- *Corresponding Author:
- R. K. Manikkam
Department of Biotechnology, Centre for Drug Discovery and Development, Sathyabama Institute of Science and Technology, Chennai, Tamil Nadu 600119
E-mail: mrkactinos@gmail.com
| Date of Received | 24 January 2021 |
| Date of Revision | 08 April 2022 |
| Date of Acceptance | 05 January 2023 |
| Indian J Pharm Sci 2023;85(1):23-30 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Facile synthesis of silver nanoparticles was attempted using soil derived Streptomyces sp. KBR3 and its antimicrobial, antioxidant and antiproliferiative properties were evaluated in vitro. The extracellular synthesis of silver nanoparticles by the biomass of Streptomyces sp. KBR3 was confirmed through visual color change and ultraviolet-visible spectral analysis. Results of transmission electron microscopy, selected area electron diffraction and X-ray diffraction analysis showed the crystalline polydispersed spherical shaped nanoparticles with the size of 36 nm. Fourier-transform infrared spectroscopy analysis suggested the role of water soluble polyols present in the Streptomyces sp. KBR3 in mediating the synthesis of silver nanoparticles. The synthesized silver nanoparticles showed significant antibacterial activity against Staphylococcus aureus and Escherichia coli. In antioxidant studies, the silver nanoparticles showed maximum scavenging effects of 84 %±1.22 %, 95.8 %±1.32 % and 57.85 %±0.54 % at 1000 μg/ml in 2,2-diphenyl-1-picrylhydrazyl, 2,2'-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) and metal chelating assay with the half-maximal inhibitory concentration value of 40 μg/ml, 15 μg/ml and 53 μg/ml, respectively. In antiproliferiative study on H357 oral cancer cell line, the silver nanoparticles showed 77.21 %±0.43 % reduction in cell viability at 250 μg/ml with the half-maximal inhibitory concentration values of 9.431 μg/ml. The present study showed that the silver nanoparticles synthesized using the Streptomyces sp. KBR3 might be a promising material for different biomedical applications.
Keywords
Streptomyces, silver nanoparticles, antimicrobial, anti-oxidant, antiproliferiative activity
Threats due to microbial pathogens have been tremendously increasing now-a-days due to the emergence of antibiotic-resistant strains and declined immune compatibility of humans. Hence many pharmaceutical companies have lost their interest in developing new antibiotic compounds due to their narrow profit margin. Despite numerous novel antibiotics and therapeutic agents available in the market, 70 % of them are inactive in treating intracellular infections because of their reduced permeability[1]. In parallel, developing new or better antibiotics and non-antibiotic substances will have a great impact on global public health. To overcome these issues, there is a dire need to develop new class of therapeutic agents with better biocompatibility and efficiency[2].
Nanoparticles are now considered as viable antimicrobial agents and seem to have tremendous potential to solve the challenges of microbial multidrug resistance[3,4]. Nanotechnology provides a platform of excellence to modify and develop the properties of metals by converting them into their nanoform (nanoparticles), that has applications in the treatment of different diseases[5]. Natural biological sources, like Streptomyces, may be used to synthesize nanoparticles with various biological activities. Nanotechnology involves the synthesis of nanoparticles (i.e., materials not exceeding 100 nm in diameter) through physical, chemical and biological approaches and the integration of the resulting nanostructures into various applications. Nanomaterial’s varying by size, distribution and particle morphology. Among them, Silver Nanoparticles (AgNPs) are the most commonly used, given their range of biological activities and good chemical stability[6,7].
Actinobacteria are gram-positive bacteria with high Guanine+Cytosine Deoxyribonucleic Acid (G+C DNA) content in their genome and it remains as one of the largest phylum in the bacterial domain[8,9]. Members of this phylum are capable of producing diffusible pigments which are considered important sources among the microorganisms for the development of secondary metabolites of industrial significance having specific applications[10,11]. Reports claim that more than 75 % of commonly available antibiotics were recovered from the actinobacteria group of microorganisms[12]. In this study, AgNPs were synthesized using the biomass of Streptomyces sp. KBR3 and further evaluated for antimicrobial, antioxidant and antiproliferiative properties.
Materials and Methods
Description and identification of actinobacterial strain KBR3:
The actinobacterial strain KBR3 used in this study was isolated from the rhizosphere soil collected from Udhagamandalam (Lat. 11º 48’N; Long. 76º 77’E), Nilgiris, Tamil Nadu, India by spread plate method using starch casein nitrate agar. Viability of the strain KBR3 was maintained in Yeast Extract Malt Extract (YEME) agar slants as well as in 20 % glycerol broth at 4° and -80°, respectively.
