- *Corresponding Author:
- Sk. Mobeen
Pharmaceutical Biotechnology Division, A. U. College of Pharmaceutical Sciences, Andhra University, Visakhapatnam-530 003, India
E-mail: mobeen.pharma@gmail.com
| Date of Submission | 12 April 2017 |
| Date of Revision | 26 February 2018 |
| Date of Acceptance | 25 August 2018 |
| Indian J Pharm Sci 2018;80(5):911-920 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Optimization of antibiotic production medium and fermentation conditions of Streptomyces parvulus strain sankarensis-A10 was studied using classical and statistical methods. After selection of the basal production medium, the effect of sea water and incubation time on antibiotic production was investigated by classical method. In the first step of optimization, factors that significantly affect antibiotic production such as starch, K2HPO4, FeSO4.7H2O, KNO3 and pH were selected based on Plackett-Burman design. In the second step, three significant factors, starch, KNO3, pH, with a positive effect were optimized with a 33 factorial Box-Behnken design and analysed using response surface analysis. The optimal values for maximum antibiotic production were as follows, starch- 20 g/l, pH- 8.0 and KNO3- 3 g/l. Under optimal conditions, the antibacterial activity was found to be 620 U/ml and 616.66 U/ml against Staphylococcus aureus and Escherichia coli, respectively, which indicated that the activity was 1.4 folds higher in contrast to antibacterial activity prior to optimization. TLC-based partial purification of crude extract and bioassay of the fractions revealed the presence of four active compounds with Rf values of 0.275, 0.2, 0.125 and 0.062. These could probably be novel bioactive compound(s) in the crude extract, which could prove to be a novel source of antibiotics.
Keywords
Antimicrobial activity, antibiotic production medium, Plackett-Burman design (PBD), response surface analysis, thin-layer chromatography
Essential or intentional stalling of novel drug discovery and development by the pharmaceutical industries due to economic and regulatory obstacles together with the worldwide escalation of emanation and spread of antimicrobial resistance (although a natural phenomenon) constitutes the greatest challenge to global public health today. Microbial natural products appear as the preponderant and promising source for developing future antibiotics [1]. Searching unexplored environments for novel actinomycetes has been the orientation of current antibiotic screening programs of natural product-based drug discovery. The marine environment in particular offers novel and chemically rich actinomycetes species [2]. Among actinomycetes, genus Streptomyces continues to be a rich source of novel bioactive compounds and many antibiotics remain unexplored. Most searches to discover novel compounds usually end up unfruitful as previously reported species and compounds repetitively get identified. In spite of taxonomic relatedness of the strains, nutritional requirements for growth and antibiotic production were strain-dependent [3] rather than species-specific, implying the possibility of different strains of the same species producing same compounds less frequently. Thus novelty of compounds among different strains is theoretically more possible. Secondary metabolomes unexplored gene clusters of ubiquitous species of actinomycetes can drive the discovery of novel lead molecules [2]. However, only a small fraction of secondary metabolomes have been investigated till date, owing to the scarcity of efficient procedures to connect genes to molecules [4], as many of the gene clusters encoded by Streptomycetes are not expressed under normal laboratory growth conditions. Minor changes in medium components pHexert a huge impact on general metabolic profile of microorganisms as well as on quality and quantity of their secondary metabolites production. Particularly in case of antibiotics, interaction between growth metabolism and product secretion is critically affected by growth-limiting nutrient concentrations constituting an important pathway for survival [5-7].
They use different metabolic pathways depending on the availability of different nutrient and fermentation conditions. The regulation of biosynthetic systems to maximally exploit the potential of the microorganisms through proper manipulation of fermentation conditions (temperature, pH, medium composition) might activate the key enzymes and/or instigate cryptic gene expressions provoking the silent or poorly expressed metabolic pathways that dominate antibiotic synthesis and can enhance the chances of discovery of novel lead molecules from their active fermentation extracts.
Secondary metabolite production differs with species and strains of microorganisms in virtue of medium components and their optimum levels, which are highly indispensable. Melioration of antibiotic production is imperative for achieving ample yield to evaluate its potentiality, novelty and ease the cost of production.
How to shorten the fermentation time of an actinomycete strain and increase the productivity of fermentation is still a challenge. However, studies on designing an appropriate production medium (PM) and fermentation conditions for the production of novel secondary metabolites from the ubiquitous species of actinomycetes are scanty.
Scientific and rational approaches of optimization of these conditions in biocontrol strain fermentation process greatly influence laboratory or industrial fermentation. Design-of-experiments (DOE) approach frequently deployed methods involving Plackett-Burman design (PBD) and response surface methodology (RSM), which lead the identification of key medium components and their optimal levels [8] were attempted in the present study to achieve improved antibiotic production with Streptomyces parvulus strain sankarensis-A10. Thin-layer chromatography (TLC)-directed partial purification and bioassay-based detection of active compounds present in the crude extract was also targeted.
Materials and Methods
The isolate S. parvulus strain sankarensis-A10 (GenBank-KT906299, Culture Deposit No: NCIM 5601) previously isolated, identified and reported from our laboratory [9] was used in the present study. All the chemicals/ingredients used in this study were of AR grade and purchased from HiMedia and Sigma- Aldrich.
Seed culture
Spore suspension of S. parvulus strain sankarensis-A10 from 8-10 d old well sporulated slant was prepared in a 5 ml of sterile distilled water and aseptically transferred into 250 ml Erlenmeyer flasks containing sterile starch casein nitrate medium (45 ml) [10] prepared in 50 % v/v sea water, cultured aerobically at 28°, 150 rpm for three days until the cells reached logarithmic phase, then the culture (10.1×107 CFU/ml) was used as inoculum in all the experiments.
Antibacterial assay
Antibacterial activity was assessed [11] against Escherichia coli (NCIM-2065) and Staphylococcus aureus (NCIM-2079) with inoculum size as per 0.5 McFarland standard (1×108 CFU/ml) using 50 μl of the clear supernatant of the fermented broth, incubated at 37° for 24 h and zones of inhibition were reported as U/ml as described by Maxwell et al. [12].
