- *Corresponding Author:
- E. Mateev
Department of Pharmaceutical Chemistry, Faculty of Pharmacy, Medical University of Sofia, Sofia 1000, Bulgaria
E-mail: e.mateev@pharmfac.mu-sofia.bg
| Date of Received | 20 June 2021 |
| Date of Revision | 23 October 2022 |
| Date of Acceptance | 16 November 2022 |
| Indian J Pharm Sci 2022;84(6):1525-1535 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Molecular docking is emerging as a frequently applied structure-based virtual technique in the drug design processes. The method could significantly reduce the time required for the development of novel and effective molecules compared to high-throughput screening. However, a major drawback of the docking simulations is the high number of false-positive ligands in the top-ranked solutions. Thus, this work focuses on the optimization of genetic optimization for GOLD, and Glide docking protocols in the active sites of crystallographic acetylcholinesterase proteins, which could be used in the processes of design and optimization of novel acetylcholinesterase inhibitors. The performance of GOLD and Glide was assessed by their ability to reproduce known inhibitor conformations of co-crystallized ligands, and to efficiently detect known active compounds seeded into a decoy set. In addition, ensemble docking and molecular mechanics with generalised Born and surface area solvation (MM/GBSA) recalculations were introduced to observe the alterations in the enrichment factors. The variances in the enrichment values between both docking softwares were significant, considering the weak performance of Glide. In all of the employed crystallographic structures, GOLD 5.3 showed drastically better results. Interestingly, the enrichment factors were not increased after utilizing the ensemble docking simulations and the free binding energy recalculations with MM/GBSA. Overall, it was noted that the application of ChemPLP (GOLD 5.3) scoring function in a single acetylcholinesterase protein structure (PDB: 1Q84) displays the most reliable docking results. The obtained data will be beneficial for future virtual screenings of novel acetylcholinesterase inhibitors.
Keywords
Molecular docking, ensemble docking, database enrichment, GOLD, Glide, molecular mechanicsss with generalised Born and surface area solvation, acetylcholinesterase
Molecular docking is emerging as a frequently applied technique in the processes of hit identifications and lead optimizations. With the rapidly progressing computing power and algorithm refinements, the application of the docking simulations is growing exponentially[1]. However, the major drawback during the virtual simulations is the high risk of incorrectly ranked inactive ligands. To decrease the number of falsepositives, a set of validation procedures is being performed[2]. Generally, re-docking, cross docking and database enrichments are being utilized in the processes of reliability assessment of the docking protocols and softwares[3]. These steps are essential prior to each virtual screening considering the lack of unified searching and scoring algorithms. Another issue is the flexibility of the receptor. Methods developed to deal with this issue are molecular dynamics, partial side-chain flexibility, induced-fit docking and simulations in several superimposed receptor conformations (ensemble docking)[4]. Recently, the utilization of ensemble docking simulations is expanding in popularity considering the reliable results[5].
Over the last 25 y, numerous docking programs have been developed and applied in the virtual screening processes. The search for minimum ligand-receptor energy serves as a foundation in the establishment of docking software. Each docking program is constructed out of one or several searching and scoring algorithms. The search algorithm places the ligands in the active site of the receptor in the search for active conformations. They are broadly classified into systematic (incremental construction, conformational search), stochastic (Monte Carlo, genetic algorithms, tabu search), and simulation algorithms (molecular dynamics, energy minimization)[6]. Genetic Optimization for Ligand Docking (GOLD) and Glide are two of the most frequently applied docking softwares. GOLD employs the genetic search algorithm and four different scoring functions[7], whereas Glide uses systematic search and an empirical-based scoring algorithm[8]. A recent study held by Wang et al.[9] has compared the sampling and the scoring abilities of ten docking programs (LigandFit, Glide, GOLD, molecular operating environment Dock, Surflex-Dock, AutoDock, AutoDock Vina, LeDock, rDock, and UCSF DOCK). The study has noted the significant robustness of Glide (XP) and GOLD.
The Acetylcholinesterase Enzyme (AChE) disables the nerve impulses as it hydrolyses the acetylcholine in the cholinergic pathway of the nervous system. Therefore, antagonists of the enzyme are part of the main drug classes which deal with memory loss and in particular with Alzheimer’s disease[10]. The first crystallographic structure of acetylcholinesterase with co-crystallized tacrine in the binding gorge, was resolved back in 1993 (Protein Data Bank (PDB) code: 1ACJ) [11]. Since then, the rapidly growing number of 3D AchE structures determined from a diverse set of sources (Homo sapiens, Torpedo californica, Mus musculus) has been deposited in the PDB. Studies have shown that the active site of AchE is located 20 Å deep into the receptor and it includes two distinct binding sites, “peripheral” and “catalytic gorge”. The peripheral site comprises a less welldefined area, located at the entrance of the catalytic pocket. The major occurring interactions between the peripheral binding site and the inhibitors are hydrophobic. Molecules such as propodium and fasciculins bind to that site[12]. Docking simulations in the anionic site of AchE have been discussed as early as 1994[13]. The catalytic site lies deeper in the binding pocket, and it consists of esteratic and anionic binding sites[14]. The active site of AChE contains a catalytic triad built of three amino acids; Ser 200, His 440, Glu 327, which is located at the bottom of the enzyme’s active site. The active amino residue Trp 84 has been reported to bind to the quaternary group of acetylcholine, decamethonium and edrophonium. Moreover, Trp 279, which is located at the entrance of the binding gorge, participates in the interactions with a second quaternary group. Such a case is the interactions of decamethonium with the active site of AChE[12].
Initially, all of the determined structures entailed mouse or electric ray (Torpedo californica) homologues. Subsequently, deposited human AChE structures were presented and some drastic differences in the sizes of the binding pockets were found[15]. In the case of typical AChE (tAChE), the binding grid is larger in size with a narrower entrance cavity. The tAChE inhibitor donepezil is an example for a ligand with considerably contrasting orientations in the active sites of the Torpedo and Human AChE crystallographic structures[16].