The presence of aerial and substrate mycelium was observed under bright field microscope. Cultural properties such as growth, colony consistency, colour of aerial mycelium, production of reverse side and soluble pigment were studied by growing the strain KBR3 on YEME agar medium for 7-14 d at 28°[13]. Total genomic DNA from the strain KBR3 was isolated using solute ready genomic DNA kit (Himedia-Hi-PurATM) according to the manufacturer’s protocol. Further, 16S ribosomal RNA (16S rRNA) gene of the strain was amplified using primers: 27F- 5´AGAGTTTGATCMTGGCTCAG3´ (forward)[14] and 1492R- 5´TACGGYTACCTTGTTACGACTT3´ (reverse)[15] and then sequenced at Eurofins Genomics, Bangalore, India. The search program Basic Local Alignment Search Tool (BLAST) was used to screen for close relatives and phylogenetic affiliations. MEGA 7 program was used to align the 16S rRNA gene sequence of strain KBR3 with close relatives and to construct the phylogenetic tree by following neighbour-joining algorithm[16]. The partial 16S rRNA nucleotide sequence of strain KBR3 was deposited to GenBank.
Biosynthesis of AgNPs:
Strain KBR3 was grown in conical flask containing sterile 25 ml YEME broth as seed culture and incubated at 28° for 48 h in a rotary shaker at 200 rpm. Later, 10 % of the inoculum was transferred into 100 ml of sterile YEME broth in 500 ml Erlenmeyer flask at 28° for 72 h in rotary shaker with 200 rpm. The mycelia biomass of the strain KBR3 was collected as pellet after washing twice with distilled water at 10 000 rpm by centrifugation. About 50 ml of Silver nitrate (AgNO3) solution (1 mm) was added to the cell pellet and incubated at 28° in a rotary shaker at 200 rpm for 48 h in dark. The reaction flask was observed intermittently for any color change. Subsequently the reaction solution was centrifuged at 10 000 rpm for 10 min and pellet was dried in an hot air oven at 100°[17].
Characterisation of synthesized AgNPs:
The optical absorption of synthesized AgNPs was studied using Ultraviolet-Visible (UV-Vis) (UV-36000, Shimadzu, Japan) and Fourier-Transform Infrared Spectroscopy (FTIR) (FTIR; IR Affinity-1, Shimadzu Corp, Tokyo, Japan) spectral analysis. UV-Vis spectroscopy analysis was carried out over wavelengths from 200 to 800 nm at a resolution of 1 nm[18]. A FTIR spectrum was recorded using Potassium bromide (KBr) pellets in the range of 500-4000 cm-1[19]. X-Ray Diffraction (XRD) analysis was performed to confirm the crystalline nature of green synthesized AgNPs. The XRD data were obtained using X'Pert Pro X-ray diffractometry (PAN analytical BV) operating at 40 kV voltage and 30 mA current with Cu K alpha radiation[18]. Morphology, size and electron diffraction pattern were examined by Scanning Electron Microscope (SEM) (SEM-JSM-7600F, Japan) and Transmission Electron Microscope ((TEM)-JEM-2100F, Japan) at a voltage of 200 kV, respectively[20]. Selected Area Electron Diffraction (SAED) patterns used graphical method to quantify distances and angles in digitalized patterns by clicking on the two shortest non-collinear vectors (spots) using user calibration data[18]. Zeta potential analysis was also performed using Malvern Zeta Sizer to evaluate the surface load of nanoparticles[21].
Evaluation of biological activities of synthesized AgNPs:
Antibacterial activity: The antibacterial activity of biosynthesized AgNPs was tested using agar well diffusion bioassay against Staphylococcus aureus (S. aureus) ATCC 29213 and Escherichia coli (E. coli) ATCC 25922. 24 h old bacterial culture was spread uniformly over Nutrient Agar (NA) plates using sterile cotton swab. 10 mm diameter wells were made on bacteria swabbed NA plates using sterile cork borer. 200 µl of AgNPs (1 mg/ml) was added to each well. After 24 h of incubation at 37°, the zone of inhibition was measured and expressed as diameter in millimetre[22].
Antioxidant activity of AgNPs:
2,2-Diphenyl-1-Picrylhydrazyl (DPPH) radical scavenging activity: A solution of DPPH (0.1 mM) was prepared using methanol and covered with aluminium foil. In five different test tubes, about 10, 50, 100, 500 and 1000 µg/ml concentration of AgNPs (2.5 ml) was taken and 500 μl of DPPH solution was added into all the tubes. The reaction mixture was vigorously shaken and permitted to stand for 30 min at room temperature[23]. After incubation, the absorbance was measured at 517 nm using spectrophotometer. Ascorbic acid was used as a standard. The Percent DPPH scavenging effect was calculated using the following equation:
% inhibition=(A0-A1/A0)×100 (1)
Where, A0 was the absorbance of control reaction and A1 was the absorbance in the presence of test or standard sample.
2, 2'-Azino-Bis (3-Ethylbenzothiazoline-6-Sulfonic Acid) (ABTS) assay: The ABTS assay of AgNPs was examined by the method of Re et al.[24]. ABTS solution was prepared with 7 mm ABTS aqueous solution and 2.4 mm of potassium per sulfate and the reaction was kept in dark for 12-16 h at room temperature. This solution was previously diluted in ethanol (around 1:89 v/v) and maintained at 30° to provide a 0.700±0.02 absorption at 734 nm. Briefly, 20 µl of AgNPs with different concentration (10, 50, 100, 500 and 1000 µg/ml) was taken in five different test tubes and 2 ml of ABTS solution was added to all the reaction tubes. The tubes were then incubated for 30 min in dark and the absorbance was taken at 734 nm. The above mentioned equation (1) was used to interpret the results.