Statistical analysis
Design Expert® 9.0 software version 9 (Stat-Ease Inc., USA) for windows was used for the experimental designs, statistical analysis and to plot threedimensional (3D) surface plots.
Optimization of PM, selection of basal medium
S. parvulus strain sankarensis-A10 was cultured on six different basal media as given below to select the appropriate basal PM by comparative studies for the formulation, subsequent non-statistical and statistical optimization of PM. PM 1: yeast extract- 0.5 %, casein- 0.5 %, starch- 2.0 %, dextrose- 1.0 %, CaCO3- 0.4 %, 50 % v/v sea water- 100 ml, pH- 7.0 ± 0.2; PM 2: glucose- 1.0 %, peptone- 0.75 %, starch- 1.0 %, meat extract- 0.75 %, NaCl- 0.3 %, 50 % v/v sea water- 100 ml, pH- 7.0 ± 0.2; PM 3: tryptone- 0.5 %, meat extract- 0.3 %, dextrose- 1.0 %, yeast extract- 0.5 %, CaCO3- 0.2 %, starch- 2.5 %, 50 % v/v sea water- 100 ml, pH- 7.0 ± 0.2; PM 4: starch- 1.0 %, KNO3- 0.2 %, casein- 0.03 %, K2HPO4- 0.2 %, NaCl- 0.2 %, CaCO3- 0.002 %, MgSO4.7H2O- 0.005 %, FeSO4.7H2O- 0.001 %, peptone- 0.2 %, 50 % v/v sea water- 100 ml, pH- 7.0 ± 0.2; PM 5: malt extract- 1.0 %, yeast extract- 0.4 %, dextrose- 0.4 %, 50 % v/v sea water- 100 ml, pHexert 7.0 ± 0.2; PM 6: peptone- 0.5 %, glycerol- 2.0 %, malt extract- 0.3 %, yeast extract- 0.3 %, CaCO3- 0.25 %, 50 % v/v sea water- 100 ml, pH- 7.0 ± 0.2. A quantity of 5 ml of the seed culture was inoculated in all the media (45 ml) contained in a 250 ml flask and incubated for 8 d at 28 ± 2°, 150 rpm on the rotary shaker, then the culture free supernatant was assayed for antibacterial activity.
Effect of sea water and incubation period on antibiotic production
Effect of sea water and incubation period on antibiotic production profile was assessed by using different concentrations of sea water (50, 60, 70, 80, 90, and 100 %, v/v) in combination with PM 4 (45 ml) inoculated with 5 ml of seed culture and cultured aerobically in a 250 ml flask at 28 ± 2°, 150 rpm on an orbital shaker for 8 d. After 72 h and thereafter at 24 h intervals each flask was withdrawn, contents of the flask were centrifuged for 15 min at 8000 rpm, 4°; filtered and antibacterial activity of the clear supernatant was determined.
Selection of significant variables by PBD
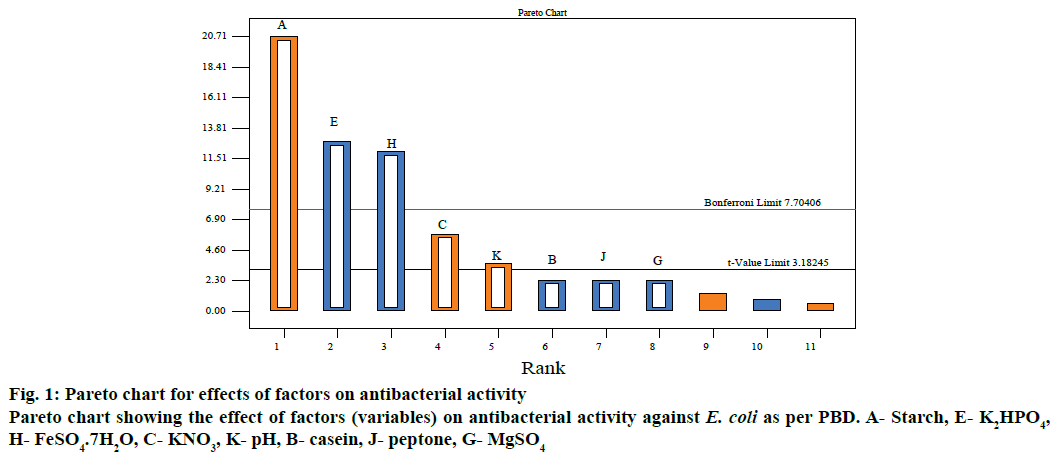
The PBD often called screening design [13] helps in selecting the most vital ingredients with significant main effect on bioactive metabolites production within a large candidate pool. Independent variables code and their selected levels were shown in Table 1. The design matrix of PBD for carrying out the experiments were shown with each column representing a single variable while each row representing one trial, either high (1) or low (-1), within each experimental trail. Antibacterial activity (U/ml) was considered as response value.
| Factor (g/l) | Code | Levels | |
|---|---|---|---|
| Low (-1) | High (1) | ||
| Starch | A | 5.0 | 15.0 |
| Casein | B | 0.15 | 0.45 |
| KNO3 | C | 1.0 | 3.0 |
| NaCl | D | 1.0 | 3.0 |
| K2HPO4 | E | 1.0 | 3.0 |
| CaCO3 | F | 0.0 | 0.2 |
| MgSO4.7H2O | G | 0.0 | 0.2 |
| FeSO4.7H2O | H | 0.0 | 0.2 |
| Peptone | J | 1.0 | 3.0 |
| pH | K | 6 | 8 |
| Level of inoculum (% v/v) | L | 5 | 15 |
Table 1: Independent Variables in the Plackett-Burman Experiment
Steepest ascent experiment
On the basis of PBD results, significant variables which are essential to effectively simulate the real situation using RSM were further optimized along the steepest ascent path that can determine a suitable direction to rapidly move towards the neighbourhood of the optimum response, by slowly increasing or decreasing the concentrations of variables until no further increase in response was observed.