To obtain reliable virtual screening results of compounds with diverse sets of chemical structures, validations processes of the docking protocols should be conducted. These methods evaluate the docking ability to distinguish the falsepositive from true-positive ligands. Furthermore, optimizing the docking parameters will result in more robust and reliable scores. Thus, the aim of our work was to achieve optimal GOLD 5.3 and Glide protocols for higher virtual enrichments in crystallographic acetylcholinesterase proteins. Ensemble docking and Molecular Mechanics with Generalised Born and Surface Area solvation (MM/GBSA) were also utilized in the processes.
Materials and Methods
Hardware:
The docking simulations were carried out on an AMD Ryzen 9 5950X 16 core CPU with GeForce RTX 3060 12 GB GPU, and 64 GB of installed RAM. The utilized operating system was 64 bit Windows 10 Pro.
Proteins preparation and refinements:
Crystallographic structures of human AChEs in complex with co-crystallized molecules: TZ4 (PDB: 1Q84; 2.45 Å), Huperzine A (PDB ID: 1VOT; 2.50 Å), Huprine derivative (PDB ID: 4A16; 2.65 Å), Huperzine A (PDB ID: 4EY5; 2.3 Å), Galantamine (PDB ID: 4EY6; 2.3 Å), Donepezil (PDB ID: 4EY7; 2.35 Å), Dihydrotanshinone I (PDB ID: 4M0E; 2 Å), Territrem B (PDB ID: 4M0F; 2.3 Å), Paraoxon and Pralidoxime (PDB ID: 5HFA; 2.20 Å), HI6 (PDB: 5HF9; 2.2 Å), HI-6 (PDB ID: 6CQU; 2.31 Å), VX(-) and HI-6 (PDB ID: 6CQW; 2.28 Å), C35 (PDB ID: 6F25; 3.05 Å), RS-170B (PDB ID: 6O5R; 2.80 Å), RS-170B (PDB ID: 6O5S; 2.80 Å), N2K (PDB ID: 6TD2; 2.80 Å), N9T (PDB ID: 6TT0; 2.80 Å), RS 194B (PDB ID: 6U34; 2.40 Å) were downloaded from the PDB (www.rcsb.org). All the target structures were further refined for docking applying the Protein Preparation Wizard available in Maestro[17]. Initially, complexes comprising a heme group or covalent interactions between protein and ligand were deleted. Subsequently, hydrogen bonds and het states were generated, and water molecules beyond 5 Å from het groups were removed. The generated het states using Epik were in pH 7.0±2.0, and the complexes were minimized utilizing the Optimized Potentials for Liquid Simulations (OPLS) force field. The H-bond assignment was carried out with PROPKA in neutral pH (default in Maestro). The grid box was generated around each co-crystallized ligand. No constraints were included in the docking protocols.
Ligands preparation:
The ligands were prepared for docking utilizing the LigPrep module (Schrödinger Release 2021-2: LigPrep, Schrödinger, LLC, New York, NY, 2021). Initially, hydrogen bonds were added, the tautomers were included, as well as the possible states at pH 7.0±2.0 were generated. Thereafter, energy minimization steps were employed.
Docking softwares:
The docking simulations were conducted utilizing GOLD 5.3 and Glide docking module in Schrödinger Maestro Suite[18].
GOLD:
GOLD 5.3[7] contains four scoring algorithms; GoldScore, ASP, ChemPLP and ChemScore. During the ligand enrichment simulations, all of the scoring functions were utilized to detect the most eminent one for docking in AChE structures. The grid space was centered on the centroid of each co-crystallized ligand with default size of 8 Å. The search efficiency for the virtual screening was set to 30 % (“Virtual Screening” option in GOLD 5.3) considering the time demanding simulations.
Glide:
Glide, (Schrödinger, LLC, New York, NY, 2021) algorithms are based on systematic searches in the active site of the receptor[8]. The docking modes consist of rigid and flexible ligand simulations. Out of the three docking options employed in Glide (Highthroughput virtual screening (HTVS)), Standard Precision (SP) and extra-precision), the HTVS mode was used for the preliminary screening of the Directory of Useful Decoys-Enhanced (DUD-E) database. The binding sites are defined by rectangular boxes positioned in the center of the co-crystallized ligand. The former process was conducted with the “Receptor Grid Generation'' application in Glide. No restraints and no rotatable bonds were established in the simulation.
Re-docking:
The main objective of the self-docking procedure is to identify the ability of the docking software to correctly predict the conformations of co-crystallized ligands. The Root Mean Square Deviation (RMSD) values were calculated to analyze the difference between the crystal ligands' poses, and the predicted coordinates by GOLD and Glide. We applied a RMSD threshold of 2 Å as opposed to the recently used 1.5 Å[19] considering the relatively high number of freely rotatable bonds in most of the applied co-crystallized ligands. 100 % search efficiency in GOLD 5.3 and the XP option in Glide were employed. For each docked ligand 20 poses were generated, and the RMSD value of the highest ranked solution was considered. No energy minimizations of the ligands were conducted prior to re-docking.
Database enrichment:
To evaluate the robustness of GOLD’s ChemPLP and Glide’s HTVS scoring functions in finding known inhibitors embedded in random decoys, we applied a modified dataset downloaded from DUD-E[20]. Considering the size of the dataset, we applied 92 actives instead of the initial 664, and a subsequent reduction in the number of the decoys from 26 373 to 8807. Calculations took an average of 10 s per run utilizing the aforementioned hardware setup.