Metal chelating assay: AgNPs in different concentrations i.e. 10, 50, 100, 500 and 1000 µg/ml were added to a solution of 0.1 mm Ferrous sulphate (FeSO4) (0.2 ml). The reaction was initiated by the addition of 0.25 mm ferrozine (0.4 ml) and the mixture was vigorously shaken and left for 10 min at room temperature. Absorbance was taken at 562 nm. Ethylenediaminetetraacetic acid was used as standard solution[25]. The percentage of metal chelation was interpreted by the above stated equation (1).
Antiproliferiative activity of AgNPs:
Antiproliferiative activity of the biosynthesised nanoparticles was assessed by 3-(4, 5-Dimethylthiazolyl-2)-2, 5-Diphenyltetrazolium Bromide (MTT) assay on normal Vero cells and oral cancer (H357) cell line. The cells were maintained in minimal essential medium supplemented with 10 % fetal bovine serum, penicillin (100 U/ml) and streptomycin (100 µg/ml) in 5 % Carbon dioxide (CO2) at 37°. The cell lines were seeded at 5000 cells/well in 96-well plate and incubated for 48 h. Various concentrations of AgNPs i.e. 1.56, 3.12, 6.25, 12.50, 25, 50, 100 and 250 µg/ml were added to the 96-well plate and incubated for 24 h at 37°. Later, the medium was removed and washed with phosphate saline solution. The cells were further incubated with 50 μl of MTT solution (5 mg/ml) at 37° for 4 h. The medium with MTT solution was then flicked off and the formed formazan crystals were dissolved in 100 µl of dimethyl sulfoxide, then the absorbance was measured at 570 nm using a micro plate reader and images were taken under fluorescence microscope. The cell inhibition percentage was determined using the formula:
Percentage inhibition=100-[(T-T0/(C-T0)]×100 (2)
Where, T is the optical density of test sample, T0 is the optical density at time zero and C is the optical density of control.
Results and Discussion
The actinobacterial strain KBR3 produced rough, powdery colonies with creamy yellow colored aerial mycelium and diffusible yellow pigment production on YEME agar plates. Under bright field microscopic observation, strain KBR3 showed presence of both aerial and substrate mycelium without any fragmentation. However, these morphological features alone are inadequate for the generic identification of strain KBR3. The further amplification of 16S rRNA gene yielded a sequence of 1477 bp in size and the same was deposited in GenBank database with the accession number MT577838.
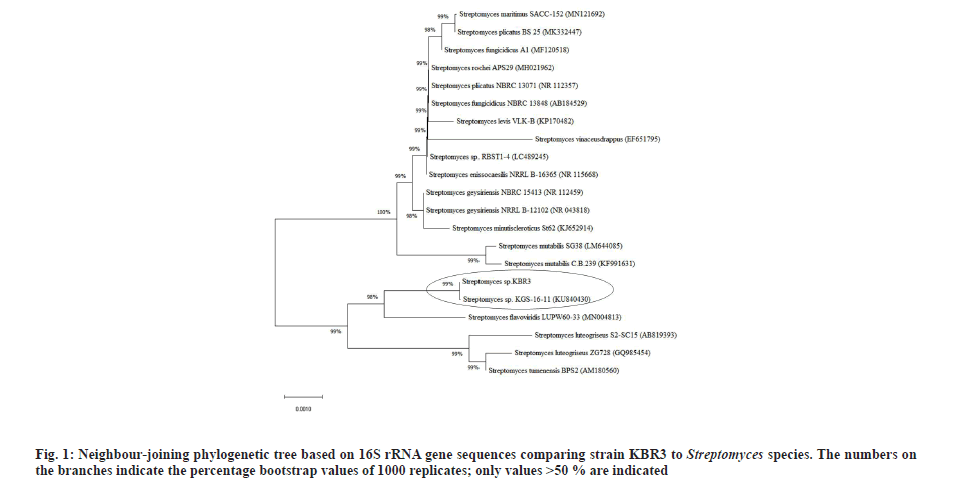
BLASTN analysis showed that the strain KBR3 shared more than 99 % similarity of 16S rRNA sequence identity with Streptomyces maritimus and Streptomyces rochei. Phylogenetic tree was constructed using the 16S rRNA gene sequences of these closely related Streptomyces sp. (fig. 1). Till now there are no reports on nanoparticle synthesis using Streptomyces maritimus or Streptomyces rochei. Hence the Streptomyces sp. KBR3 reported in this study would be a newly added source for the synthesis of AgNPs.
