 | -7.0 | -6.4 | |
| Cyanidin | -8.6 | -9.0 | -7.3 | -8.1 | |
| Cyanidin-3-5-diglucoside | -7.1 | -10.7 | -7.7 | -6.0 | |
| Cyanidin-3-glucoside | -8.3 | -10.3 | -7.7 | -8.3 | |
| Cyanidin-3-rutinoside | -8.2 | -10.8 | -8.5 | -9.1 | |
| Cyanin | -7.1 | -10.6 | -7.3 | -9.0 | |
| Cycloartenol acetate | -7.0 | -10.2 | -6.1 | -9.5 | |
| Cynaroside | -7.9 | -10.3 | -8.6 | -9.4 | |
| Daidzein | -7.6 | -8.8 | -6.7 | -7.5 | |
| Daucosterol | -6.8 | -9.8 | -8.0 | -8.2 | |
| Delphinidin | -8.2 | -9.2 | -7.4 | -8.0 | |
| Delphinidin-3-5-di-o-glucoside | -7.8 | -10.9 | -7.3 | -9.2 | |
| Ellagic acid | -8.0 | -9.6 | -7.6 | -8.1 | |
| Epicatechin gallate | -8.9 | -10.9 | -7.9 | -8.6 | |
| Epicatechin | -8.5 | -8.8 | -7.3 | -7.8 | |
| Epigallocatechin-3-o-gallate | -9.1 | -11.0 | -8.1 | -8.2 | |
| Eschweilenol C | -8.8 | -11.2 | -8.7 | -9.6 | |
| Esterone | -7.6 | -8.8 | -7.4 | -8.6 | |
| Estradiol | -7.2 | -8.7 | -7.0 | -8.4 | |
| Estriol | -7.4 | -9.0 | -6.8 | -8.4 | |
| Ethyl-brevifolin-carboxylate | -7.5 | -8.9 | -7.6 | -7.7 | |
| Ferulic acid | -6.4 | -7.1 | -6.1 | -6.2 | |
| Flavan-3-ol | -8.0 | -8.9 | -6.3 | -7.6 | |
| Flavogallol | -8.4 | -10.3 | -8.1 | -10.1 | |
| Gallic acid 3-O-Beta-D-(6'-O-galloyl)-glucopyranoside | -8.4 | -9.8 | -7.7 | -8.2 | |
| Gallocatechin (4 β →8)-catechin | -6.7 | -11.6 | -8.2 | -10.8 | |
| Gamma Sitosterol | -6.7 | -10.1 | -7.1 | -8.9 | |
| Gemin D | -6.7 | -7.3 | -8.2 | -8.6 | |
| Genistein | -7.8 | -9.2 | -6.8 | -7.6 | |
| Hirsutrin | -8.9 | -11.1 | -8.3 | -9.0 | |
| Hovetrichoside C | -8.0 | -10.6 | -6.9 | -8.6 | |
| Icariside D1 | -9.3 | -9.6 | -7.3 | -8.5 | |
| Isocorilagin | -7.7 | -10.0 | -8.2 | -10.0 | |
| Isohydroxymatairesinol | -8.0 | -9.3 | -6.7 | -7.8 | |
| Isolariciresinol | -6.9 | -8.6 | -6.9 | -8.0 | |
| Isoquercetrin | -8.9 | -10.6 | -8.1 | -8.9 | |
| Kaempferol | -8.7 | -9.3 | -7.2 | -7.7 | |
| Kaempferol-3-o-glycoside | -8.6 | -9.3 | -8.0 | -8.9 | |
| Kaempferol-3-o-rhamnoglucoside | -8.6 | -10.2 | -8.7 | -10.8 | |
| Lupenone | -7.0 | -11.3 | -8.5 | -11.2 | |
| Luteolin 3'-o-β-xylopyranoside | -8.8 | -11.0 | -7.3 | -8.8 | |
| Luteolin | -8.9 | -9.5 | -7.2 | -8.0 | |
| luteolin-3’-o-β-D-glucoside | -9.3 | -10.9 | -8.8 | -8.8 | |
| Luteolin-4'-o- β -glucopyranoside | -8.1 | -10.5 | -8.2 | -9.0 | |
| Matairesinol | -7.4 | -9.5 | -7.4 | -8.0 | |
| Medioresinol | -7.5 | -9.4 | -7.3 | -8.7 | |
| Melatonin | -7.3 | -7.6 | -6.6 | -6.8 | |
| Mirtillin | -8.3 | -10.5 | -7.8 | -8.7 | |
| Myricetin | -8.6 | -9.5 | -7.4 | -8.2 | |
| Naringin | -8.2 | -10.3 | -7.1 | -9.2 | |
| Neochlorogenic acid | -8.0 | -9.1 | -7.2 | -7.7 | |
| Oleanolic acid | -6.0 | -11.0 | -7.4 | -6.8 | |
| Oxandrolone | -7.4 | -9.6 | -7.1 | -8.8 | |
| Pelargonidin | -8.4 | -8.9 | -7.2 | -7.4 | |
| Phellatin | -8.2 | -10.3 | -7.5 | -8.6 | |
| Phenethyl rutinoside | -7.7 | -9.8 | -7.3 | -8.0 | |
| Phloretin | -7.6 | -8.5 | -6.8 | -6.9 | |
| Phlorizin | -8.0 | -10.2 | -8.2 | -8.6 | |
| Phyllanthusiin E | -7.7 | -9.9 | -7.9 | -7.7 | |
| Phylligenin | -7.7 | -9.5 | -7.1 | -8.1 | |
| Pinocembrin | -8.6 | -9.3 | -7.3 | -8.1 | |
| Pinoresinol | -8.2 | -9.1 | -6.2 | -8.3 | |
| Pomegralignan | -8.3 | -9.8 | -8.6 | -8.8 | |
| Pomegranatate | -9.5 | -11.6 | -7.7 | -10.3 | |
| Procyanidin B1 | -7.8 | -11.8 | -7.7 | -10.3 | |
| Procyanidin B3 | -7.5 | -11.3 | -8.2 | -9.0 | |
| Procyanidin-B2 | -7.8 | -11.6 | -8.6 | -10.1 | |
| Prodelphinidin B | -6.9 | -9.2 | -8.6 | -6.6 | |
| Prodelphinidin C | -7.0 | -9.7 | -8.3 | -9.6 | |
| Punicafolin | -6.1 | -10.2 | -7.4 | -6.3 | |
| Punicanolic acid | -6.1 | -10.3 | -6.8 | -7.8 | |
| Punigluconin | -8.3 | -10.5 | -8.6 | -9.7 | |
| Quercetin 3-O-rhamnoside | -9.0 | -10.8 | -8.1 | -9.1 | |