Enrichment Factor (EF):
The validation of the docking protocol was conducted through calculations of the EF. It considers the number of active molecules located in the top positions[21]. The enrichment values are enhanced when more actives are located in the examined fraction. The formula of the classic EF is given below:
EF=(Hitssampled/Hitstotal)/(Nsampled/Ntotal)
In the above formula, Hitssampled stands for the active compounds which are detected in the chosen percentage of the dataset. Hitstotal is the count of all active structures seeded in the decoys, which in our case equals 92. Ntotal is the value of the docking dataset 8899, while Nsampled is the percentage of the dataset which is being observed. To examine the early enrichment, 1 % of the benchmark set was considered which in our case equaled to 89. Percentages over five are used only when HTVS could be conducted for the observed ligands.
A major drawback of the classical EF is that it does not consider the rankings of the active ligands. Therefore, we included calculations of a modified EF`, which contemplates the fitness scores of the obtained active molecules. EF`(x) is defined below:
EF`(N)=(50 %/APRsampled)×(Hitssampled/Hitstotal)
Here N is the percent of the explored active compounds; ARP stands for “Average Percentile Rank” of Hitstotal. We calculated the EF` value of 10 % of the seeded active compounds, therefore the Hitssampled corresponds to 9; Hitstotal equals 92. The maximum number of EF`(10) is 90.1, which could be achieved when the top 9 rankings of the docking are occupied by 9 of the seeded actives.
Virtual screening:
An AChE database freely available in the DUD-E was used to validate and assess the performance of GOLD 5.3 and Glide[20]. For the current study, 92 active AChE inhibitors were seeded in 8807 decoys. The decoys contain similar molecular weight, LogP and rotatable bonds compared to the actives, however, the topological characteristics are unlike. It should be noted that most of the decoys are not experimentally tested, therefore they could possess in vitro activity.
MM/GBSA:
In this study, MM-GBSA recalculations with Prime were employed to assess the free binding energies of the obtained complexes. The calculations were performed by the incorporation of the OPLS3 force field and VSGB dissolvable model[22].
Results and Discussion
13 human, two Torpedo and three mouse AChE receptors were included in the re-docking simulations to compare the crystallographic proteins resolved from Homo sapiens, Tetronarce californica and Mus musculus organisms (Table 1). Most of the ligands were successfully redocked with both docking softwares. The lowest RMSD values were observed when Territrem B was placed back in the active site of 4M0F. However, none of the searching algorithms were able to correctly locate the active co-crystallized ligands of 5HFA, 6CQU, 6F25, 6O5R and 6O5S. After the redocking of the highly rotatable co-crystallized ligand situated in the 6F25 AChE crystallographic structure, significantly elevated RMSD values were obtained. In the former case, both softwares failed to correctly place the ligand back into the active gorge. High values of over 5 Å were detected for the dual inhibitor C-35[23].
| PDB Code | Organism | Docking Software | Docking Scores | RMSD | Co-crystallized ligand | Molecular weight | Rotatable bonds |
|---|---|---|---|---|---|---|---|
| 1Q84 | Mus musculus | GOLD 5.3 | 51.23 PLP.Fitness | 10.0 Å | TZ4 | 661.86 | 12 |
| Glide | -19.622 kcal/mol | 0.55 Å | |||||
| 1VOT | Tetronarce californica | GOLD 5.3 | 69 PLP.Fitness | 0.43 Å | Huperzine A | 242.32 | 0 |
| Glide | -11.29 kcal/mol | 0.31 Å | |||||