RSM [13] is a powerful compilation of efficient mathematical and statistical techniques widely applied in designing and analysis of fermentation process optimization in a multi-factor system to find the optimum conditions resulting enhanced yield. Box- Behnken design (BBD), a more efficient, convenient and economical among the other RSM methods best suited for the study. The most significant positively affecting independent variables obtained for antibiotic production in the PBD studies were selected for further optimization by RSM. The coded values of the BBD experiments are illustrated in Table 2. A, B, C represented starch, KNO3, pH, respectively and the response value was represented by antibacterial activity (U/ml) and their levels were determined by steepest ascent experiment. Their optimum levels and interaction effects were further investigated by 33 factorial BBD. A total of 17 experimental trials were carried out, 5 trials were used as controls to estimate experimental error while 12 trials used different factors.
| Factors | Code | Coded level | ||
|---|---|---|---|---|
| -1 | 0 | +1 | ||
| Starch (g/l) | A | 15.0 | 17.5 | 20.0 |
| KNO3 (g/l) | B | 3.0 | 3.5 | 4.0 |
| pH | C | 8.0 | 8.5 | 9.0 |
Table 2: Coded Values of the BBD Experimental Variables
Experimental validation
S. parvulus strain sankarensis-A10 was cultured in non-optimized and optimized production media, which yielded the maximum response for experimentally validation. The clear supernatant of the fermented broths were assayed for antibacterial activity.
Extraction, purification and antibacterial activity of metabolites
The cell-free culture broth was extracted with ethyl acetate (1:1) twice, mycelium was washed twice with ethyl acetate followed by extraction with acetone once, and then samples were mixed, concentrated by rotary evaporation and the crude product was evaporated to dryness by pouring in to a Petri dish and kept inside a fume hood until dry. The crude extract was fractionated on TLC plate (silica gel 60 F254 aluminium sheets (Merck)) using chloroform and methanol (95:5) as mobile phase. Rf values of the spots were calculated using one TLC plate as a reference under UV transillumination. The partially purified individual fractions recovered by scratching the TLC plates were tested for antibacterial activity.
Results and Discussion
To improve the antibiotic production by actinomycetes, basal PM selection is a mandatory and significant step for formulation and further optimization of PM. Among the six different basal production media, S. parvulus strain sankarensis-A10 showed highest antibacterial activity (440 ± 0.00 and 440 ± 10.00 U/ml against E. coli and S. aureus, respectively) in PM 4 at 168 h and the results were shown in Table 3. Therefore, PM 4 was chosen for further optimization studies by classical and statistical methods.
| Media | 120 h | 144 h | 168 h | 192 h | ||||
|---|---|---|---|---|---|---|---|---|
| E. coli | S. aureus | E. coli | S. aureus | E. coli | S. aureus | E. coli | S. aureus | |
| PM 1 | 263.33 ± 5.771,2 | 280.00 ± 10.00 | 296.66 ± 11.54 | 300.00 ± 0.00 | 346.66 ± 5.77 | 360.00 ± 10.00 | 316.66 ± 5.77 | 323.33 ± 15.27 |
| PM 2 | - | - | 273.33 ± 5.77 | 286.66 ± 15.27 | 293.33 ± 11.54 | 300.00 ± 10.00 | 330.00 ± 0.00 | 336.66 ± 15.27 |
| PM 3 | 280.00 ± 0.00 | 276.66 ± 5.77 | 383.33 ± 15.27 | 376.66 ± 5.77 | 320.00 ± 0.00 | 310.00 ± 10.00 | 266.66 ± 11.54 | 260.00 ± 0.00 |
| PM 4 | 360.00 ± 0.00 | 350.00 ± 10.00 | 393.33 ± 5.77 | 386.66 ± 5.77 | 440.00 ± 0.00 | 440.00 ± 10.00 | 366.66 ± 5.77 | 380.00 ± 10.00 |
| PM 5 | - | - | 246.66 ± 11.54 | 256.66 ± 11.54 | 283.33 ± 15.27 | 300.00 ± 0.00 | 260.00 ± 10.00 | 280.00 ± 0.00 |
| PM 6 | - | - | - | - | 256.66 ± 5.77 | 243.33 ± 20.81 | 306.66 ± 15.27 | 303.33 ± 5.77 |
Table 3: Antibacterial Activity in Different Production Media
The highest antibacterial activity was obtained at 70 % v/v sea water concentration (Table 4), which indicates that the production of antibiotic compounds by S. parvulus strain sankarensis-A10 was sea waterdependent. The antibacterial activity of the culture filtrate started after 72 h of fermentation, reached its highest after 168 h of incubation, so the fermentation was terminated after 168 h. Hence, modified PM (PM 4 prepared in 70 % v/v sea water) was used for further optimization studies.