| Quercetin | -8.7 | -9.5 | -7.5 | -7.9 | |
| Quercetin-3-o-rutinoside | -8.3 | -11.6 | -7.8 | -8.8 | |
| Quercimeritrin | -8.3 | -10.5 | -6.4 | -8.7 | |
| Rutin | -8.3 | -11.0 | -7.8 | -8.9 | |
| Secoisolariciresinol | -7.9 | -8.3 | -6.9 | -7.2 | |
| Serraten | -6.1 | -12.0 | -7.5 | -8.6 | |
| Stigmasterol | -7.6 | -10.1 | -7.6 | -9.4 | |
| Strictinin | -8.0 | -11.0 | -8.1 | -10.0 | |
| Terminalin | -8.9 | -6.5 | -7.8 | -10.9 | |
| Tricetin | -8.7 | -9.3 | -7.3 | -8.1 | |
| Tricin | -8.5 | -9.3 | -7.0 | -8.0 | |
| Ursolic acid | -6.9 | -11.1 | -6.6 | -8.4 | |
| Valoneic acid dilactone | -8.3 | -11.2 | -8.7 | -9.6 | |
| 1-2-benzenedicarboxylic acid -mono(2-ethylhexyl) ester | -6.6 | -7.2 | -6.3 | -6.7 | |
Table 2: Compounds Qualified as Hit Molecules (Binding Energy ≤-6 kcal/mol) from Punica granatum Against All the Selected Targets such as AG85C, DPRE1, FTSZ and PANK.
To observe the molecular interaction and to avoid the selection of false positive ranked molecules as lead, the docked structures were analyzed using the Hbind tool which can precisely demonstrate H-bond interaction with residues, its accurate distance and angle and donor-acceptor criterion by avoiding false scoring. Raschka et al.[15] reported that generally natural compounds avoid the presence of chemical groups bearing both H-bond donor and acceptor capacity in the binding site of protein or ligand lead to non-selective ligand binding and false positive scoring/ranking. But proteins selectively donate H-bonds (instead of donate and accept) to small molecules avoid false positive scoring/ranking and better binding. The Hbind tool follows the analysis of molecular interaction based on the forgoing principle. The analysis of the docked structures using Hbind tool revealed that among the selected top ranked hit molecules against Ag85C except epigallocatechin-3-o-gallate and luteolin-3-o- β-glucoside all others gave true positive scoring while against FtsZ only two compounds such as tercatain and 1,2,4-tri-o-galloyl-β-gluco-pyranose showed true positive scoring. In the case of PanK, except the compound terminalin, all other top ranked hit molecules showed positive scoring and against DprE1 only two compounds viz catechin-(4β→8)-gallo-catechin and 1,2,4,6-tetra-o-galloyl-β-D-glucose showed positive scoring.
It was noted that although 126 phytochemicals have inhibitory effect on all the selected targets none of the lead molecule identified from top ranked five hit molecules through the forgoing process could inhibit all targets. Therefore, to identify common lead molecule against all the four selected targets the phytochemicals with free energy of binding ≤-8.6 kcal/mol against Ag85C, <-7.7 kcal/mol against FtsZ, <-9.7 kcal/mol against PanK and ≤-10.2 kca/mol against DprE1 were further analysed using Hbind tool since the number of active molecules with free energy of binding level depend on each target protein (Table 3). Analysis of molecular interaction revealed that pomegranatate showed hydrogen bonding with one of the catalytic triad, Ser124 whereas the compound showed hydrogen bonding with His132, the residue in which the natural inhibitor bound with the protein, DprE1. In case of 1,2,4-tri-o-galloyl-β- gluco-pyranose, two hydrogen bonds were formed with the critical residue, Tyr235 of PanK protein whereas with DprE1 protein, one hydrogen formed with critical residue, His 132. Kaempferol-3-o-rhamnoglucoside showed interaction with critical residue, Tyr235 with PanK protein only. Thus the compounds pomegranatate, kaempferol-3-o-rhamnoglucoside and 1,2,4-tri-o- galloyl-β-gluco-pyranose could inhibit all the selected target proteins.
