- *Corresponding Author:
- V. Raphael
Department of Chemistry, Government Engineering College, Affiliated to APJ Abdul Kalam Technological University, Thiruvananthapuram, Kerala 680009, India
E-mail: vinodpraphael@gectcr.ac.in
| Date of Received | 31 January 2022 |
| Date of Revision | 13 May 2023 |
| Date of Acceptance | 29 September 2023 |
| Indian J Pharm Sci 2023;85(5):1367-1372 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Coronavirus disease-19 disease originated in China by the end of 2019 and has spread all over the world, with many casualties reported since then. Research for the discovery of effective pharmaceuticals is going on around the world. Pharmaceutical scientists are keenly interested in revealing the capacities of a large number of already existing molecules in molecular and drug databases to fight against coronavirus. We identified several triazole and quinazolinone derivatives from molecular databases that share key pharmacophore features with oxadiazole, a known drug-like compound, and conducted computational analysis to evaluate their ability to bind with the envelope protein of Severe Acute Respiratory Syndrome Coronavirus 2. The molecules were screened for their absorption, distribution, metabolism, excretion properties and drug-likeness using the SwissADME webserver. Out of a large number of molecules investigated, six were reported in this work that showed appreciable binding energy values. In vitro, in vivo toxicological studies and clinical trials have to be conducted to get the complete picture of their efficiency and toxicity.
Keywords
Coronavirus disease-19, absorption, distribution, metabolism, excretion, quinazolinone, envelope protein
The dreadful pandemic of the 21st century, Coronavirus Disease-19 (COVID-19), caused by Severe Acute Respiratory Syndrome coronavirus-2 (SARS-CoV-2) originated in Wuhan, China, in the last months of 2019 and spread all over the world within a few months. Infected people may have symptoms like headache, running nose, fever, throat pain, etc. In severely affected patients, respiratory problems and pneumonia may lead to fatalities[1,2]. Till date, more than 50 lakh people around the world have lost their lives due to COVID-19[3]. Though vaccination drives are progressing around the world to combat the coronavirus, the lack of effective drugs increases the casualties in many countries. Pharmaceutical scientists and medical practitioners are performing rigorous research to find effective therapeutics to alleviate the effects of COVID-19. Some doctors do clinical trials with already-existing antivirals[4,5]. Researchers use various drug targets, like structural and nonstructural proteins of coronaviruses, to bind the small molecules that are already present in chemical databases. Membrane protein (M), Nucleocapsid protein (N), Envelope protein (E) and Spike protein (S) are the important structural proteins of coronaviruses[6-8].
E protein is the smallest structural protein of SARS-CoV-2. E is a viroporin consisting of 75 amino acid residues and acts as a cation selective channel. The inner pore of the ion channel contains hydrophobic amino acid residues such as Asn, Leu, Val, Ala and Thr. E-protein consists of five alpha helical strands that are stabilised by the stacking interaction of Phe residues. Solid state nuclear magnetic resonance studies showed that protein conformation is highly homogeneous. It is also established that the N-terminal segment of E-protein is more dynamic and hydrophilic than the C-terminal segment, which is considered a rigid part. The Transmembrane Domain (TMD) of E proteins is hydrophobic in nature and consists of approximately 25 amino acids. This domain contains five strands that act as the ion channel. Due to the dynamic nature of the N-terminus, it will be more perturbed by drug molecules. Deletion of the coronavirus's E protein (SARS-CoV-1 mouse model) or disruption of ion channel activity can reduce viral activity. Thus, the E protein of the COVID-19 virus is a potential drug target[9-12].
In this work, we took the TMD of the envelope protein of COVID-19 as the drug target to bind molecules obtained from the chEMBL and Zinc databases. Pharmacophore and in silico studies were done to determine structurally similar molecules and evaluate their binding affinity with the envelope proteins.