| Bacteria | (v/v) S.W. (%) | 72 h | 96 h | 120 h | 144 h | 168 h | 192 h |
|---|---|---|---|---|---|---|---|
| E. coli | 50 % | - | 266.66 ± 5.772 | 360.00 ± 0.00 | 393.33 ± 5.77 | 440.00 ± 0.00 | 366.66 ± 5.77 |
| 60 % | - | - | 236.66 ± 5.77 | 283.33 ± 11.54 | 243.33 ± 15.27 | 186.66 ± 5.77 | |
| 70 % | 313.33 ± 11.54 | 356.66 ± 5.77 | 416.66 ± 11.54 | 460.00 ± 10.00 | 483.33 ± 5.77 | 410.00 ± 10.00 | |
| 80 % | - | 263.33 ± 11.54 | 296.66 ± 5.77 | 360.00 ± 17.32 | 343.33 ± 5.77 | 316.66 ± 5.77 | |
| 90 % | - | 240.00 ± 10.00 | 306.66 ± 5.77 | 323.33 ± 11.54 | 360.00 ± 10.00 | 303.33 ± 11.54 | |
| 100% | - | - | 223.33 ± 5.77 | 286.66 ± 5.77 | 353.33 ± 11.54 | 326.66 ± 5.77 | |
| S. aureus | 50 % | - | 276.66 ± 11.54 | 350.00 ± 10.00 | 386.66 ± 5.77 | 440.00 ± 10.00 | 380.00 ± 10.00 |
| 60 % | - | - | 243.33 ± 5.77 | 296.66 ± 15.27 | 246.66 ± 5.77 | 203.33 ± 5.77 | |
| 70 % | 320.00 ± 10.00 | 366.66 ± 11.54 | 423.33 ± 11.54 | 463.33 ± 5.77 | 486.66 ± 15.27 | 416.66 ± 5.77 | |
| 80 % | - | 250.00 ± 10.00 | 283.33 ± 5.77 | 353.33 ± 5.77 | 333.33 ± 5.77 | 310.00 ± 0.00 | |
| 90 % | - | 226.66 ± 5.77 | 293.33 ± 5.77 | 330.00 ± 10.00 | 376.66 ± 5.77 | 306.66 ± 11.54 | |
| 100 % | - | - | 240.00 ± 0.00 | 303.33 ± 15.27 | 363.33 ± 5.77 | 336.66 ± 5.77 |
Table 4: Effect of Sea Water and Incubation Period on Antibacterial Activity
Levels selection and evaluation of experimental variables effects were summarized in Tables 5 and 6. As shown in Table 6, the confidence level of factors starch (A), KNO3 (C), K2HPO4 (E), FeSO4.7H2O (H) and pH (K) were shown to be above 95 % and were considered to be significant. The factors casein (B), MgSO4.7H2O (G) and peptone (J) had a confidence level below 95 % in antibiotic production hence were considered insignificant. The data revealed that the factors NaCl (D), CaCO3 (F) and level of inoculum (L) had the low confidence level with no obvious effects on antibiotic production, so they too were considered insignificant. Figure 1 illustrated the relation between t-value (effect) vs. ranks according to their significance with a horizontal reference line to indicate the statistical significance (t=3.18245). Any effect that extends past this reference line is considered to be significant or vice versa with respect to E. coli. The main effect of each variable on antibacterial metabolites production against E. coli and S. aureus, respectively are shown in Table 6. The significant variables (main effect) with a positive effect (A, C, K) were fixed at high level and the variables, which exerted a negative effect (E, H) on antibacterial antibiotic production were maintained at low level for further optimization by steepest ascent design and RSM-BBD. Similar results were reported by El-Naggar et al. [14].
| Coded levels of independent variables | Antibacterial activity (U/ml) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Run | A | B | C | D | E | F | G | H | J | K | L | E. coli | S. aureus |
| 1 | 1 | 1 | -1 | 1 | 1 | 1 | -1 | -1 | -1 | 1 | -1 | 466.66 ± 11.541 | 463.33 ± 5.771 |
| 2 | -1 | 1 | 1 | -1 | 1 | 1 | 1 | -1 | -1 | -1 | 1 | 363.33 ± 5.77 | 366.66 ± 11.54 |
| 3 | 1 | -1 | 1 | 1 | -1 | 1 | 1 | 1 | -1 | -1 | -1 | 480.00 ± 10.00 | 476.66 ± 15.27 |
| 4 | -1 | 1 | -1 | 1 | 1 | -1 | 1 | 1 | 1 | -1 | -1 | 280.00 ± 0.00 | 280.00 ± 10.00 |
| 5 | -1 | -1 | 1 | -1 | 1 | 1 | -1 | 1 | 1 | 1 | -1 | 340.00 ± 17.32 | 336.66 ± 15.27 |
| 6 | -1 | -1 | -1 | 1 | -1 | 1 | 1 | -1 | 1 | 1 | 1 | 416.66 ± 15.27 | 410.00 ± 17.32 |
| 7 | 1 | -1 | -1 | -1 | 1 | -1 | 1 | 1 | -1 | 1 | 1 | 400.00 ± 10.00 | 416.66 ± 11.54 |
| 8 | 1 | 1 | -1 | -1 | -1 | 1 | -1 | 1 | 1 | -1 | 1 | 433.33 ± 5.77 | 440.00 ± 10.00 |
| 9 | 1 | 1 | 1 | -1 | -1 | -1 | 1 | -1 | 1 | 1 | -1 | 520.00 ± 20.00 | 523.33 ± 15.27 |
| 10 | -1 | 1 | 1 | 1 | -1 | -1 | -1 | 1 | -1 | 1 | 1 | 396.66 ± 15.27 | 400.00 ± 20.00 |
| 11 | 1 | -1 | 1 | 1 | 1 | -1 | -1 | -1 | 1 | -1 | 1 | 470.00 ± 10.00 | 480.00 ± 10.00 |
| 12 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | 416.66 ± 11.54 | 406.66 ± 5.77 |
Table 5: Design Matrix and Results of Plackett-Burman Design
| Terms | E. coli | S. aureus | ||||||
|---|---|---|---|---|---|---|---|---|
| Coefficients | Effect | F-value | P-value (Prob.>F) |