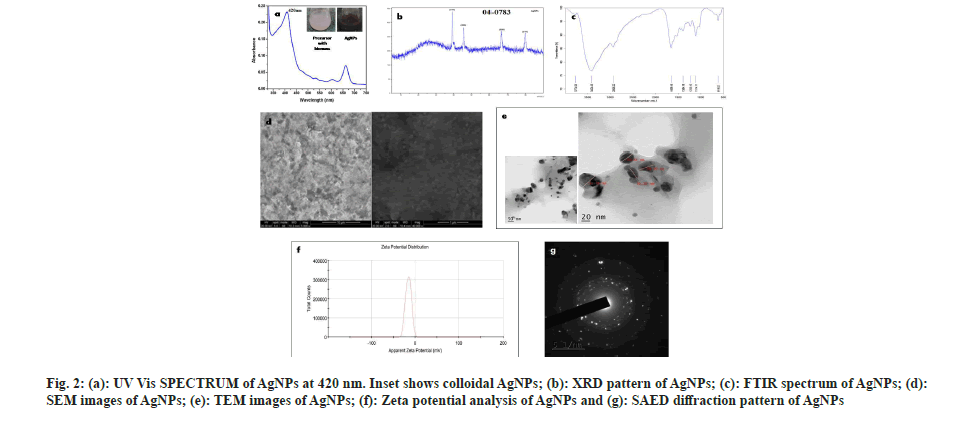
The brown color appeared on the reaction mixture clearly indicated the formation of AgNPs[26]. This color shift is due to the reduction of Ag+ ions and the formation of Surface Plasmon Resonance (SPR) in the reaction mixture, whereas no color change was observed in cell pellet without silver nitrate. This observation correlated with the previous reports by Narasimha et al.[27] and Zarina et al.[28]. The biosynthesis of AgNPs was confirmed by observing the colour change from light blue to dark brown (fig. 2a).
Every element has free electrons, giving rise to the peak of SPR. It is well known that the SPR peak is dependent on the size and shape of the nanoparticles formed. The peak appeared at 420 nm (fig. 2a) was correlated with others findings that indicate the formation of AgNPs[29-31]. Hence, our result from UV spectrophotometer clearly shows that the λmax of biosynthesized AgNPs occurs at 420 nm[7].
The regular XRD pattern of the biosynthesized AgNPs was shown in fig. 2b. The XRD peaks were seen located at 2θ=38.1, 44.6, 64.6 and 77.5 and are assigned to (111), (200), (220), (311) and (222) plane orientation of AgNPs. From the above result, it is clear that the most dominant orientation of Streptomyces synthesized silver nano particles must be crystal plane (111). This outcome demonstrates that the crystallinity of AgNPs was upgraded by adequate nucleation vitality from warm treatment. XRD pattern which shows a similar pattern, observed by Skladanowski et al.[32].
The size, shape and dispersion of biosynthesized AgNPs were confirmed by SEM. It was clear from SEM analysis that the AgNPs were present in nano form. The white spherical shaped bodies represent the AgNPs which are highly dispersed and small sized (fig. 2d) shows that each element is uniformly distributed throughout the material which correlate with Mohanta et al.[33]. Abd-Elnaby et al.[34] reported that Streptomyces synthesized AgNPs show the size of nanoparticles between 22 and 85 nm. Similarly, others also found that the extracellular biosynthesized AgNPs to be spherical and ranging in the size from 10 to 100 nm[35-37].
TEM images of AgNPs that were scanned up to 10 nm are shown in fig. 2e, which indicates the formation of a uniformly distributed polydispersed spherical structures with 36.5 nm in diameter. The shape and size of synthesized AgNPs were mostly spherical which correlates with Sk?adanowski et al.[32] and Zhou et al.[38].
The Face-Centered Cubic (FCC) crystalline nature of Streptomyces cell pellet associated AgNPs was also confirmed by SAED (fig. 2g). Three well resolved circular rings were indicated by SAED pattern which confirms the (111), (200) and (220) Braggs' reflection planes of XRD pattern. Besides, the circular fringes also indicated the highly crystalline structure of AgNPs and FCC was also confirmed by the report of Zhou et al.[38].
The FTIR analysis was used to determine the possible functional groups present in the biosynthesized AgNPs. FTIR spectra show major peaks at 618.52, 1114.11, 1235.15, 1394.18, 1658.16, 2936.22, 3424.43 cm-1 which are compared to the bond angles extending of C-S linkage, C-O-C group, alkyl ketone, alkane (C-H) group, aromatics, alkanes (C-H) group, O-H stretching. Similar results were reported by Zarina et al.[39], Singh et al.[40], Prakasham et al.[41] and Lakhsmi et al.[42]. The present study showed FTIR spectroscopy peaks at 1658.16, 2936.22 and 3424.43 cm-1 which might be the reason in the reduction of silver ions and stabilization of AgNPs (fig. 2c) which also correlates with Sholkamy et al.[7].