Materials and Methods
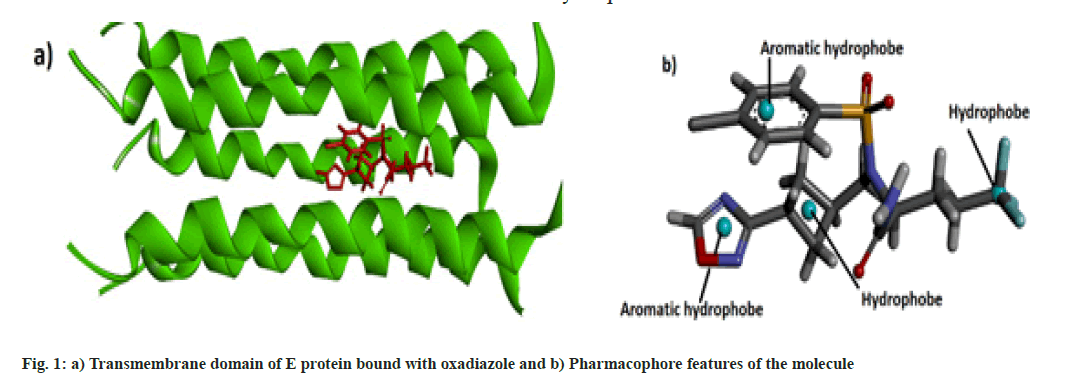
Molecular data bases such as chEMBL and Zinc were used to determine the lead compounds for computational investigations. Biovia Discovery Studio 2020[13] and UCSF Chimera[14] software were used for the visualization of the docked structure and checking the binding sites respectively. Online web server SwissDock[15] was used for the molecular docking and the physicochemical and pharmacokinetics of the molecules were determined by the SwissADME web server[16]. Pharmacophore studies of the molecules were conducted with the help of Pharmagist[17,18] and Zincpharmer[19] web servers. The protein data bank structure of the envelope protein (TMD) of Covid-19 was downloaded from Protein Data Bank and docked with the oxadiazole molecule is shown in fig.1a[10].
Results and Discussion
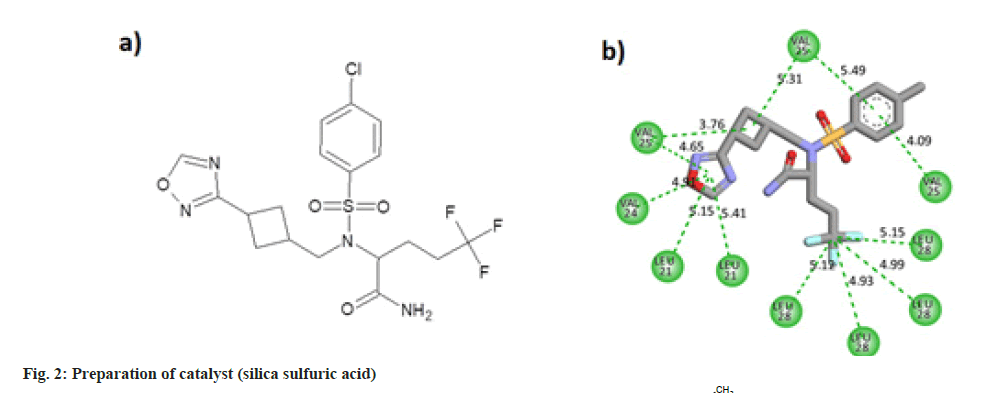
Some pharmaceutically important compounds obtained from the chEMBL data were screened computationally to identify their binding efficacies on the envelope protein of SARS-CoV-2. A molecule having chEMBL id: 2059020 (oxadiazole derivative) showed a good binding score (-9.34 kcal/mol) on the TMD of the envelope protein using the Swissdock webserver. According to the ChemBL database, target classes of this oxadiazole molecule consist of enzymes and ion channels. According to the database, this molecule has a 50 % chance of binding to ion channels. After docking with the TMD of the E protein, the poses of the molecule were analysed using UCSF Chimaera software. The main pose of this molecule appeared near the middle region of the hydrophobic ion channel of the E protein. The main sites of the oxadiazole derivative (aromatic, cyclobutane, trifluoromethyl, oxadiazole ring) which interacted with the binding pocket of the envelope protein are mapped from the best ligand-receptor pose and shown in fig. 1b. The pharmacophore feature of the molecule consisted of four hydrophobic sites. Out of four sites, three were located in the "head" region of the molecule and one was located at the end of the "tail". Two aromatic rings (benzene and oxadiazole) and one alicyclic ring (cyclobutane) system constituted the head region. The terminal aliphatic moiety of the molecule acted as the hydrophobic tail end.
The oxadiazole interacted with the TMD of the envelope protein using 12 strong hydrophobic bonds. The cyclobutane ring of the molecule made bonds with Valine residues of the protein. Oxadiazole ring of the ligand interacted with Valine and Leucine segments of the receptor. Benzene ring of the molecule also made pi-alkyl bonds with the valine residues. Leu28 amino acid residues of four protein chains made hydrophobic alkyl interactions with the trifluoro methyl carbon atom. The molecular structure and two dimensional interaction plot of the oxadiazole with the amino acid residues are shown in fig. 2.