| H bond interactions – Punica granatum-1DQY | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sl. No |
Name of the ligand & target protein | Atom No. | Atom Type Ligand |
Residue & No. | Atom Type Protein |
Distance | Angle | Interaction | Hydro-phobic interact-tions |
| 1 | Pomegranatate & Ag85C/FbpC |
8 | O.3 | TRP262 | NE1 | 2.998 | 150.0 | Acceptor - Donor | 42 |
| 10 | O.3 | ARG41 | NE | 2.944 | 156.5 | Acceptor - Donor | |||
| 10 | O.2 | ARG41 | NH2 | 3.132 | 146.2 | Acceptor - Donor | |||
| 16 | O.2 | LEU40 | N | 3.221 | 147.1 | Acceptor - Donor | |||
| 16 | O.2 | SER124 | OG | 3.051 | 170.3 | Acceptor - Donor | |||
| 17 | O.2 | MET125 | N | 3.028 | 120.4 | Acceptor - Donor | |||
| 24 | O.3 | ARG41 | NH2 | 2.806 | 156.5 | Acceptor - Donor | |||
| 2 | Icariside D1& Ag85C/FbpC | 6 | O.2 | LEU40 | N | 3.117 | 152.1 | Acceptor - Donor | 74 |
| 11 | O.2 | ARG41 | N | 3.085 | 171.3 | Acceptor - Donor | |||
| 14 | O.2 | TRP262 | NE1 | 3.052 | 153.8 | Acceptor - Donor | |||
| 17 | O.3 | ASN52 | ND2 | 3.139 | 127.4 | Acceptor - Donor | |||
| 17 | O.3 | TRP262 | NE1 | 3.097 | 128.6 | Acceptor - Donor | |||
| 20 | O.3 | ASN52 | ND2 | 2.916 | 155.0 | Acceptor - Donor | |||
| 21 | O.2 | ARG41 | NH2 | 3.069 | 122.0 | Acceptor - Donor | |||
| 3 | 1,2,4-tri-o-galloyl-β-gluco-pyranose & Ag85C/FbpC | 16 | O.3 | ASN52 | ND2 | 3.233 | 139.0 | Acceptor - Donor | 67 |
| 18 | O.3 | ASN52 | ND2 | 3.193 | 131.8 | Acceptor - Donor | |||
| 20 | O.3 | ARG41 | NE | 2.591 | 172.7 | Acceptor - Donor | |||
| 20 | O.3 | ARG41 | NH2 | 3.255 | 131.9 | Acceptor - Donor | |||
| 34 | O.3 | ARG41 | NH2 | 3.232 | 138.6 | Acceptor - Donor | |||
| 4 | Apigenin & Ag85C/FbpC | 4 | O.2 | TRP262 | NE1 | 2.961 | 158.1 | Acceptor - Donor | 54 |
| 11 | O.2 | ARG41 | N | 3.351 | 158.7 | Acceptor - Donor | |||
| 20 | O.3 | ASN52 | ND2 | 2.892 | 132.5 | Acceptor - Donor | |||
| 20 | O.3 | TRP262 | NE1 | 3.060 | 125.5 | Acceptor - Donor | |||
| 5 | Kaempferol 3-o-rhanoglucoside & Ag85C/FbpC | 21 | O.3 | GLN43 | N | 3.115 | 126.3 | Acceptor - Donor | 51 |
| 32 | O.2 | ARG41 | NE | 2.856 | 155.3 | Acceptor - Donor | |||
| 32 | O.2 | ARG41 | NH2 | 3.196 | 139.4 | Acceptor - Donor | |||
| 41 | O.2 | LEU40 | N | 2.877 | 140.1 | Acceptor - Donor | |||
| 6 | Pinocembrin_ Ag85C/FbpC | 1 | O.3 | ARG41 | N | 3.368 | 156.5 | Acceptor - Donor | 45 |
| 2 | O.2 | TRP262 | NE1 | 3.162 | 157.1 | Acceptor - Donor | |||
| 18 | O.3 | ASN52 | ND2 | 2.854 | 134.7 | Acceptor - Donor | |||
| 7 | 1,2,4 tri-o-galloyl-β-glucopyranose & FtsZ | 10 | O.2 | ARG140 | NH2 | 3.150 | 139.9 | Acceptor - Donor | 63 |
| 16 | O.3 | THR130 | OG1 | 2.828 | 178.0 | Acceptor - Donor | |||
| 16 | O.3 | ASN163 | ND2 | 2.831 | 161.4 | Acceptor - Donor | |||
| 18 | O.3 | THR130 | OG1 | 3.123 | 155.7 | Acceptor - Donor | |||
| 33 | O.3 | ARG140 | NE | 2.918 | 163.9 | Acceptor - Donor | |||
| 33 | O.3 | ARG 140 | NH2 | 3.227 | 145.0 | Acceptor - Donor | |||
| 8 | Kaempferol-3-o rhamnoglucoside & FtsZ | 39 | O.2 | ASN163 | ND2 | 2.801 | 141.2 | Acceptor - Donor | 70 |
| 42 | O.2 | ARG140 | NH1 | 2.756 | 159.1 | Acceptor - Donor | |||
| 42 | O.2 | ARG140 | NH2 | 3.074 | 141.2 | Acceptor - Donor | |||
| 9 | Tercatain & FtsZ | 16 | O.2 | ARG140 | NE | 3.007 | 122.9 | Acceptor - Donor | 70 |
| 27 | O.2 | ARG139 | NH1 | 3.200 | 143.3 | Acceptor - Donor | |||
| 30 | O.2 | ARG139 | NH1 | 3.014 | 123.9 | Acceptor - Donor | |||
| 42 | O.3 | ASN22 | ND2 | 3.042 | 151.1 | Acceptor - Donor | |||
| 10 | 1,2,4-tri-o-galloyl-β-gluco-pyranose & PanK | 2 | O.2 | LYS103 | NZ | 3.225 | 169.0 | Acceptor - Donor | 61 |
| 7 | O.2 | TYR153 | OH | 2.738 | 144.4 | Acceptor - Donor | |||
| 22 | O.2 | HIS179 | NE2 | 2.942 | 134.2 | Acceptor - Donor | |||
| 28 | O.3 | TYR235 | OH | 3.116 | 135.3 | Acceptor - Donor | |||