The stability of biosynthesized AgNPs was determined by Zeta potential analysis. It is mostly used for dispersion stability of nanoparticles. The Zeta potential value of the calculated average was noted as -15.9 mV indicating the stability of synthesized nanoparticles (fig. 2f). This shows a negative value, meaning that nanoparticles are capped with negatively charged constituents that make nanoparticles repulsive and therefore increase stability[43]. This electrostatic repulsion among the negatively charged AgNPs prevents aggregation, which are responsible for their constant stability. Similarly, other researchers also found negative zeta potential values of synthesized nanoparticles of -8.5 mV and -5.08 mV, respectively[44,45]. Our findings more closely correlate with Mohanta et al.[46] where the zeta potential of AgNPs synthesized by Streptomyces sp. was found to be -17.7 mV.
In antimicrobial assay, the biosynthesized AgNPs showed 23 mm and 20 mm zone of inhibition against S. aureus ATCC 29213 and E. coli ATCC 25922,respectively. Similarly, other reports also proved that AgNPs synthesized by Streptomyces sp. was highly susceptible to Gram-positive bacteria and showed moderate activity against Gram-negative bacteria[47]. Moreover, Vennila et al.[48] reported that AgNPs synthesized Streptomyces observed highest inhibition against S. aureus, Klebsiella pneumoniae, Salmonella typhi, Streptococcus pyogenes, Pseudomonas aeruginosa and Candida albicans.
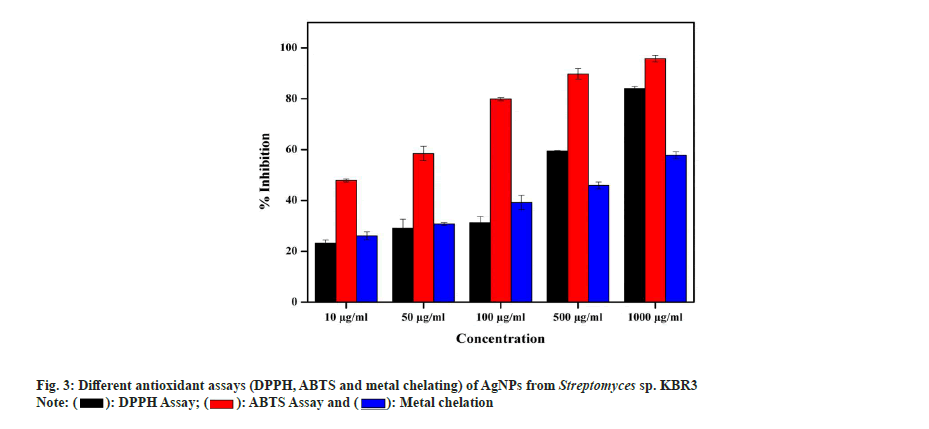
In DPPH radical scavenging assay, AgNP showed a maximum of 84 %±1.22 % and 59.52 %±0.45 % scavenging activity at 1000 µg/ml and 500 µg/ml concentrations, respectively, with a Half-Maximal Inhibitory Concentration (IC50) value of 40 µg/ml. Similarly, Kumar et al.[49] also reported that the AgNPs exhibited increased radical scavenging activity with respect to increase in concentration. In ABTS assay, the activity of AgNPs is in concentration dependent manner and the maximum radical scavenging activity is 95.8 %±1.32 % inhibition at 1000 µg/ml whereas 58.5 %±0.78 % observed in 500 µg/ml with IC50 value of 15 µg/ml. In the metal chelating assay, AgNPs showed a maximum of 57.85 %±0.54 % at 1000 µg/ml and 45.93 %±2.11 % inhibition at 500 µg/ml with an IC50 value of 53 µg/ml (fig. 3). Also, Shanmugasundam et al.,[17] reported that actinobacteria strain used to synthesize AgNPs showed 68.9 % chelating power at 500 µl (1:1 w/v) concentration.