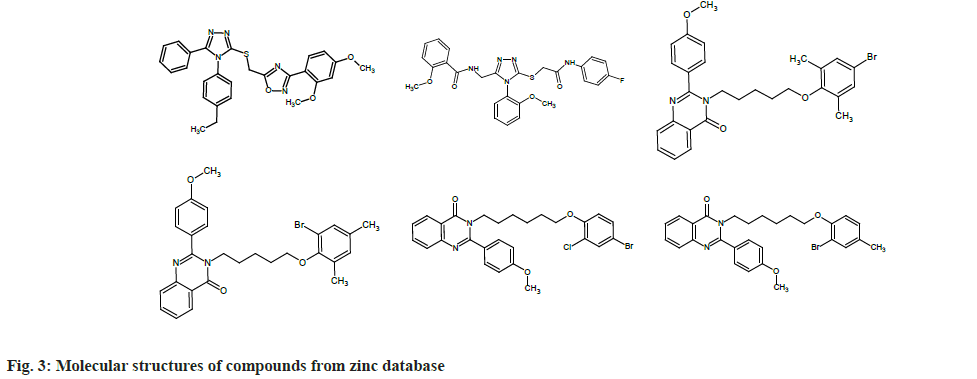
To find out which molecules had pharmacophore features similar to that of chEMBL2059020, we uploaded the same to the Zinpharmer web server. Out of 146 molecules displayed by the server, we filtered six by fixing the conditions of the molecular mass range 500-600 Da and Root Mean Square Deviation (RMSD)<0.15 at the pharmacophore sites. The molecules were docked with the TMD of the envelope protein using the Swissdock webserver and analyzed with UCSF Chimera. In this work, we present six molecules that exhibit binding energy >9 kcal/mol (magnitude) on the E protein of SARS-CoV-2. Six compounds obtained from the zinc database are named from A to F for the sake of discussion (ZINC09185092:A, ZINC09278965:B, ZINC09014307: C, ZINC02418731: D, ZINC2401500: E, and ZINC9282093: F). The molecular structures of these compounds are given in fig. 3.
Out of the six compounds selected from the zinc database, two are heterocyclic in nature and are derivatives of 1,2,4-triazole. The triazole derivatives A and B displayed -9.61 and 9.0 kcal/mol binding energy on the envelope protein of the coronavirus, respectively. Other four compounds are also heterocyclic molecules and showed more or less similar structural behavior. These four heterocyclics belonged to the class 4-quinozolinone, which is an important group in the field of medicinal chemistry. All four molecules showed good binding affinity with the transmembrane part of envelope protein (-9.32 to -9.61 kcal/mol). Compounds A and E showed maximum binding energy (-9.61 kcal/mol) with the receptor (Table 1).
| Molecule | Molecular mass (D) | RMSD | Binding energy kcal/mol |
|---|---|---|---|
| A | 500 | 0.144 | -9.61 |
| B | 522 | 0.136 | -9.00 |
| C | 521 | 0.123 | -9.39 |
| D | 521 | 0.128 | -9.32 |
| E | 541.87 | 0.126 | -9.61 |
| F | 521.45 | 0.128 | -9.49 |
Table 1: Binding Energies of Zinc Molecules on the Envelope Protein of Sars-Cov-2
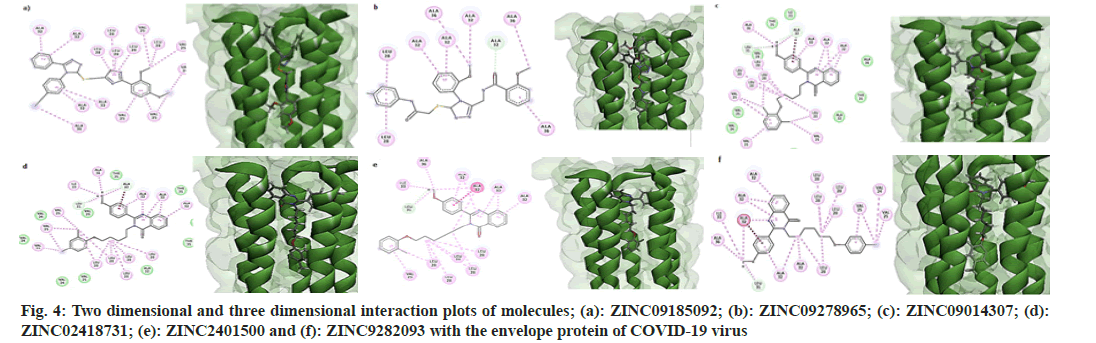
All molecules possessed the features of a pharmacophore shown in fig. 2, i.e., two aromatic rings and one hydrophobe in the "head" region of the molecules and one hydrophobe in the "tail" part. In addition to these features, all the molecules consisted of hydrogen bond donors and acceptor sites. But, due to the non-polar nature of the inner core of the TMD of the protein, only hydrophobic interactions were established between the receptor and ligands. Fig. 4 shows the two dimensional and three dimensional interaction diagrams of hit compounds with the TMD of the envelope protein of SARS-CoV-2.