| 4A16 | Mus musculus | GOLD 5.3 | 118 PLP.Fitness | 0.51 Å | H34 | 423.94 | 6 |
| Glide | -16.27 kcal/mol | 0.63 Å | |||||
| 4EY5 | Homo sapiens | GOLD 5.3 | 78.02 PLP.Fitness | 0.13 Å | Huperzine A | 242.32 | 0 |
| Glide | -9.878 kcal/mol | 0.04 Å | |||||
| 4EY6 | Homo sapiens | GOLD 5.3 | 71.25 PLP.Fitness | 0.21 Å | (-)-Galantamine | 287.35 | 1 |
| Glide | -10.196 kcal/mol | 0.21 Å | |||||
| 4EY7 | Homo sapiens | GOLD 5.3 | 107.45 PLP.Fitness | 0.27 Å | Donepezil | 379.49 | 6 |
| Glide | -19.536 kcal/mol | 0.42 Å | |||||
| 4M0E | Homo sapiens | GOLD 5.3 | 78.61 PLP.Fitness | 0.20 Å | Dihydrotanshinone I | 278.3 | 0 |
| Glide | -12.2 kcal/mol | 0.40 Å | |||||
| 4M0F | Homo sapiens | GOLD 5.3 | 104.52 PLP.Fitness | 0.63 Å | Territrem B | 526.58 | 4 |
| Glide | -13.44 kcal/mol | 0.09 Å | |||||
| 5HFA | Homo sapiens | GOLD 5.3 | 57.21 PLP.Fitness | 3.94 Å | FP1 | 138.17 | 1 |
| Glide | -9.468 kcal/mol | 3.86 Å | |||||
| 5HF9 | Homo sapiens | GOLD 5.3 | 81.04 PLP.Fitness | 1.91 Å | HI6 | 288.3 | 6 |
| Glide | -13.197 kcal/mol | 1.39 Å | |||||
| 6CQU | Homo sapiens | GOLD 5.3 | 90.64 PLP.Fitness | 2.46 Å | HI6 | 288.3 | 6 |
| Glide | -11.176 kcal/mol | 3.26 Å | |||||
| 6CQW | Homo sapiens | GOLD 5.3 | 115.95 PLP.Fitness | 1.99 Å | HI6 | 288.3 | 6 |
| Glide | -16.165 kcal/mol | 8.07 Å | |||||
| 6F25 | Homo sapiens | GOLD 5.3 | 117.59 PLP.Fitness | 5.00 Å | CVZ | 620.74 | 17 |
| Glide | -13.904 kcal/mol | 13.7 Å | |||||
| 6O5R | Homo sapiens | GOLD 5.3 | 73.29 PLP.Fitness | 2.91 Å | LND | 274.3 | 5 |
| Glide | -10.629 kcal/mol | 2.49 Å | |||||
| 6O5S | Homo sapiens | GOLD 5.3 | 73.42 PLP.Fitness | 2.83 Å | LND | 274.3 | 5 |
| Glide | -6.78 kcal/mol | 7.14 Å | |||||
| 6TD2 | Mus musculus | GOLD 5.3 | 80.32 PLP.Fitness | 3.23 Å | *N2K | 338.39 | 8 |
| Glide | -15.17 kcal/mol | 1.04 Å | |||||
| 6TT0 | Tetronarce californica | GOLD 5.3 | 102.96 PLP.Fitness | 2.71 Å | N9T | 436.5 | 7 |
| Glide | -13.88 kcal/mol | 0.71 Å | |||||
| 6U34 | Homo sapiens | GOLD 5.3 | 54.08 PLP.Fitness | 1.65 Å | PQV | 213.28 | 4 |
| Glide | -7.371 kcal/mol | 5.45 Å |
Table 1: PDB Codes, Re-Docking Scores, RMSD Values, Co-Crystallized Ligand, Molecular Weight, and Rotatable Bonds of the Applied Crystallographic Structures
Interestingly, several PDB structures comprising small co-crystallized ligands with low count of rotatable bonds, such as 5HFA and 6CQU, demonstrated poor results with RMSD values over 2 Å when GOLD and Glide were employed. The number of rotatable bonds was in the range of 5-7, however both docking softwares failed in the process of recreating the active conformations. Theoretically, Glide displayed lower RMSD values when the TZ4 ligand, which comprises 12 rotatable bonds, was docked back into the active site of 1Q84 (0.55 Å). Furthermore, simulations in the active center of 6TD2 also demonstrated more reliable results when Glide was employed. A RMSD value of 1.042 Å was obtained for a ligand with eight rotatable bonds.
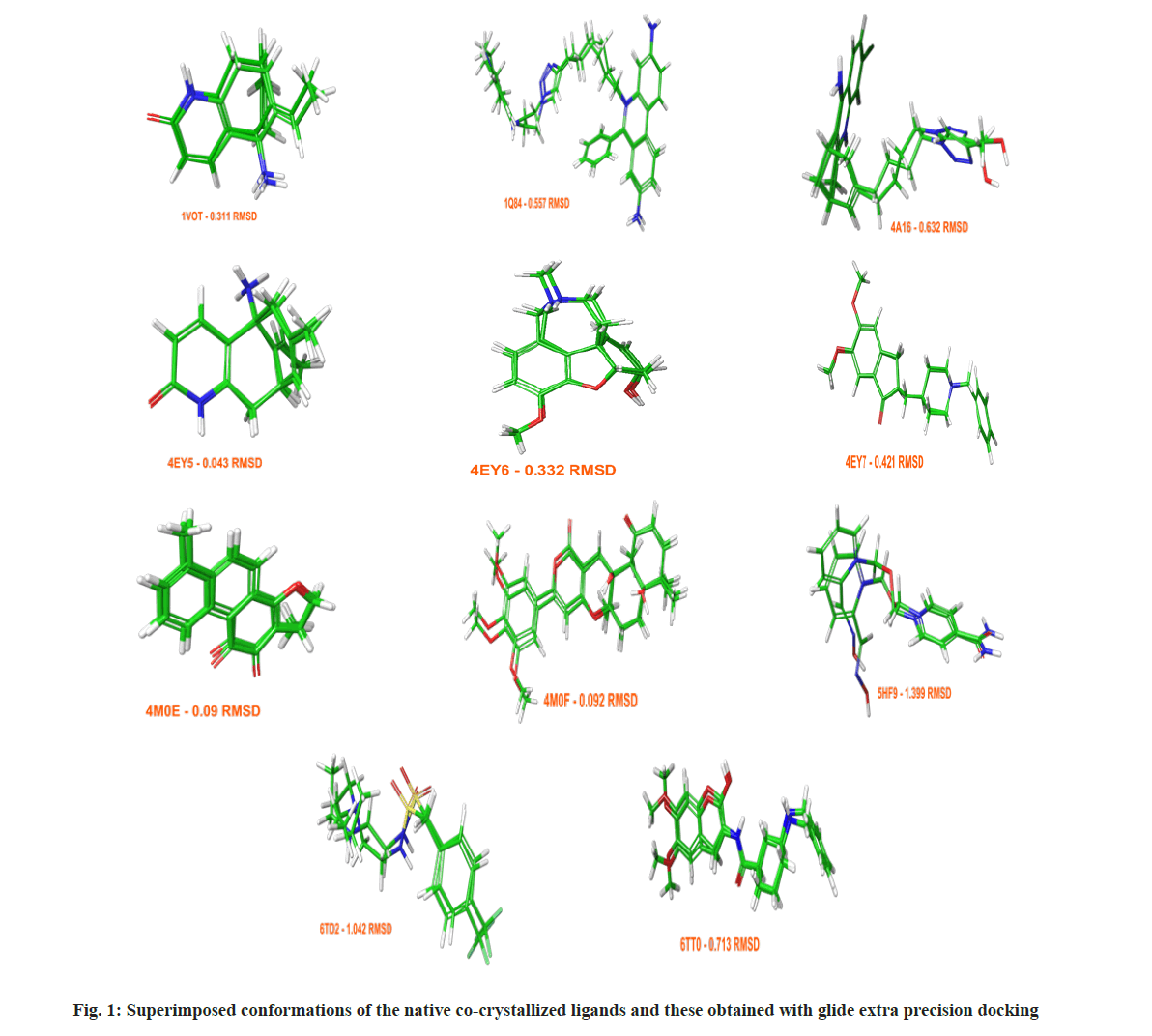
Assessing the docking poses with RMSD values under 2 Å, Glide achieved better results with a success rate of 61 %, while GOLD 5.3 succeeded in 56 % of the cases. The obtained docking conformations from Glide with RMSDs under 2 Å are given in fig. 1. To validate and optimize an optimal docking protocol for a future virtual screening of novel AChE inhibitors, and to evaluate the accuracy of GOLD 5.3 and Glide both a standard enrichment factor (EF (1 %)) and a modified version of it (EF`(10)), which takes into account the ranks of the active ligands were applied. Detailed information about the enrichment calculations is given in the materials and methods section. The benchmarking set consisted of 8807 decoys and 92 active AChE inhibitors.