| 30 | O.3 | TYR235 | OH | 2.935 | 154.9 | Acceptor - Donor | |||
| 33 | O.3 | THR128 | OG1 | 3.208 | 172.6 | Acceptor - Donor | |||
| 36 | O.2 | TYR153 | OH | 3.172 | 166.6 | Acceptor - Donor | |||
| 44 | O.3 | TYR182 | OH | 2.715 | 171.1 | Acceptor - Donor | |||
| 11 | Betulic_acid & PanK | 28 | O.2 | TYR235 | OH | 2.746 | 156.7 | Acceptor - Donor | 97 |
| 12 | Betulinic_acid & PanK | 28 | O.2 | TYR235 | OH | 2.738 | 156.5 | Acceptor - Donor | 97 |
| 13 | Cycloartenol acetate & PanK | 31 | O.3 | HIS179 | NE2 | 2.854 | 156.6 | Acceptor - Donor | 32 |
| 14 | Gallocatechin-(4β→ 8)-catechin & PanK | 10 | O.3 | TYR153 | OH | 2.695 | 145.8 | Acceptor - Donor | 70 |
| 20 | O.3 | ARG238 | NH1 | 3.269 | 153.5 | Acceptor - Donor | |||
| 15 | Kaempferol3-o-rhamnoglucoside& PanK | 14 | O.2 | TYR257 | OH | 3.126 | 178.1 | Acceptor - Donor | 70 |
| 18 | O.2 | TYR257 | OH | 2.788 | 147.1 | Acceptor - Donor | |||
| 39 | O.2 | LYS103 | NZ | 2.962 | 179.9 | Acceptor - Donor | |||
| 16 | Pomegranatate & PanK | 7 | O.3 | THR128 | OG1 | 3.021 | 123.8 | Acceptor - Donor | 31 |
| 7 | O.3 | TYR153 | OH | 2.802 | 162.3 | Acceptor - Donor | |||
| 16 | O.2 | TYR153 | OH | 2.807 | 126.1 | Acceptor - Donor | |||
| 17 | 1,2,4-tri-o-galloyl -β-gluco-pyranose & DprE1 |
18 | O.2 | ARG58 | NE | 3.265 | 124.6 | Acceptor - Donor | 74 |
| 18 | O.2 | ARG58 | NH2 | 2.785 | 135.5 | Acceptor - Donor | |||
| 28 | O.3 | HIS 132 | N | 3.079 | 168.7 | Acceptor - Donor | |||
| 28 | O.3 | TYR415 | OH | 2.936 | 179.4 | Acceptor - Donor | |||
| 30 | O.3 | TYR415 | OH | 3.001 | 170.7 | Acceptor - Donor | |||
| 36 | O.2 | LYS 418 | NZ | 2.821 | 175.1 | Acceptor - Donor | |||
| 18 | 1,2,4,6- Tetra_O_galloyl-beta-D-glucose & DprE1 | 12 | O.2 | HIS132 | N | 3.088 | 147.5 | Acceptor - Donor | 94 |
| 12 | O.2 | TYR415 | OH | 3.123 | 160.1 | Acceptor - Donor | |||
| 32 | O.3 | ARG58 | NE | 3.078 | 142.7 | Acceptor - Donor | |||
| 32 | O.3 | ARG58 | NH2 | 3.101 | 142.0 | Acceptor - Donor | |||
| 19 | Catechin-(4β→8)-gallocatechin & DprE1 | 12 | O.2 | HIS132 | N | 3.088 | 147.5 | Acceptor - Donor | 94 |
| 12 | O.2 | TYR415 | OH | 3.123 | 160.1 | Acceptor – Donor | |||
| 32 | O.3 | ARG58 | NE | 3.078 | 142.7 | Acceptor - Donor | |||
| 32 | O.3 | ARG58 | NH2 | 3.101 | 142.0 | Acceptor - Donor | |||
| 43 | O.3 | TYR60 | N | 3.409 | 128.9 | Acceptor - Donor | |||
| 20 | Kaempferol 3-o-rhamnoglucoside & DprE1 | 39 | O.2 | ASN163 | ND2 | 2.801 | 141.2 | Acceptor - Donor | 64 |
| 42 | O.2 | ARG140 | NH1 | 2.756 | 159.1 | Acceptor - Donor | |||
| 42 | O.2 | ARG140 | NH2 | 3.074 | 141.2 | Acceptor - Donor | |||
| 21 | Pomegranatate & DprE1 |
7 | O.3 | TYR415 | OH | 2.965 | 161.8 | Acceptor - Donor | 38 |
| 10 | O.2 | TYR60 | OH | 3.216 | 174.8 | Acceptor - Donor | |||
| 17 | O.2 | HIS132 | NE2 | 3.321 | 130.4 | Acceptor - Donor | |||
| 24 | O.3 | TYR60 | OH | 3.006 | 146.9 | Acceptor - Donor | |||
| 22 | L-malic acid & DprE1 | 4 | O.2 | SER228 | OG | 2.794 | 122.8 | Acceptor - Donor | 18 |
| 5 | O.3 | LYS134 | NZ | 3.252 | 164.7 | Acceptor - Donor | |||
| 5 | O.3 | PHE313 | O | 2.855 | 138.1 | Donor - Acceptor | |||
| 5 | O.3 | HIS 315 | O | 3.118 | 162.3 | Donor - Acceptor | |||
| 9 | O.3 | ALA244 | O | 2.712 | 159.6 | Donor - Acceptor | |||
| 11 | O.3 | SER228 | O | 3.020 | 128.3 | Donor - Acceptor | |||
saltb 1 4 O.2 -- LYS A 134 NZ 3.705 N/A Acceptor - Donor
Table 3: Hydrogen Bond and Hydrophobic Interaction of the Top Five Lead Molecules against Selected Targets
True hit molecules against each target and the three common hit molecules identified against all the four targets were subjected to molecular property analysis using the tool OSIRIS property explorer which indicated that the compound tercatain showed all kind of tested toxicity such as tumerogenic, irritant, mutagenic and reproductive effect (Table 4).