Coefficients | Effect | F-value | P-value (Prob.>F) | |
| Model | 100.08 | 0.0015* | 173.16 | 0.0007* | ||||
| A- Starch | 46.39 | 92.78 | 429.07 | 0.0002* | 50.00 | 100.00 | 837.82 | <0.0001* |
| B- Casein | -5.28 | -10.56 | 5.55 | 0.0997 | -4.44 | -8.89 | 6.62 | 0.0823 |
| C- KNO3 | 13.06 | 26.11 | 33.99 | 0.0101* | 13.89 | 27.78 | 64.65 | 0.0040* |
| E- K2HPO4 | -28.61 | -57.22 | 163.22 | 0.0010* | -26.11 | -52.22 | 228.49 | 0.0006* |
| G-MgSO4.7H2O | -5.28 | -10.56 | 5.55 | 0.0997 | -4.44 | -8.89 | 6.62 | 0.0823 |
| H- FeSO4.7H2O | -26.94 | -53.89 | 144.76 | 0.0012* | -25.00 | -50.00 | 209.46 | 0.0007* |
| J- Peptone | -5.28 | -10.56 | 5.55 | 0.0997 | -5.00 | -10.00 | 8.38 | 0.0682 |
| K- pH | 8.06 | 16.11 | 12.94 | 0.0368* | 8.33 | 16.67 | 23.27 | 0.0170* |
| E. coli | S. aureus | E. coli | S. aureus | |||||
| SD | 7.76 | 5.98 | R-squared | 0.9963 | 0.9978 | |||
| Mean | 415.28 | 416.67 | Adj. R-squared | 0.9863 | 0.9921 | |||
| CV (%) | 1.87 | 1.44 | Pred. R-squared | 0.9403 | 0.9654 | |||
| PRESS | 2888.84 | 1718.74 | Adeq. precision | 36.632 | 47.599 | |||
Table 6: Anova of Experimental Variables Using Plackett-Burman Design
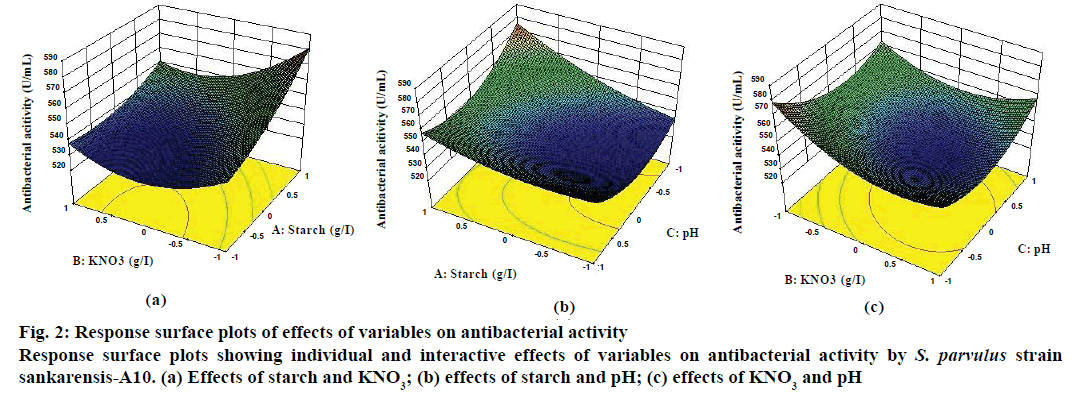
PBD results indicated starch, KNO3 and pH as significant positively affecting variables, thus increasing their concentrations should result in a higher antibiotic production. As per the steepest ascent path designed by considering the centre point of the PBD as the origin of the path shown in Table 7, optimal fermentation conditions should be around run 4 and was hence considered as a centre point in BBD. A quadratic polynomial Eqn. was established to identify the relationship between response and variables based on the experimental results of BBD (Table 8) and regression analysis (Table 9). The mathematical model of coded units can be expressed as: Y1 [yield (E. coli)]= 532.67+12.92 A–8.75 B–1.67 C–6.67 AB–7.50 AC+5.83 BC+6.17 A2+14.50 B2+15.33 C2; Y1 [yield (S. aureus)] = 536.67+15.00 A–5.42 B–1.25 C–5.83 AB–14.17 AC+6.67 BC+5.00 A2+14.17 B2+10.83 C2, where Y1 is the yield/response of antibacterial metabolites produced and A, B, C are starch (g/l), KNO3 (g/l) and pH, respectively. ANOVA summary was shown in Table 10. The F-values of 38.86, 42.89 for E. coli and S. aureus, respectively implied that the model was significant. The significance of model was also supported by statistically insignificant lack-of-fit, as was evident from lower calculated lackof- fit F-value of 0.34 (E. coli) and 0.39 (S. aureus). Values of prob>F less than 0.05 indicated significant model terms. A p-value of <0.0001 indicated that there was only a chance of 0.01 % that model’s large F-value could be due to noise. The mathematical model was found reliable with R2 values of 0.9804 and 0.9822. It is a clear proof that statistical experimental designs offer an efficient and feasible approach for optimization of PM. Response surface 3D plots illustrated the pairwise combination of the three factors, starch, KNO3 and pH, while the rest was held at the middle level (Figure 2). It is obvious from the plots that higher concentration of starch and lower concentration of KNO3, and pH favour higher antibacterial production. With the increase in the concentration of starch from 15 to 20 g/l (coded values, –1 to +1), the antibacterial activity gradually increased to a maximum at a low concentration of KNO3 (Figure 2a). However, the antibacterial activity significantly decreased when the concentration of KNO3 was increased to higher levels in the PM. When the concentration of pH was near to middle level (coded values, +1 to 0.0), the antibacterial activity increased with increasing concentration of starch from 15 to 20 g/l (coded values, –1 to +1). Further reduction in pH from 8.5 to 8, its low level (coded values, 0.0 to –1) resulted in significant increase in antibacterial activity (Figure 2b). Decreasing the concentration of KNO3 from 4 to 3 g/l (coded values, +1 to –1) resulted in a gradual increase in the antibacterial activity at lower pH (coded values, –0.5 to –1; Figure 2c). Thus it is evident that the medium composition significantly affected fermentation process.