The PDB crystallographic structures with RMSD values under 2 Å from the redocking simulations were employed in the virtual enrichment. Interestingly, Glide demonstrated inadequate docking sampling, considering the elevated binding scores of the decoys (Table 2). It was observed that docking with Glide in the active gorges of 5HFA, 6CQW and 6TT0 led to zero enrichment when 1 % of the dataset was inspected. One active ligand was found in the top 89 positions when 6CQU, 6CQW, 6O5S, 6TD2 and 6U34 were employed as AChE proteins. Less than four active ligands were observed when 4EY5, 4EY6, 4M0F, 4M0E and 6O5R were introduced into the simulations. The modified enrichment factors for these receptors were not calculated considering the insufficient values of the classical EF.
| PDB code | EF (1 %) | EF`(10) | ||
|---|---|---|---|---|
| GOLD 5.3 | Glide | GOLD 5.3 | Glide | |
| 1Q84 | ChemPLP=35.49 | 21.58 | ChemPLP=89.96 | 24.24 |
| ChemScore=29 | ChemScore=89.96 | |||
| GoldScore=24.74 | GoldScore=53 | |||
| ASP=24.74 | ASP=55 | |||
| 1VOT | 14.8 | 3.22 | 37.3 | N/A |
| 4A16 | 6.45 | 2.15 | N/A | N/A |
| 4EY5 | 1.07 (modified-20.4) | 2.15 | N/A (modified-56.23) | N/A |
| 4EY6 | 24.74 | 2.15 | 69.81 | N/A |
| 4EY7 | 11.03 | 5.37 | 14.78 | N/A |
| 4M0E | 21.1 | 3.22 | 32.13 | N/A |
| 4M0F | 29 | 2.15 | 43 | N/A |
| 5HFA | 0 | 0 | 0 | N/A |
| 5HF9 | 7.53 | 0 | N/A | N/A |
| 6CQU | 3.22 | 1.07 | N/A | N/A |
| 6CQW | 12.9 | 1.07 | 15.99 | N/A |
| 6F25 | 15.06 | 6.44 | 16.07 | N/A |
| 6O5R | 17.2 | 2.15 | 36.47 | N/A |
| 6O5S | 16.13 | 1.07 | 30.45 | N/A |
| 6TD2 | 9 | 1.07 | 11.56 | N/A |
| 6TT0 | 16.13 | 0 | 25.46 | N/A |
| 6U34 | 1.07 | 1.07 | N/A | N/A |
Table 2: Ligand Enrichments of Gold 5.3 and Glide
Interestingly, only one receptor demonstrated good EF (1 %) value when the DUD-E dataset was docked with Glide. In the active site of 1Q84, 20 of the seeded active molecules were situated in the top 89 ranks. Moreover, the top 9 actives were placed in 7, 8, 12, 14, 19, 24, 25, 28 and 29 positions, respectively, which delivered EF`(10) of 24.24. To re-analyze the aforementioned failure of Glide, we carried out more detailed and therefore computationally demanding simulations in the active sites of 4M0E and 4M0F applying SP as more precise option, together with a fixation of the binding gorge at 12 Å. However, the former docking protocols did not lead to significant improvements in the enrichment values, therefore it was not considered for a future evaluation.
Initially, all four scoring functions of GOLD 5.3 (ChemPLP, ChemScore, GoldScore, ASP), were utilized to detect the most prominent one. ChemPLP demonstrated the best capacity of identifying the true active molecules, thus it was applied in the rest of the simulations.
Overall, it was evident that when GOLD 5.3 was employed for the database enrichments of AChE, the reliability of the docking simulations was significantly enhanced. Only in six of the utilized PDBs, less than nine actives were observed in the top 1 % of the dataset (Table 2).
The highest possible modified enrichment factor (EF`(10)=max) was acquired after virtual simulations in the active site of 1Q84. In the former case, the program was able to situate 10 % of the active inhibitors in the first nine ranks. Moreover, 33 actives were found in the top 1 % of the scored database which led to an EF (1 %) of 35.49. The fitness scores acquired with ChemPLP were in the range from 122.56 to 135.52.
The human AChE receptor with PDB code 4EY5, acquired the lowest value of 1.07 after docking simulations with GOLD 5.3, which corresponds to only one active placed among the top 89 positions. To further examine the incapability of GOLD’s aforementioned settings to detect true positive ligands in the active cleft of 4EY5, we carried out simulations employing 10 Å binding grid and 200 % search efficiency in the active site of the former crystallographic structure. The obtained data showed that the enrichment increased significantly at the cost of computational time. We detected 19 of the seeded actives in the top 89 docking solutions, which led to enhanced EF of 20.43. Furthermore, nine of the seeded active ligands were located in top 20 ranks and the value of the modified EF`(10) was calculated to be 56.23.