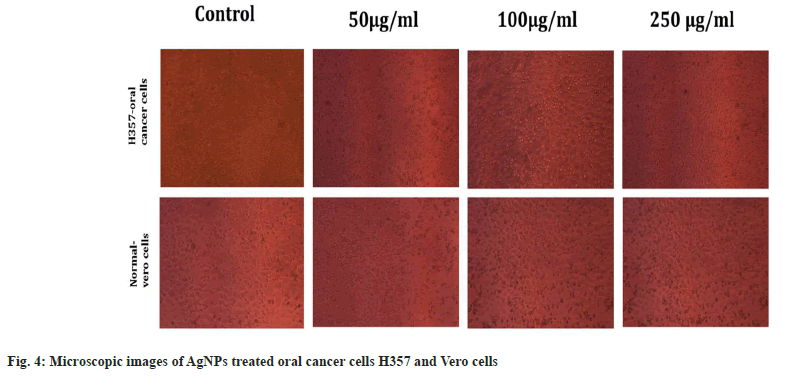
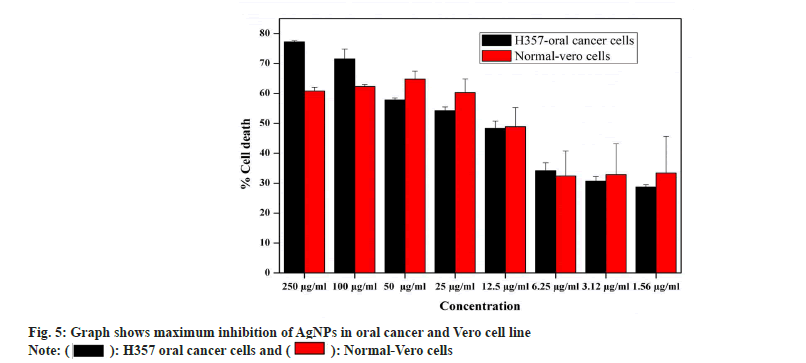
Antiproliferiative activity of biosynthesized AgNPs against the normal Vero cell line and H357 oral cancer cell line was given in fig. 4. The biosynthesized AgNPs by Streptomyces sp. KBR3 was accessed for cytotoxicity (MTT assay) against H357-oral cancer and Vero cell lines (fig. 4). The results revealed that the synthesized AgNPs had maximum inhibition of 77.21 %±0.43 % at 250 μg/ml concentration whereas 54.22 %±1.28 % inhibition was observed in 25 μg/ml with the IC50 values of 9.431 μg/ml (fig. 5). The results from this % study indicated that the biosynthesized AgNPs have anticancer activity in desirable concentration and minimal toxicity to non-cancerous cells. Similar to this result, Subbaiya et al.[50] reported that AgNPs were highly cytotoxic to the MCF-7 breast cancer cell lines at very low concentrations and IC50 dosage differ with time and dose-dependent manner.
Facile synthesis of AgNPs using Streptomyces is a simple, inexpensive and efficient method for obtaining nanoparticles in a non-toxic and eco-friendly approach. Interestingly, the biosynthesis of AgNPs by Streptomyces sp. KBR3 documented a broad range of promising antimicrobial, antioxidant and antiproliferiative activity. Therefore, the promising Streptomyces strain KBR3 could be an ideal model for the development of nano-based medicine for the treatment of various diseases in future.
Acknowledgements
Authors thank the management of Sathyabama Institute of Science and Technology, Chennai for the research facilities provided.
Conflict of interests:
The authors declared no conflict of interests.
References
- Sánchez-López E, Gomes D, Esteruelas G, Bonilla L, Lopez-Machado AL, Galindo R, et al. Metal-based nanoparticles as antimicrobial agents: An overview. Nanomaterials 2020;10(2):292.
[Crossref] [Google Scholar] [PubMed]
- Wypij M, Czarnecka J, ?wiecimska M, Dahm H, Rai M, Golinska P. Synthesis, characterization and evaluation of antimicrobial and cytotoxic activities of biogenic silver nanoparticles synthesized from Streptomyces xinghaiensis OF1 strain. World J Microbiol Biotechnol 2018;34(2):23.
[Crossref] [Google Scholar] [PubMed]
- Rai M, Yadav A, Gade A. Silver nanoparticles as a new generation of antimicrobials. Biotechnol Adv 2009;27(1):76-83.
[Crossref] [Google Scholar] [PubMed]
- Franci G, Falanga A, Galdiero S, Palomba L, Rai M, Morelli G, et al. Silver nanoparticles as potential antibacterial agents. Molecules 2015;20(5):8856-74.
[Crossref] [Google Scholar] [PubMed]
- Ebrahiminezhad A, Bagheri M, Taghizadeh SM, Berenjian A, Ghasemi Y. Biomimetic synthesis of silver nanoparticles using microalgal secretory carbohydrates as a novel anticancer and antimicrobial. Adv Nat Sci Nanosci Nanotechnol 2016;7(1):015018.
- Gopinath PM, Ranjani A, Dhanasekaran D, Thajuddin N, Archunan G, Akbarsha MA, et al. Multi-functional nano silver: A novel disruptive and theranostic agent for pathogenic organisms in real-time. Sci Rep 2016;6(1):1-6.
[Crossref] [Google Scholar] [PubMed]
- Sholkamy EN, Ahamd MS, Yasser MM, Eslam N. Anti-microbiological activities of bio-synthesized silver Nano-stars by Saccharopolyspora hirsuta. Saudi J Biol Sci 2019;26(1):195-200.
- Shivlata L, Satyanarayana T. Thermophilic and alkaliphilic Actinobacteria: Biology and potential applications. Front Microbiol 2015;15(6):1014.
[Crossref] [Google Scholar] [PubMed]
- Jose PA, Jha B. New dimensions of research on actinomycetes: Quest for next generation antibiotics. Front Microbiol 2016;7:1295.
[Crossref] [Google Scholar] [PubMed]
- Das R, Romi W, Das R, Sharma HK, Thakur D. Antimicrobial potentiality of actinobacteria isolated from two microbiologically unexplored forest ecosystems of Northeast India. BMC Microbiol 2018;18(1):1-6.