| Run | Starch (g/l) A |
KNO3 (g/l) B |
pH C |
Antibacterial activity (U/ml) | |
|---|---|---|---|---|---|
| E. coli | S. aureus | ||||
| 1 | 10.0 | 2.0 | 7 | 480.00 ± 10.001 | 483.33 ± 15.271 |
| 2 | 12.5 | 2.5 | 7.5 | 503.33 ± 15.27 | 506.66 ± 11.54 |
| 3 | 15.0 | 3.0 | 8 | 526.66 ± 5.77 | 523.33 ± 5.77 |
| 4 | 17.5 | 3.5 | 8.5 | 540.00 ± 10.00 | 540.00 ± 0.00 |
| 5 | 20.0 | 4.0 | 9 | 440.00 ± 20.00 | 436.66 ± 5.77 |
Table 7: Design and Results of the Steepest Ascent Search Experiment
| Run | Code A |
Code B |
Code C |
Antibiotic production (U/ml)/response | |||
|---|---|---|---|---|---|---|---|
| E. coli | S. aureus | ||||||
| Observed | Predicted | Observed | Predicted | ||||
| 1 | -1 | -1 | 0 | 543.33 ± 5.771 | 542.50 | 540.00 ± 0.001 | 540.41 |
| 2 | 1 | -1 | 0 | 580.00 ± 0.00 | 581.68 | 580.00 ± 0.00 | 582.08 |
| 3 | -1 | 1 | 0 | 540.00 ± 0.00 | 538.33 | 543.33 ± 5.77 | 541.25 |
| 4 | 1 | 1 | 0 | 550.00 ± 10.00 | 550.83 | 560.00 ± 0.00 | 559.58 |
| 5 | -1 | 0 | -1 | 533.33 ± 11.54 | 535.41 | 523.33 ± 5.77 | 524.58 |
| 6 | 1 | 0 | -1 | 576.66 ± 5.77 | 576.25 | 583.33 ± 5.77 | 582.91 |
| 7 | -1 | 0 | 1 | 546.66 ± 11.54 | 547.08 | 550.00 ± 10.00 | 550.41 |
| 8 | 1 | 0 | 1 | 560.00 ± 0.00 | 557.91 | 553.33 ± 5.77 | 552.08 |
| 9 | 0 | -1 | -1 | 580.00 ± 0.00 | 578.75 | 576.66 ± 5.77 | 575.00 |
| 10 | 0 | 1 | -1 | 550.00 ± 10.00 | 549.58 | 550.00 ± 10.00 | 550.83 |
| 11 | 0 | -1 | 1 | 563.33 ± 5.77 | 563.75 | 560.00 ± 0.00 | 559.16 |
| 12 | 0 | 1 | 1 | 556.66 ± 15.27 | 557.91 | 560.00 ± 20.00 | 561.66 |
| 13 | 0 | 0 | 0 | 530.00 ± 10.00 | 532.66 | 536.66 ± 5.77 | 536.66 |
| 14 | 0 | 0 | 0 | 540.00 ± 0.00 | 532.66 | 536.66 ± 5.77 | 536.66 |
| 15 | 0 | 0 | 0 | 533.33 ± 11.54 | 532.66 | 540.00 ± 0.00 | 536.66 |
| 16 | 0 | 0 | 0 | 530.00 ± 10.00 | 532.66 | 530.00 ± 10.00 | 536.66 |
| 17 | 0 | 0 | 0 | 530.00 ± 10.00 | 532.66 | 540.00 ± 0.00 | 536.66 |
Table 8: Design and Results of BBD Experiments
| Model Terms | Coefficient estimate | Standard error | F- value | P-value (Prob>F) | ||||
|---|---|---|---|---|---|---|---|---|
| E. coli | S. aureus | E. coli | S. aureus | E. coli | S. aureus | E. coli | S. aureus | |
| A | 12.92 | 15.00 | 1.30 | 1.24 | 98.35 | 146.33 | <0.0001 | <0.0001 |
| B | -8.75 | -5.42 | 1.30 | 1.24 | 45.13 | 19.08 | 0.0003 | 0.0033 |
| C | -1.67 | -1.25 | 1.30 | 1.24 | 1.64 | 1.02 | 0.2414 | 0.3470 |
| AB | -6.67 | -5.83 | 1.84 | 1.75 | 13.10 | 11.06 | 0.0085 | 0.0127 |
| AC | -7.50 | -14.17 | 1.84 | 1.75 | 16.58 | 65.26 | 0.0047 | <0.0001 |
| BC | 5.83 | 6.67 | 1.84 | 1.75 | 10.03 | 14.45 | 0.0158 | 0.0067 |
| A2 | 6.17 | 5.00 | 1.80 | 1.71 | 11.80 | 8.56 | 0.0109 | 0.0222 |
| B2 | 14.50 | 14.17 | 1.80 | 1.71 | 65.23 | 68.70 | <0.0001 | <0.0001 |
| C2 | 15.33 | 10.83 | 1.80 | 1.71 | 72.94 | 40.17 | <0.0001 | 0.0004 |
Table 9: Regression Coefficients for the Fitted Quadratic Polynomial Model of BBD
| Source | Sum of Squares | Degree of freedom | Mean Square | F-value | Prob>F | Significance | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| E. coli | S. aureus | E. coli, S. aureus | E. coli | S. aureus | E. coli | S. aureus | E. coli | S. aureus | ||
| Model | 4746.80 | 4747.88 | 9 | 527.42 | 527.54 | 38.86 | 42.89 | < 0.0001 | Significant | |
| Residual | 95.00 | 86.11 | 7 | 13.57 | 12.30 | |||||
| Lack of Fit | 19.45 | 19.44 | 3 | 6.48 | 6.48 | 0.34 | 0.39 | 0.7969 0.7681 | Not-significant | |
| Pure Error | 75.56 | 66.67 | 4 | 18.89 | 16.67 | |||||
| Total | 4841.81 | 4833.99 | 16 | |||||||
| R2 | 0.9804 | 0.9822 | CV % | 0.67 | 0.64 | |||||
Table 10: Analysis of Variance (ANOVA) for the Selected Quadratic Model
Figure 2: Response surface plots of effects of variables on antibacterial activity
Response surface plots showing individual and interactive effects of variables on antibacterial activity by S. parvulus strain sankarensis-A10. (a) Effects of starch and KNO3; (b) effects of starch and pH; (c) effects of KNO3 and pH
In order to validate the model and regression equation, validation experiments were performed in triplicate with the optimal (predicted) medium. A quadratic model predicted that the maximum antibacterial production was 612.01 U/ml (E. coli), 615.01 U/ml (S. aureus), when optimum values of independent variables were at starch- 20.0 (g/l), KNO3- 3.0 (g/l) and pH- 8. The maximum antibacterial activity was found to be 616.66 ± 5.773 U/ml (E. coli), 620.00 ± 10.00 U/ml (S. aureus), suggesting that the predicted and experimental values were in good agreement. By means of optimizing the medium composition, the antibacterial activity was enhanced from 440 ± 0.00 U/ml (E. coli), 440 ± 10.00 U/ml (S. aureus) to 616.66 ± 5.773 U/ml (E. coli), 620.00 ± 10.00 U/ml (S. aureus).