| Name | Molecular weight | LOGP | Rotatable Bonds | Acceptors | Donors | Total Polar Surface Area | Mutagenic | Tumerogenic | Irrirant | Reproductive effective | Drug likeness score | Drug score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1,2,4-tri-o-galloyl-β-gluco-pyranose | 636.471 | -0.28 | 7 | 18 | 11 | 249.5 | N | N | N | N | 1.8 | 0.48 |
| Apigenin | 270.248 | 2.69 | 1 | 4 | 3 | 112.519 | Y | N | N | N | 1.21 | 0.47 |
| Icariside D1 | 410.375 | 0.63 | 8 | 10 | 6 | 165.315 | N | N | N | N | -1.26 | 0.41 |
| Kaempferol-3-o-rhamnoglucoside | 586.458 | 4.03 | 5 | 15 | 10 | 234.318 | N | N | N | N | -1.34 | 0.24 |
| Pinocembrin | 256.257 | 2.80 | 1 | 4 | 2 | 109.441 | N | N | N | N | 1.95 | 0.83 |
| Pomegranatate | 392.316 | 0.54 | 0 | 9 | 4 | 154.646 | N | N | N | Y | -9.4 | 0.26 |
| Tercatain | 812.768 | -0.08 | 4 | 19 | 15 | 319.296 | Y | Y | Y | Y | -0.66 | 0.15 |
| Betulic acid | 456.711 | 7.09 | 2 | 2 | 2 | 201.354 | N | N | N | N | -21.49 | 0.15 |
| Cycloartenolacetate | 468.766 | 8.74 | 5 | 2 | 0 | 209.609 | N | N | Y | N | -4.2 | 0.07 |
| 1,2,4,6-Catechin-(4 β→8)-gallaocatechin | 592.509 | 4.62 | 2 | 13 | 11 | 241.371 | N | N | N | N | -0.17 | 0.67 |
| L malic_acid | 134.087 | -1.09 | 3 | 3 | 3 | 50.54 | N | N | N | N | 0.71 | 0.83 |
Table 4: Molecular Property Analysis Using Osiris Datawarrior
The ADMET analysis using the tool pkCSM (Table 5 & Table 6) revealed that the phytochemical, pomegranatate can be considered as the best lead against transferase antigen 85C protein and pantothenate kinase protein whereas 1,2,4-tri-o-galloyl-β-glucopyranose can be considered as the best lead molecule against filamentous temperature-sensitive protein Z and decaprenyl phosphoryl β-D-ribose 2’-epimerase protein. Both phytochemicals act as common lead against all the target proteins. In addition, kaempferol-3-o-rhamnoglucoside present in fruit[31] can be recommended as a common lead since its binding energy level and ADMET properties were on par with other two molecules.
| Absorption | Distribution | Metabolism | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Compound | Caco2 permeability | Intestinal absorption (human) | Skin Permeability | P-glycoprotein substrate | P-glycoprotein I inhibitor | P-glycoprotein II inhibitor | VDss (human) | BBB permeability | CNS permeability | CYP2D6 substrate | CYP3A4 substrate | CYP1A2 inhibitior | CYP2C19 inhibitior | CYP2C9 inhibitior | CYP2D6 inhibitior | CYP3A4 inhibitior |
| 1,2,4-tri-o-galloyl-β-gluco-pyranose | -1.99 | 11.66 | -2.74 | Y | Y | Y | 0.60 | -3.22 | -5.38 | N | N | N | N | N | N | N |
| Apigenin | 1.05 | 91.02 | -2.74 | Y | N | N | 0.09 | -0.90 | -2.10 | N | N | Y | Y | Y | N | N |
| Icariside D1 | -0.55 | 22.47 | -2.74 | Y | Y | N | 0.07 | -1.68 | -4.03 | N | N | N | N | N | N | N |
| Pinocembrin | 0.95 | 92.30 | -3.00 | Y | N | N | -0.26 | 0.42 | -2.12 | N | Y | Y | Y | N | N | N |