To further validate the acquired enrichments in the AChE protein 1Q84 with GOLD 5.3, we applied the whole DUD-E database constructed out of 664 active ligands and 26 373 decoys. The top scored 22 ranks were occupied with true positive ligands which demonstrated the highest modified EF. The classical EF (1 %) was calculated to be 17.7 which correspond to 120 actives in the first 270 positions. When EF of 0.5 % of the database was considered, the obtained enrichment was equivalent to 27.44. Therefore, 1Q84 is the most promising PDB structure for future docking investigations with the docking software GOLD.
In the search for higher enrichments, several modifications in the docking protocols of 1Q84 were made. Ensemble docking, and rescoring with MM/ GBSA were employed. To analyze the effects of the protein superimposition in the virtual screening of AChE receptors, we conducted superimpositions, and docking simulations in GOLD 5.3. The classical and the modified enrichment factors were calculated to quantify the reliability of the ensemble docking (Table 3).
| Ensemble | EF (1 %) | EF`(10) |
|---|---|---|
| 1VOT-4EY6-4M0E | 37.64 | 59.54 |
| 4M0E-4M0F-4EY6 | 33.34 | 64.26 |
| 1Q84-4EY6-4M0E | 37.64 | 73.61 |
| 1Q84-4EY6-4M0E-4M0F-6O5R | 37.64 | 57.02 |
| 1Q84-4EY6-4M0E-4M0F-6O5R-6O5S-6F25-1VOT-6TT0-4EY7 | 38.72 | 67.48 |
Table 3: Ensemble Docking Simulations of Several Ache Proteins.
The docking simulations with an ensemble constructed out of 1VOT, 4EY6 and 4M0E AChE PDB structures located 35 actives in the top 89 solutions. Moreover, nine of the actives were placed within the top 15 positions, which unambiguously demonstrate the prominent ability of the former ensemble in redeeming early enrichments. When superimposing the crystallographic structures of 4M0E, 4M0F and 4EY6, 31 of the seeded actives were found in the top 1 % of the dataset which led to an enrichment factor of 33.34. However, nine of the highest scored active ligands were situated in 1, 2, 4, 5, 7, 8, 10, 12 and 14 ranks, respectively, therefore the EF`(10) was higher than the previous complex. The simulation demonstrated that the employment of only the classical enrichment is not sufficient for assessing the full capability of the docking protocol. The utilization of five or ten superimposed AChE receptors did not lead to better enrichments when compared to the single target docking. Furthermore, the computational cost during these simulations was significantly increased.
The free binding energies (MM/GBSA) of the top complexes acquired with ChemPLP (GOLD 5.3) in the active site of 1Q84 were recalculated. Compared to the results obtained with the ChemPLP docking scoring function, MM/GBSA did not demonstrate significantly better results. We found that MMGBSA is not able to completely distinguish the actives from the decoys. Interestingly, several decoys were rescored with high free binding energies which decreased the overall enrichments.
Each docking program relies on scoring algorithms to evaluate the binding affinities of the dataset. As for now, there is no universal docking scoring function for all available receptors, thus it is necessary to apply benchmarking sets for each target prior to virtual screening[3]. A confirmation to that is a comparative assessment of eleven scoring functions, which are part of a diverse set of docking software, by Wang et al.[24]. The paper has unambiguously concluded that by no means are the current scoring functions perfect, herein further work in that field needs to be conducted.
A study held by Halim et al.[25] investigated the reliability of six docking softwares towards AChE, however several limitations in that work were observed. Firstly, the employed 3D acetylcholinesterase receptors are resolved from Tetronarce californica, not from Homo sapiens. The differences between the two types of receptors are considerably significant with more reliable in silico results being obtained by docking into the crystallographic structures resolved from Homo sapiens[16]. Secondly, Glide (Schrodinger inc.) has not been utilized in the virtual screening even though the former is considered as one of the most robust docking softwares[26]. Finally, three of the GOLD scoring algorithms have been applied; Chemscore, GoldScore and ASP. However, the most recent scoring function of GOLD, introduced as a default scoring algorithm after version 5.1-ChemPLP, has demonstrated better results[27]. Thus, we introduced the drawbacks of the aforementioned work, and carried out an exhaustive benchmarking study.
Firstly, re-docking simulations were carried out. Initial docking validation is a necessity considering the approximate character of all scoring functions, and the steadily growing count of PDB structures and docking softwares[28]. The simplest and most frequently applied internal validation procedure is the self-docking methodology. During the simulation, an experimentally resolved conformation of a cocrystallized ligand is docked back into the active site of the receptor. The final docking solutions are compared to the primary poses of the co-crystallized ligand, and the RMSD values are calculated[19]. The reliability of Glide and GOLD 5.3 to regenerate the bioactive poses of AChE co-crystallized ligands was evaluated. Docking poses with RMSD values under 2 Å were defined as acceptable. The presence of active waters was considered in all cases, taking into account the importance of conserved waters in the active site[29]. It is unambiguous, however, that docking simulations with large ligand molecules, comprising a high number of rotatable bonds, could drastically affect the robustness of the virtual results. In such cases the conformational flexibility increases significantly, which leads to wrong ligand poses in the active site and poor docking scores[30]. Overall, most of the obtained redocking results were in good agreement with recent findings[25,31,32].
The conducted benchmarking simulations employing Glide as docking software revealed the major drawback of some searching and scoring algorithms[33]. Most of the decoys were ranked in the top positions which drastically decreased the enrichment factors. In contrast, the simulations with GOLD 5.3 acquired more reliable data which could be related to the different searching algorithm implemented in the software, the genetic algorithm[7]. Moreover, after initial evaluation of all scoring functions employed in GOLD 5.3, ChemPLP demonstrated the highest enrichments. The potential of the former algorithm has been reported by Li et al.[34].