- Siddharth S, Vittal RR, Wink J, Steinert M. Diversity and bioactive potential of actinobacteria from unexplored regions of Western Ghats, India. Microorganisms 2020;8(2):225.
[Crossref] [Google Scholar] [PubMed]
- Sharma P, Thakur D. Antimicrobial biosynthetic potential and diversity of culturable soil actinobacteria from forest ecosystems of Northeast India. Sci Rep 2020;10(1):4104.
[Crossref] [Google Scholar] [PubMed]
- Shirling ET, Gottlieb D. Methods for characterization of Streptomyces species. Int J Syst Evol Bacteriol 1966;16(3):313-40.
- Wilmotte A, Van der Auwera G, De Wachter R. Structure of the 16 S ribosomal RNA of the thermophilic cyanobacterium Chlorogloeopsis HTF (‘Mastigocladus laminosus HTF’) strain PCC7518, and phylogenetic analysis. FEBS Lett 1993;317(1-2):96-100.
[Crossref] [Google Scholar] [PubMed]
- Lane D1. 16S/23S rRNA sequencing. Nucl Acid Tech Bacterial Syst 1991:115-75.
- Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol 1987;4(4):406-25.
[Crossref] [Google Scholar] [PubMed]
- Shanmugasundaram T, Radhakrishnan M, Gopikrishnan V, Kadirvelu K, Balagurunathan R. Biocompatible silver, gold and silver/gold alloy nanoparticles for enhanced cancer therapy: In vitro and in vivo perspectives. Nanoscale 2017;9(43):16773-90.
- Yang X, Li Q, Wang H, Huang J, Lin L, Wang W, et al. Green synthesis of palladium nanoparticles using broth of Cinnamomum camphora leaf. J Nanopart Res 2010;12(5):1589-98.
- Ghosh S, Jagtap S, More P, Shete UJ, Maheshwari NO, Rao SJ, et al. Dioscorea bulbifera mediated synthesis of novel AucoreAgshell nanoparticles with potent antibiofilm and antileishmanial activity. J Nanomater 2015;2015:1-12.
- Tahir K, Nazir S, Ahmad A, Li B, Shah SA, Khan AU, et al. Biodirected synthesis of palladium nanoparticles using Phoenix dactylifera leaves extract and their size dependent biomedical and catalytic applications. RSC Adv 2016;6(89):85903-16.
- Dauthal P, Mukhopadhyay M. In vitro free radical scavenging activity of biosynthesized gold and silver nanoparticles using Prunus armeniaca (apricot) fruit extract. J Nanoparticle Res 2013;15(1):1366.
- Priyaragini S, Sathishkumar SR, Bhaskararao KV. Biosynthesis of silver nanoparticles using actinobacteria and evaluating its antimicrobial and cytotoxicity activity. Int J Pharm Pharm Sci 2013;5(2):709-12.
- Zou Y, Chang SK, Gu Y, Qian SY. Antioxidant activity and phenolic compositions of lentil (Lens culinaris var. Morton) extract and its fractions. J Agric Food Chem 2011;59(6):2268-76.
[Crossref] [Google Scholar] [PubMed]
- Re R, Pellegrini N, Proteggente A, Pannala A, Yang M, Rice-Evans C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radical Biol Med 1999;26(9-10):1231-7.
[Crossref] [Google Scholar] [PubMed]
- Madhanraj R, Eyini M, Balaji P. Antioxidant assay of gold and silver nanoparticles from edible basidiomycetes mushroom fungi. Free Radic Antioxidants 2017;7(2):137-42.
- Sastry M, Ahmad A, Khan MI, Kumar R. Biosynthesis of metal nanoparticles using fungi and actinomycete. Curr Sci 2003;85(2):162-70.
- Narasimha G, Alzohairy M, Khadri H, Mallikarjuna K. Extracellular synthesis, characterization and antibacterial activity of Silver nanoparticles by Actinomycetes isolative. Int J Nano Dimens 2013;4(1):77-83.
- Zarina A, Nanda A. Combined efficacy of antibiotics and biosynthesised silver nanoparticles from Streptomyces albaduncus. Int J Pharm Technol Res 2014a;6(6):1862-9.
- Kathiresan K, Manivannan S, Nabeel MA, Dhivya B. Studies on silver nanoparticles synthesized by a marine fungus, Penicillium fellutanum isolated from coastal mangrove sediment. Colloids Surf B Biointerfaces 2009;71(1):133-7.
[Crossref] [Google Scholar] [PubMed]
- Alani F, Moo-Young M, Anderson W. Biosynthesis of silver nanoparticles by a new strain of Streptomyces sp. compared with Aspergillus fumigatus. World J Microbiol Biotechnol 2012;28(3):1081-6.
[Crossref] [Google Scholar] [PubMed]
- Sosa IO, Noguez C, Barrera RG. Optical properties of metal nanoparticles with arbitrary shapes. J Phys Chem B. 2003;107(26):6269-75.