| Pomegranatate | 0.51 | 59.53 | -2.74 | Y | N | N | 1.18 | -1.17 | -3.44 | N | N | N | N | N | N | N |
| Kaempferol 3-o- rhamnoglucoside |
-0.31 | 46.16 | -2.74 | Y | Y | N | 2.10 | -2.58 | -4.10 | N | N | N | N | N | N | N |
| Tercatain | -2.37 | 0.00 | -2.74 | Y | N | N | -0.61 | -4.28 | -6.27 | N | N | N | N | N | N | N |
| Betulic_acid | 1.26 | 99.23 | -2.74 | N | N | N | -1.32 | -0.27 | -1.30 | N | Y | N | N | N | N | N |
| Cycloartenol acetate | 1.20 | 97.98 | -2.74 | N | Y | Y | -0.14 | 0.72 | -1.75 | N | Y | N | N | N | N | N |
| 1,2,4,6-Catechin(4 β→8)- gallocatechin | -1.48 | 44.82 | -2.74 | Y | Y | Y | 0.36 | -2.70 | -3.75 | N | N | N | N | N | N | N |
| L-malic acid | -0.39 | 14.72 | -2.74 | N | N | N | -0.88 | -0.77 | -3.52 | N | N | N | N | N | N | N |
Table 5: Adsorption, Distribution and Metabolism Analysis of Selected Molecules Using PKCSM Server
| Excretion | Toxicity | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Compound | Caco2 permeability | Intestinal absorption (human) | Renal OCT2 substrate | Max. tolerated dose (human) | AMES toxicity | hERG I inhibitor | hERG II inhibitor | Oral Rat Acute Toxicity (LD50) | Oral Rat Chronic Toxicity (LOAEL) | Hepatotoxicity | Skin Sensitisation |
| 1,2,4-tri-o-galloyl-β-gluco-pyranose | -1.99 | 11.66 | N | 0.44 | N | N | Y | 2.48 | 6.11 | N | N |
| Apigenin | 1.05 | 91.02 | N | 0.34 | N | N | N | 2.46 | 1.57 | N | N |
| Icariside D1 | -0.55 | 22.47 | N | 0.50 | N | N | N | 3.21 | 4.36 | N | N |
| Pinocembrin | 0.95 | 92.30 | N | 0.07 | N | N | N | 1.75 | 1.97 | N | N |
| Pomegranatate | 0.51 | 59.53 | N | 0.09 | N | N | N | 2.88 | 2.64 | N | N |
| Kaempferol 3-o- rhamnoglucoside |
-0.31 | 46.16 | N | 0.49 | N | N | Y | 2.51 | 3.48 | N | N |
| Tercatain | -2.37 | 0.00 | N | 0.53 | N | N | Y | 2.58 | 3.72 | N | N |
| Betulic acid | 1.26 | 99.23 | N | 0.14 | N | N | N | 2.31 | 2.15 | Y | N |
| Cycloartenol acetate | 1.20 | 97.98 | N | -0.32 | N | N | Y | 2.61 | 2.32 | N | N |
| 1,2,4,6-Catechin(4 β→8)- gallocatechin | -1.48 | 44.82 | N | 0.44 | N | N | Y | 2.49 | 3.64 | N | N |
| L-malic acid | -0.39 | 14.72 | N | 0.57 | N | N | N | 1.49 | 3.10 | N | N |
Table 6: Excretion and Toxicity Analysis of Selected Molecules Using PKCSM Server
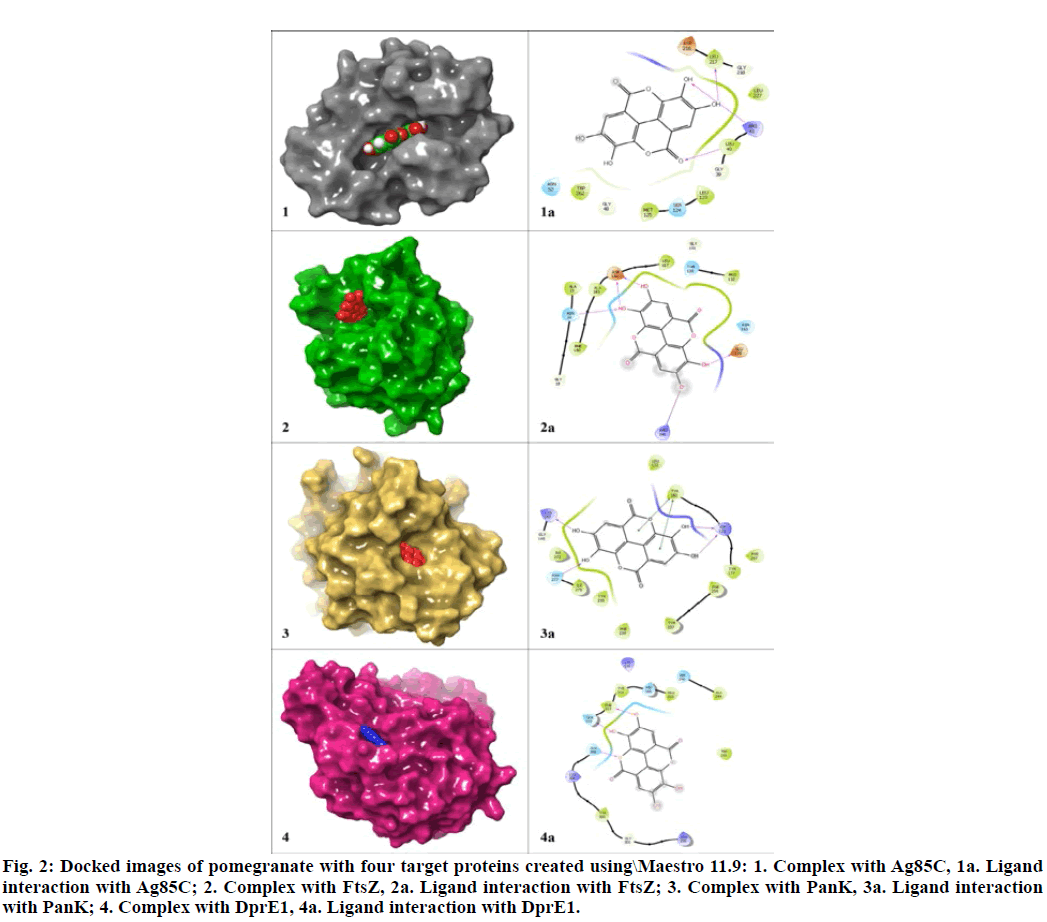
In order to check the accuracy and determining the best lead, the selected three lead molecules were docked with each of the four targets using Glide (Table 7) and ADMET properties of these molecules were analysed using QikProp (Schrödinger, LLC, New York, NY, USA) (Table 8). The comparative analysis of the results revealed that among the three common leads, the compound pomegranatate derived from the flowers[32] could be considered as the best one since it showed least violation in both QikProp and pkCSM analysis. The docked structure of pomogranatate with its target proteins are depicted in fig. 2. Besides, the difference in binding interaction and binding energy level with all the four targets was insignificant.