The highest enrichment achieved in the crystallographic structure 1Q84 could be explained with the solvent-accessible and highly mobile active amino residue Trp 286[35]. After the utilization of the available DUD-E dataset 1Q84 acquired the most promising results out of all applied AChE PDBs when GOLD 5.3 was used. Out of the crystallographic structures resoluted from Homo sapiens, 4M0F demonstrated the best ability to differentiate trueactive inhibitors from decoys.
To observe the alteration in the enrichment we incorporated the protein’s flexibility in the docking simulations, as well as MM/GBSA recalculations[36]. Numerous papers have reported higher enrichments when ensemble docking and free binding energy recalculations have been introduced in the docking simulations[5,37]. Interestingly, the implementation of several AChE crystallographic structures with dissimilar active site conformations led to higher EF value, however, the rankings of the active ligands were not ideal which was observed after calculating the modified EF`. Thus, the employment of a single AChE structure (1Q84) in the virtual screenings with GOLD 5.3 is more reliable than using a superimposed complex of crystallographic enzymes. Furthermore, the MM/GBSA method was applied to calculate the free binding energies of the top scored docking complexes. Interestingly, the implementation of the former method did not achieve higher enrichment in the current work which might be due to the limitations of MM/GBSA[38].
In conclusion, exhaustive docking simulations in numerous AChE receptors were conducted utilizing two of the most robust docking softwares-GOLD 5.3 and Glide. In all of the employed crystallographic structures, GOLD 5.3 showed better results when compared to Glide. The best docking protocol was constructed out of the ChemPLP scoring function with 8 Å binding grid in the active site of 1Q84. Moreover, out of the crystallographic structures resoluted from Homo sapiens, 4M0F demonstrated the best ability to differentiate true-active inhibitors from decoys. From the obtained data the inability of Glide to correctly score most of the actives, in all of the crystallographic AChE receptors, should be pointed out. Furthermore, the ensemble docking, and the free binding energy recalculations with MM/ GBSA did not demonstrate improvements in the enrichment values. This study will be beneficial for a future virtual screening of novel AChE inhibitors.
Acknowledgements
This work was supported by the Council of Medical Sciences at the Medical University of Sofia (Contract No. 160/04.06.2022, Grand No. 7418/19.11.2021).
Confict of Interest
The authors declare no conflicts of interest.
References
- Maia EH, Assis LC, de Oliveira TA, da Silva AM, Taranto AG. Structure-based virtual screening: From classical to artificial intelligence. Front Chem 2020;8:343.
[Crossref] [Google Scholar] [PubMed]
- Sieg J, Flachsenberg F, Rarey M. In need of bias control: Evaluating chemical data for machine learning in structure-based virtual screening. J Chem Inf Model 2019;59(3):947-61.
- Kumar A, Zhang KY. A cross docking pipeline for improving pose prediction and virtual screening performance. J Comput Aided Mol Des 2018;32(1):163-73.
[Crossref]
- Saikia S, Bordoloi M. Molecular docking: Challenges, advances and its use in drug discovery perspective. Curr Drug Targets 2019;20(5):501-21.
[Crossref] [Google Scholar] [PubMed]
- Rao S, Sanschagrin PC, Greenwood JR, Repasky MP, Sherman W, Farid R. Improving database enrichment through ensemble docking. J Comput Aided Mol Des 2008;22(9):621-7.
[Crossref] [Google Scholar] [PubMed]
- Pagadala NS, Syed K, Tuszynski J. Software for molecular docking: A review. Biophys Rev 2017;9(2):91-102.
[Crossref] [Google Scholar] [PubMed]
- Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J Mol Biol 1997;267(3):727-48.
[Crossref] [Google Scholar] [PubMed]
- Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 2004;47(7):1739-49.
- Wang Z, Sun H, Yao X, Li D, Xu L, Li Y, et al. Comprehensive evaluation of ten docking programs on a diverse set of protein-ligand complexes: The prediction accuracy of sampling power and scoring power. Phys Chem Chem Phys 2016;18(18):12964-75.
[Crossref] [Google Scholar] [PubMed]
- Lazarevic-Pasti T, Leskovac A, Momic T, Petrovic S, Vasic V. Modulators of acetylcholinesterase activity: From Alzheimer's disease to anti-cancer drugs. Curr Med Chem 2017;24(30):3283-309.
[Crossref] [Google Scholar] [PubMed]
- Harel M, Schalk I, Ehret-Sabatier L, Bouet F, Goeldner M, Hirth C, et al. Quaternary ligand binding to aromatic residues in the active-site gorge of acetylcholinesterase. Proc Natl Acad Sci USA 1993;90(19):9031-5.
[Crossref] [Google Scholar] [PubMed]
- Castro A, Martinez A. Peripheral and dual binding site acetylcholinesterase inhibitors: Implications in treatment of Alzheimer's disease. Mini Rev Med Chem 2001;1(3):267-72.
[Crossref] [Google Scholar] [PubMed]
- Pang YP, Kozikowski AP. Prediction of the binding sites of huperzine A in acetylcholinesterase by docking studies. J Comput Aided Mol Des 1994;8(6):669-81.
[Crossref] [Google Scholar] [PubMed]
- Harel M, Kasher R, Nicolas A, Guss JM, Balass M, Fridkin M, et al. The binding site of acetylcholine receptor as visualized in the X-ray structure of a complex between α-bungarotoxin and a mimotope peptide. Neuron 2001;32(2):265-75.
[Crossref] [Google Scholar] [PubMed]
- Wiesner J, Kříž Z, Kuča K, Jun D, Koča J. Acetylcholinesterases-the structural similarities and differences. J Enzyme Inhib Med Chem 2007;22(4):417-24.
[Crossref] [Google Scholar] [PubMed]
- Cheung J, Rudolph MJ, Burshteyn F, Cassidy MS, Gary EN, Love J, et al. Structures of human acetylcholinesterase in complex with pharmacologically important ligands. J Med Chem 2012;55(22):10282-6.