- Sk?adanowski M, Golinska P, Rudnicka K, Dahm H, Rai M. Evaluation of cytotoxicity, immune compatibility and antibacterial activity of biogenic silver nanoparticles. Med Microbiol Immunol 2016;205(6):603-13.
[Crossref] [Google Scholar] [PubMed]
- Mohanta YK, Panda SK, Bastia AK, Mohanta TK. Biosynthesis of silver nanoparticles from Protium serratum and investigation of their potential impacts on food safety and control. Front Microbiol 2017;8:626.
[Crossref] [Google Scholar] [PubMed]
- Abd-Elnaby HM, Abo-Elala GM, Abdel-Raouf UM, Hamed MM. Antibacterial and anticancer activity of extracellular synthesized silver nanoparticles from marine Streptomyces rochei MHM13. Egyp Aquatic Res 2016;42(3):301-12.
- Deepa S, Kanimozhi K, Panneerselvam A. Antimicrobial activity of extracellularly synthesized silver nanoparticles from marine derived actinomycetes. Int J Curr Microbiol App Sci 2013;2(2):223-30.
- Devika R, Elumalai S, Manikandan E, Eswaramoorthy D. Biosynthesis of silver nanoparticles using the fungus Pleurotus ostreatus and their antibacterial activity. Sci Rep 2012;1:557.
- Zonooz NF, Salouti M. Extracellular biosynthesis of silver nanoparticles using cell filtrate of Streptomyces sp. ERI-3. Sci Iran 2011;18(6):1631-5.
- Zhou M, Wei Z, Qiao H, Zhu L, Yang H, Xia T. Particle size and pore structure characterization of silver nanoparticles prepared by confined arc plasma. J Nanomater 2009;2009:1-5.
- Zarina A, Nanda A. Green approach for synthesis of silver nanoparticles from marine Streptomyces-MS 26 and their antibiotic efficacy. J Pharm Sci Res 2014;6(10):321-7.
- Singh D, Rathod V, Fatima L, Kausar A, Vidyashree NA, Priyanka B. Biologically reduced silver nanoparticles from Streptomyces sp. VDP-5 and its antibacterial efficacy. Int J Pharm Pharm Sci Res 2014;4(2):31-6.
- Prakasham RS, Kumar BS, Kumar YS, Kumar KP. Production and characterization of protein encapsulated silver nanoparticles by marine isolate Streptomyces parvulus SSNP11. Indian J Microbiol 2014;54(3):329-36.
[Crossref] [Google Scholar] [PubMed]
- Lakhsmi SY, Lakhsmi H, Sharmila S. Isolation, screening, identification, characterization and applications of green synthesized silver nanoparticle from marine Actinomycetes- Streptomyces althioticus. World J Pharm Res 2015;4(7):1592-611.
- Majumdar R, Tantayanon S, Bag BG. Synthesis of palladium nanoparticles with leaf extract of Chrysophyllum cainito (Star apple) and their applications as efficient catalyst for C–C coupling and reduction reactions. Int Nano Lett 2017;7(4):267-74.
- Fayaz AM, Balaji K, Girilal M, Yadav R, Kalaichelvan PT, Venketesan R. Biogenic synthesis of silver nanoparticles and their synergistic effect with antibiotics: A study against gram-positive and gram-negative bacteria. Nanomedicine 2010;6(1):103-9.
[Crossref] [Google Scholar] [PubMed]
- Kupryashina MA, Vetchinkina EP, Burov AM, Ponomareva EG, Nikitina VE. Biosynthesis of gold nanoparticles by Azospirillum brasilense. Microbiology 2013;82(6):833-40.
- Mohanta YK, Behera SK. Biosynthesis, characterization and antimicrobial activity of silver nanoparticles by Streptomyces sp. SS2. Bioproc Biosyst Eng 2014;37(11):2263-9.
[Crossref] [Google Scholar] [PubMed]
- Narayana KJ, Prabhakar P, Vijayalakshmi MU, Venkateswarlu Y, Krishna PS. Biological activity of phenylpropionic acid isolated from a terrestrial Streptomycetes. Polish J Microbiol 2007;56(3):191-7.
[Google Scholar] [PubMed]
- Vennila K, Chitra L, Balagurunathan R, Palvannan T. Comparison of biological activities of selenium and silver nanoparticles attached with bioactive phytoconstituents: Green synthesized using Spermacoce hispida extract. Adv Nat Sci Nanosci Nanotechnol 2018;9(1):015005.
- Kumar S, Sandhir R, Ojha S. Evaluation of antioxidant activity and total phenol in different varieties of Lantana camara leaves. BMC Res Notes 2014;7(1):1-9.
[Crossref] [Google Scholar] [PubMed]
- Subbaiya R, Saravanan M, Priya AR, Shankar KR, Selvam M, Ovais M, et al. Biomimetic synthesis of silver nanoparticles from Streptomyces atrovirens and their potential anticancer activity against human breast cancer cells. IET Nanobiotechnol 2017;11(8):965-72.
[Crossref] [Google Scholar] [PubMed]