The culture supernatant was extracted with each solvent separately (1:1) and both the aqueous and organic layers of all the solvents were tested for antibacterial activity. Ethyl acetate extract gave highest antibacterial activity when compared to hexane, butanol and chloroform extracts and hence ethyl acetate was used for the extraction process. The brown to dark reddishorange colored crude extract partial purification on TLC plate revealed many bands in UV transilluminator out of which four fractions with Rf values of 0.275, 0.2, 0.125 and 0.062 recovered from TLC plates exhibited antibacterial activity (Table 11).
| S. No. | Fractions | Rf value | Antibacterial activity (U/ml) | |
|---|---|---|---|---|
| E. coli | S. aureus | |||
| 1 | Compound 1 | 0.275 | 700.00 ± 0.001 | 733.33 ± 5.771 |
| 2 | Compound 2 | 0.200 | 114.66 ± 11.54 | 121.33 ± 5.77 |
| 3 | Compound 3 | 0.125 | 692.00 ± 20.00 | 720.66 ± 11.54 |
| 4 | Compound 4 | 0.062 | 366.00 ± 11.54 | 400.00 ± 10.00 |
Table 11: Antibacterial Activity of TLC Purified Fractions
The selection of basal PM (PM 4) among different production media, optimum incubation period (168 h) and sea water dependence (70 % v/v sea water) of antibiotic production were done by a series of traditional approach of optimization studies. Complex media are used rather than defined media for optimization studies since antibiotic producing organisms usually produce limited quality and quantities of their antibiotics in defined media [15]. Generally, Streptomycetes exhibit variability in growth and antibiotic production in complex media [16]. Like many other complex medium components, sea water has non-defined composition [17] that provides complex ionic sources, which make the medium suitable for marine microorganisms and also buffer the medium. It has been reported that 30 % sea water is optimum to produce maximum antimicrobial metabolites by S. rochei [18]. Significant positive variables, as well as their levels, picked up by PBD revealed that starch, KNO3 and pH are the important factors and assisted in moving towards the neighbourhood of the optimum response by steepest ascent path. RSM is a paramount statistical technique that disclosed optimum levels of significant individual variables and their interaction effects. BBD analysis of the quadratic regression model coefficients indicated that the pH had a significant effect showing maximum antibacterial production at pH 8.0, speculating the probability of degeneration or inactivation of the key enzymes that dominate antibiotic synthesis at high and/or low pH. This result was found to be in good agreement with the previous reports [19-21]. Starch proved to be the best for both cell growth as well as antibiotic production at 20.0 g/l as a sole carbon source. Similar results were reported by Jose et al., and Hamza et al. [22,23] specifying starch as a significant medium component for antibiotic production. The optimum level of KNO3 for antibacterial production was found to be 3.0 g/l. Ali et al. [19] reported that S. mesioensis produced maximum antibiotic with KNO3 at 2.5 g/l which is in near agreement with our results. It could be concluded that starch and KNO3 were the best carbon and nitrogen sources for maximum antibacterial production. After statistical optimization, the optimum levels of starch, pH, KNO3, 70 % v/v sea water for a period of 168 h of incubation under laboratory scale submerged fermentation process, 1.4 fold increase in antibacterial activity was obtained. The current study served as an excellent reminder that traditional and statistical approach of optimization of medium and fermentation conditions continue to be very effective in improving the yield of antibacterial metabolites by S. parvulus strain sankarensis-A10. TLC-based partial purification and bioassay of the recovered fractions revealed the presence of four active compounds, which gave rise to a probability of new bioactive compound(s) existing in the crude extract. A previous study reported also active compounds with relatively lower Rf values of 0.11 and 0.29 from S. parvulus ICN698 [24]. Genome sequence and antiSMASH analysis predicted 21 (chromosome) and 3 (plasmid) gene clusters related to secondary metabolites in S. parvulus 2297 [25] revealing its hidden potential for biosynthesizing surplus secondary metabolites more than the actual number of compounds reported from it. It is also reported that “one strain many compounds” approach represented an effective avenue for activating silent or poorly expressed genes and metabolic pathways [2]. Hence, it could be predicted that multiple compounds responsible for improved antibacterial activity (additive) during the course of optimization of fermentation process by PBD and RSM.
Temporal nature of antibiotic formation is certainly genetic, but their expression can be influenced greatly by environmental manipulation. The present work primarily focused on studying the potential of S. parvulus strain sankarensis-A10 through antibacterial activity, PM optimization, partial purification by TLC and bioassay-based detection of active compounds of the crude extract. Further studies on purification and structural elucidation of active compounds are currently in progress. It is expected that the current attempt to optimize antibiotic production from the ubiquitous species of actinomycetes, S. parvulus strain sankarensis-A10 from Visakhapatnam coastal marine ecosystem would help in the isolation and identification of novel antibiotics and/or could be a novel source of antibiotics previously reported. Furthermore, upon the structural characterization of these lead molecules, the present information could be considered fundamental and useful for subjecting the biosynthetic machinery of S. parvulus strain sankarensis-A10 to scale-up fermentation process for obtaining a single lead molecule on large scale and/or subjecting to combinatorial biosynthetic strategies for more than one antibiotic production.
Target-based high-throughput screening and optimization of PM for actinomycin D using traditional approach by various strains of S. parvulus have been reported till date. This is the first report on enhanced antibiotic production using S. parvulus strain sankarensis-A10 by optimization of medium and fermentation conditions employing traditional and DOE approaches, which ultimately lead to the production of more than one bioactive compounds (partially purified) with antibacterial activity revealing its hidden potential for biosynthesizing surplus secondary metabolites and could be a source for supply of natural products-based novel lead molecules to the pharmaceutical pipeline.