[Crossref] [Google Scholar] [PubMed]
- Madhavi Sastry G, Adzhigirey M, Day T, Annabhimoju R, Sherman W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des 2013;27(3):221-34.
[Crossref] [Google Scholar] [PubMed]
- Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, et al. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein ligand complexes. J Med Chem 2006;49(21):6177-96.
[Crossref] [Google Scholar] [PubMed]
- Hevener KE, Zhao W, Ball DM, Babaoglu K, Qi J, et al. Validation of molecular docking programs for virtual screening against dihydropteroate synthase. J Chem Inf Model 2009;49(2):444-60.
[Crossref] [Google Scholar] [PubMed]
- Mysinger MM, Carchia M, Irwin JJ, Shoichet BK. Directory of useful decoys, enhanced (DUD-E): Better ligands and decoys for better benchmarking. J Med Chem 2012;55(14):6582-94.
[Crossref] [Google Scholar] [PubMed]
- Mateev EM, Valkova IV, Georgieva MA, Zlatkov AL. Database enrichments of Mao-B through ensemble docking. Int J Pharm Pharm Sci 2021;13:32-5.
- Sahakyan H. Improving virtual screening results with MM/GBSA and MM/PBSA rescoring. J Comput Aided Mol Des 2021;35(6):731-6.
[Crossref] [Google Scholar] [PubMed]
- Zueva I, Dias J, Lushchekina S, Semenov V, Mukhamedyarov M, Pashirova T, et al. New evidence for dual binding site inhibitors of acetylcholinesterase as improved drugs for treatment of Alzheimer's disease. Neuropharmacol 2019;155:131-41.
[Crossref] [Google Scholar] [PubMed]
- Wang R, Lu Y, Wang S. Comparative evaluation of 11 scoring functions for molecular docking. J Med Chem 2003;46(12):2287-303.
[Crossref] [Google Scholar] [PubMed]
- Halim SA, Uddin R, Madura JD. Benchmarking docking and scoring protocol for the identification of potential acetylcholinesterase inhibitors. J Mol Graph Model 2010;28(8):870-82.
[Crossref] [Google Scholar] [PubMed]
- Chaput L, Martinez-Sanz J, Saettel N, Mouawad L. Benchmark of four popular virtual screening programs: Construction of the active/decoy dataset remains a major determinant of measured performance. J Cheminform 2016;8(1):1-7.
[Crossref] [Google Scholar] [PubMed]
- Liebeschuetz JW, Cole JC, Korb O. Pose prediction and virtual screening performance of GOLD scoring functions in a standardized test. J Comput Aided Mol Des 2012;26(6):737-48.
[Crossref] [Google Scholar] [PubMed]
- Shivanika C, Kumar D, Ragunathan V, Tiwari P, Sumitha A. Molecular docking, validation, dynamics simulations, and pharmacokinetic prediction of natural compounds against the SARS-CoV-2 main-protease. J Biomol Struct Dyn 2020:40(2):585-611.
[Crossref] [Google Scholar] [PubMed]
- Atanasova M, Yordanov N, Dimitrov I, Berkov S, Doytchinova I. Molecular docking study on galantamine derivatives as cholinesterase inhibitors. Mol Inform 2015;34(6‐7):394-403.
[Crossref] [Google Scholar] [PubMed]
- Lang PT, Brozell SR, Mukherjee S, Pettersen EF, Meng EC, Thomas V, et al. DOCK 6: Combining techniques to model RNA-small molecule complexes. RNA 2009;15(6):1219-30.
[Crossref] [Google Scholar] [PubMed]
- Lopes JP, Silva L, Ceschi MA, Lüdtke DS, Zimmer AR, Ruaro TC, et al. Synthesis of new lophine–carbohydrate hybrids as cholinesterase inhibitors: Cytotoxicity evaluation and molecular modeling. Medchemcomm. 2019;10(12):2089-101.
- Zhang L, Li D, Cao F, Xiao W, Zhao L, Ding G. Identification of human acetylcholinesterase inhibitors from the constituents of EGb761 by modeling docking and molecular dynamics simulations. Comb Chem High Throughput Screen 2018;21(1):41-9.
[Crossref] [Google Scholar] [PubMed]
- García-Sosa AT, Hetényi C, Maran U. Drug efficiency indices for improvement of molecular docking scoring functions. J Comput Chem 2010;31(1):174-84.
[Crossref ] [ Google Scholar] [PubMed]
- Li Y, Han L, Liu Z, Wang R. Comparative assessment of scoring functions on an updated benchmark: 2. Evaluation methods and general results. J Chem Inf Model 2014;54(6):1717-36.
[Crossref] [Google Scholar] [PubMed]
- Bourne Y, Kolb HC, Radić Z, Sharpless KB, Taylor P, Marchot P. Freeze-frame inhibitor captures acetylcholinesterase in a unique conformation. Proc Natl Acad Sci 2004;101(6):1449-54.
[Crossref] [Google Scholar] [PubMed]
- Ricci-Lopez J, Aguila SA, Gilson MK, Brizuela CA. Improving structure-based virtual screening with ensemble docking and machine learning. J Chem Inform Model 2021;61(11):5362-76.
[Crossref] [Google Scholar] [PubMed]
- Sgobba M, Caporuscio F, Anighoro A, Portioli C, Rastelli G. Application of a post-docking procedure based on MM-PBSA and MM-GBSA on single and multiple protein conformations. Eur J Med Chem 2012;58:431-40.
[Crossref] [Google Scholar] [PubMed]
- Wang E, Sun H, Wang J, Wang Z, Liu H, Zhang JZ, et al. End-point binding free energy calculation with MM/PBSA and MM/GBSA: strategies and applications in drug design. Chem Rev 2019;119(16):9478-508.
[Crossref] [Google Scholar] [PubMed]