| Phytochemicals | |||
|---|---|---|---|
| Name of the target protein | Kaempferol-3-o-rhamnoglucoside (kcal/mol) |
Pomegranatate (kcal/mol) |
1,2,4-tri-o-galloyl-β-gluco-pyranose (kcal/mol) |
| Mycolyl transferase antigen 85C protein (Ag85C) | -6.394 | -5.304 | -5.747 |
| Filamentous temperature sensitive protein Z (FtsZ) | -6.046 | -5.723 | -6.337 |
| Pantothenate kinase (PanK) | -6.799 | -6.845 | -6.843 |
| Decaprenyl phosphoryl beta D-ribose 2’-epimerase (DprE1) | -6.171 | -8.8 | -5.405 |
Table 7: Glide XP Docking Scores of the Top Hit Molecules
| Variant | Kaempferol-3-o rhamnoside | Pomegranatate | 1,2,4-tri-o-galloyl-β-gluco-pyranose |
|---|---|---|---|
| #stars | 7 | 1 | 10 |
| #amine | 0 | 0 | 0 |
| #amidine | 0 | 0 | 0 |
| #acid | 0 | 0 | 0 |
| #amide | 0 | 0 | 0 |
| #rotor | 14 | 4 | 18 |
| #rtvFG | 2 | 2 | 4 |
| CNS | -2 | -2 | -2 |
| mol MW | 594.525 | 392.316 | 636.476 |
| dipole | 8.098 | 0.001 | 10.121 |
| SASA | 735.895 | 449.818 | 909.134 |
| FOSA | 152.343 | 0 | 63.095 |
| FISA | 394.923 | 327.304 | 612.828 |
| PISA | 188.629 | 122.514 | 233.211 |
| WPSA | 0 | 0 | 0 |
| Volume-1 | 1502.62 | 766.726 | 1673.1 |
| doNrHB | 8 | 4 | 11 |
| accptHB | 19.8 | 8 | 17.85 |
| dip^2/V | 0.04364 | 0 | 0.06123 |
| QPpolrz | 46.793 | 23.132 | 51.473 |
| QPlogPC16 | 17.92 | 9.574 | 22.596 |
| QPlogPoct | 39.676 | 18.332 | 45.393 |
| QPlogPw | 33.591 | 16.717 | 37.384 |
| QPlogPo/w | -2.08 | -1.342 | -2.861 |
| QPlogS | -1.91 | -1.85 | -3.26 |
| CIQPlogS | -4.208 | -3.214 | -5.44 |
| QPlogHERG | -4.662 | -3.724 | -6.694 |
| QPPCaco | 1.782 | 7.8 | 0.015 |
| QPlogBB | -3.822 | -2.368 | -7.791 |
| QPPMDCK | 0.528 | 2.606 | 0.003 |
| QPlogKp | -6.789 | -6.736 | -10.264 |
| #metab | 9 | 4 | 11 |
| QPlogKhsa | -1.088 | -0.662 | -1.273 |
| HumaOralAbsorption | 1 | 2 | 1 |
| PercentHumaOralAbsorption | 0 | 35.052 | 0 |
| SAfluorine | 0 | 0 | 0 |
| SAamideO | 0 | 0 | 0 |
| PSA | 249.574 | 164.745 | 345.451 |
| #NandO | 15 | 8 | 18 |
| RuleOfFive | 3 | 0 | 3 |
| RuleOfThree | 2 | 1 | 2 |
Table 8: Analysis of Top Leads Using QIKPROP
Figure 2: Docked images of pomegranatate with four target proteins created using\Maestro 11.9: 1. Complex with Ag85C, 1a. Ligand interaction with Ag85C; 2. Complex with FtsZ, 2a. Ligand interaction with FtsZ; 3. Complex with PanK, 3a. Ligand interaction with PanK; 4. Complex with DprE1, 4a. Ligand interaction with DprE1.
The in silico screening results revealed that phytochemiccals present in Punica granatum have significant inhibitory activity on the growth and multiplication of Mycobacterium tuberculosis. Among such compounds, pomegranatate can be recommended as the best one for further investigation. However, the compound kaempferol-3-o-rhamnoglucoside also known as nicotiflorin or kaempferol 3-rutinoside an equally competent compound determined through this investigation has already been approved by Food and Drug Administration (FDA) as a druggable compound[33]. In this backdrop, both compounds have been recommended for further study leading to the discovery of novel drug against tuberculosis.
Acknowledgments:
We authors thank UGC for providing financial assistance through Maulana Azad National Fellowship Scheme. We also thank the Director, Jawaharlal Nehru Tropical Botanic Garden and Research Institute and Dr. T. Madhan Mohan, former advisor, Department of Biotechnology, Government of India for their immense support and providing facilities.
Conflict of interest:
The author(s) confirm that this article content has no conflict of interest.