Acknowledgement
The authors would like to acknowledge Maulana Azad National Fellowship for Minority Students, Ministry of Minority Affairs, UGC, New Delhi, Government of India for granting the fellowship.
Conflict of interest
There is no conflict of interest among authors.
References
- Sharma D, Kaur T, Chadha BS, Manhas RK. Antimicrobial activity of actinomycetes against multidrug resistant Staphylococcus aureus, E. coli and various other pathogens. Trop J Pharm Res 2011;10(6):801-8.
- Abdelmohsen UR, Grkovic T, Balasubramanian S, Kamel MS, Quinn RJ, Hentschel U. Elicitation of secondary metabolism in actinomycetes. Biotechnol Adv 2015;33(6):798-811.
- Genilloud O, González I, Salazar O, Martín J, Tormo JR, Vicente F. Current approaches to exploit actinomycetes as a source of novel natural products. J Ind Microbiol Biotechnol 2011;38(3):375-89.
- Yamanaka K, Reynolds KA, Kersten RD, Ryan KS, Gonzalez DJ, Nizet V, et al. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc Natl Acad Sci USA 2014;111(5):1957-62.
- Seipke RF. Strain-level diversity of secondary metabolism in Streptomyces albus. PLoS One 2015;10(1):1-14.
- Rajeswari P, Jose PA, Amiya R, Jebakumar SRD. Characterization of saltern based Streptomyces sp. and statistical media optimization for its improved antibacterial activity. Front Microbiol 2015;10(1):e0116457.
- Sircar A, Sridhar P, Das PK. Optimization of solid state medium for the production of clavulanic acid by Streptomyces clavuligerus. Process Biochem 1998;33(3):283-9.
- Kim HU, Charusanti P, Lee SY, Weber T. Metabolic engineering with systems biology tools to optimize production of prokaryotic secondary metabolites. Nat Prod Rep 2016;33:933-41.
- Shaik M, Sankar GG, Iswarya M, Rajitha P. Isolation and characterization of bioactive metabolites producing marine Streptomyces parvulus strain sankarensis-A10. J Genet Eng Biotechnol 2017;15(1):87-94.
- Sharma D, Manhas RK. Application of Plackett – Burman experimental design and Box and Wilson design to improve broad-spectrum antimicrobial compound. Int J Biotechnol 2013;12(3):386-94.
- Balouiri M, Sadiki M, Ibnsouda SK. Methods for in vitro evaluating antimicrobial activity: A review. J Pharm Anal 2016;6(2):71-9.
- Maxwell PW, Chen G, Webster JM, Al MET. Stability and activities of antibiotics produced during infection of the insect Galleria mellonella by two isolates of Xenorhabdus nematophilus. Appl Environ Microbiol 1994;60(2):715-21.
- Latha S, Sivaranjani G, Dhanasekaran D. Response surface methodology: A non-conventional statistical tool to maximize the throughput of Streptomyces species biomass and their bioactive metabolites. Crit Rev Microbiol 2017;43(5):567-82.
- El-Naggar NEA, Ragaa AH. Antimicrobial potentialities of Streptomyces lienomycini NEAE-31 against human pathogen multidrug-resistant Pseudomonas aeruginosa. Int J Pharmacol 2016;12(8):769-88.
- Bundale S, Begde D, Nashikkar N, Kadam T, Upadhyay A. Optimization of culture conditions for production of bioactive metabolites by Streptomyces spp. isolated from soil. Adv Microbiol 2015;5(5):441-51.
- Dekleva ML, Titus JA, Strohl WR. Nutrient effects on anthracycline production by Streptomyces peucetius in a defined medium. Can J Microbiol 1985;31(3):287-94.
- Tsueng G, Teisan S, Lam KS. Defined salt formulations for the growth of Salinispora tropica strain NPS21184 and the production of salinosporamide A (NPI-0052) and related analogues. Appl Microbiol Biotechnol 2008;78(5):827-32.
- Reddy NG, Ramakrishna DPN, Rajagopal SV. Optimization of culture conditions of Streptomyces rochei (MTCC 10109) for the production of antimicrobial metabolites. Egypt J Biol 2011;13(1):21-9.
- Ali AM, El Gammal AA, Rifaat HM. Influence of nutrition and culturing conditions on antimicrobial metabolite production by Streptomyces mesioensis isolated from Egyptian soil. Int J ChemTech Res 2014;6(4):2552-7.
- Singh LS, Mazumder S, Bora TC. Optimisation of process parameters for growth and bioactive metabolite produced by a salt-tolerant and alkaliphilic actinomycete, Streptomyces tanashiensis strain A2D. J Med Mycol 2009;19(4):225-33.
- Wang YH, Feng JT, Zhang Q, Zhang X. Optimization of fermentation condition for antibiotic production by Xenorhabdus nematophila with response surface methodology. J Appl Microbiol 2008;104(3):735-44.
- Arul Jose P, Sivakala KK, Jebakumar SRD. Formulation and statistical optimization of culture medium for improved production of antimicrobial compound by Streptomyces sp. JAJ06. Int J Microbiol 2013;2013:526260.
- Hamza AA, Ali HA, Clark BR, Murphy CD, Elobaid EA. Isolation and characterisation of actinomycin D producing Streptomyces spp. from Sudanese soil Afr J Biotechnol 2013;12(19):2624-32.
- Subakaran M, Joshua SA, Jansi M, Vincent SGP. Isolation and Identification of antibacterial compound producing Streptomyces parvulus ICN698 from a Wetland Ecosystem of Kanyakumari. Indian J Pharm Pharmacol 2015;2(4):217.
- Nishizawa T, Miura T, Harada C, Guo Y, Narisawa K, Ohta H, et al. Complete genome sequence of Streptomyces parvulus 2297, integrating site-specifically with actinophage R4. Genome Announc 2016;4(4):e00875-16.