References
- Global tuberculosis report. Geneva, Switzerland: WHO; 2019.
- da Silva JA, Rana TS, Narzary D, Verma N, Meshram DT, Ranade SA. Pomegranate biology and biotechnology: A review. Sci Hortic 2013;160:85-107.
- Wang R, Ding Y, Liu R, Xiang L, Du L. Pomegranate: constituents, bioactivities and pharmacokinetics. Fruit Veg Cereal Sci Biotech 2010;4(2):77-87.
- Wang D, Özen C, Abu-Reidah IM, Chigurupati S, Patra JK, Horbanczuk JO, et al. Vasculoprotective effects of pomegranate (Punica granatum L.). Front Pharmacol 2018;9:544.
- Rahimi HR, Arastoo M, Ostad SN. A comprehensive review of Punica granatum (pomegranate) properties in toxicological, pharmacological, cellular and molecular biology researches. Iran J Pharm Res 2012;11(2):385.
- Khwairakpam AD, Bordoloi D, Thakur KK, Monisha J, Arfuso F, Sethi G, et al. Possible use of Punica granatum (Pomegranate) in cancer therapy. Pharmacol Res 2018;133:53-64.
- Bhowmik D, Gopinath H, Kumar BP, Kumar K. Medicinal uses of Punica granatum and its health benefits. J Pharmacogn Phytochem 2013;1(5):28-35.
- Negi PS, Jayaprakasha GK. Antioxidant and antibacterial activities of Punica granatum peel extracts. J Food Sci 2003;68(4):1473-7.
- Chaudhary A, Rahul SN. Antibacterial activity of Punica granatum (pomegranate) fruit peel extract against pathogenic and drug resistance bacterial strains. Int J Current Microbiol Appl Sci 2017;6(12):3802-7.
- Willard L, Ranjan A, Zhang H, Monzavi H, Boyko RF, Sykes BD, et al. VADAR: a web server for quantitative evaluation of protein structure quality. Nucleic Acid Res 2003;31(13):3316-9.
- Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy server. The Protein Protocols Handbook 2005:571-607.
- Laskowski RA. PDBsum: summaries and analyses of PDB structures. Nucleic Acids Res 2001;29(1):221-2.
- O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: An open chemical toolbox. J Cheminformatics 2011;3(1):33:1-4.
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem 2010;31(2):455-61.
- Raschka S, Wolf AJ, Bemister-Buffington J, Kuhn LA. Protein–ligand interfaces are polarized: discovery of a strong trend for intermolecular hydrogen bonds to favor donors on the protein side with implications for predicting and designing ligand complexes. J Comput Aid Mol Des 2018;32(4):511-28.
- Ippolito JA, Alexander RS, Christianson DW. Hydrogen bond stereochemistry in protein structure and function. J Mol Biol 1990;215(3):457-71.
- McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J Mol Biol 1994;238(5):777-93.
- Sander T, Freyss J, von Korff M, Rufener C. DataWarrior: an open-source program for chemistry aware data visualization and analysis. J Chem Inf Model 2015;55(2):460-73.
- Schrödinger Release 2019-2: Maestro, Schrödinger, LLC, New York, NY, 2019.
- Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 2004;47(7):1739-49.
- Speck-Planche A, V Kleandrova V, Luan F, Cordeiro ND. In silico discovery and virtual screening of multi-target inhibitors for proteins in Mycobacterium tuberculosis. Comb Chem High Throughput Screen 2012;15(8):666-73.
- Brennan PJ, Nikaido H. The envelope of mycobacteria. Annu Rev Biochem 1995;64(1):29-63.
- Liu J, Nikaido H. A mutant of Mycobacterium smegmatis defective in the biosynthesis of mycolic acids accumulates meromycolates. Proc Natl Acad Sci 1999;96(7):4011-6.
- Bi E, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature 1991;354(6349):161-4.
- Hong W, Deng W, Xie J. The structure, function, and regulation of Mycobacterium FtsZ. Cell Biochem Biophys 2013;65(2):97-105.
- Awasthy D, Ambady A, Bhat J, Sheikh G, Ravishankar S, Subbulakshmi V, et al. Essentiality and functional analysis of type I and type III pantothenate kinases of Mycobacterium tuberculosis. Microbiology 2010;156(9):2691-701.
- Mikusova K, Makarov V, Neres J. DprE1–from the discovery to the promising tuberculosis drug target. Curr Pharm Des 2014;20(27):4379-403.
- Nisha NC, Sreekumar S, Biju CK, Krishnan PN. Identification of lead compounds with cobra venom neutralizing activity in three Indian medicinal plants. Int J Pharm Pharm Sci 2014;6:536-41.
- Shefin B, Sreekumar S, Biju CK. Identification of lead molecules with anti-hepatitis B activity in Bacopa monnieri (L.) Wettst. and Cassia fistula L. through in silico method. J Pharm Biol Sci 2016;11(5):16-21.
- Shityakov S, Förster C. In silico predictive model to determine vector-mediated transport properties for the blood–brain barrier choline transporter. Adv Appl Bioinforma Chem. 2014;7:23.
- Mena P, Calani L, Dall'Asta C, Galaverna G, García-Viguera C, Bruni R, et al. Rapid and comprehensive evaluation of (poly) phenolic compounds in pomegranate (Punica granatum L.) juice by UHPLC-MSn. Molecules 2012;17(12):14821-40.
- Wang R, Wang W, Wang L, Liu R, Ding Y, Du L. Constituents of the flowers of Punica granatum. Fitoterapia 2006;77(7-8):534-7.
- https://pubchem.ncbi.nlm.nih.gov/substance/198937591