On close examination of the protein-ligand interactions, it is evident that the aromatic rings of the head region of the molecules made hydrophobic interactions with Alanine 32 and Alanine 36 segments of the five strands of the envelope protein. For molecule B, the methyl group of Alanine 32 in one stand made a non-conventional hydrogen bond with the carbonyl group of the amide linkage. The hydrophobic region of the head region in all molecules (except B) interacted with the Leucine28 amino acid segment of the protein, while the hydrophobic part of the tail made bonds with Valine28 of the envelope protein. All the 4-quinozolinone class of molecules displayed a non-conventional hydrogen bond interaction with the methoxy (-OCH3) group, which leads to the increased binding of these ligands with the receptor pocket.
Drug likeness of the ligands predicted by SwissADME showed that only ZINC 09185092 and ZINC 09278965 (triazoles) perfectly obeyed Lipinski rule[20]. Other four molecules showed some violations of the rule. But according to the Veber rule, all molecules except Zinc 09278965 have drug-like behavior. According to computational studies, the bioavailability of triazole derivatives was 0.55. Oxazole and quinozolinone derivatives exhibited a comparatively lower bioavailability score compared to triazole (Table 2).
| Molecule | M. wt (g/mol) | TPSA (Å2) | Consensus of logPo/w | Water solubility | Molar refractivity |
|---|---|---|---|---|---|
| A | 499.58 | 113.39 | 4.88 | Poorly soluble | 139.05 |
| B | 521.56 | 132.67 | 3.51 | Moderately soluble | 137.34 |
| C | 521.45 | 53.35 | 6.22 | Poorly soluble | 142.07 |
| D | 521.45 | 53.35 | 6.13 | Poorly soluble | 142.07 |
| E | 541.86 | 53.35 | 6.31 | Poorly soluble | 141.95 |
| F | 521.45 | 53.35 | 6.05 | Poorly soluble | 141.91 |
Table 2: Physical Properties of The Molecules Predicted by Swissadme
The Topological Surface Area (TPSA) of a molecule is important in predicting intestinal permeability. In general, absorption of the drug by the intestinal tract will be lowered if it has a high TPSA. Modifications such as lowering the number of hydrogen bond donors and acceptors were found to increase intestinal permeability. The TPSA value of most drugs will be between 0 and 1402. Since the TPSA values of the investigated molecules ranged between 53 and 133 Ų in this work, we can infer that all molecules possess appreciable permeability. Poor solubility in water was noted for all molecules except ZINC 09014307. This is an indication of the enhanced lipophilic nature of molecules.
The log Po/w of a molecule is very important in determining the extent of hydrophilicity or lipophilicity[21]. Highly hydrophilic molecules show poor permeability and blood plasma binding capacity. Out of the different models predicted by SwissADME, consensus LogP values for the molecules are reported here. Molecules ZINC 09185092 and ZINC 09185092 showed 4.88 and 3.11 logP values, respectively, and fell in the range predicted by Lipinski. Other molecules displayed slightly higher log p values than Lipinski rules.
The Molar Refractivity (MR) of the six molecules deviated slightly from the standard range (40-130). This parameter is an indicator of the polarizability of a molecule, and it is very important when considering drug-receptor interaction[22]. The MR values of the investigated molecules varied from 139 to 142.
In modern pharmaceutical science, a major concern is the potential for drug-drug interaction in multi-drug therapies. One drug may induce or inhibit Cytochrome P450 (CYP450) enzymes and may alter the metabolism of the other drug[23]. Six cytochrome enzymes, namely CYP1A2, CYP2C19, CYP2C9, CYP2D6, CYP3A4 and CYP2E1, play an important role in the oxidation of drugs. Understanding the pharmacology of drugs that inhibit CYP450 enzymes is very important in multi-drug therapies. This study also reveals the risk of giving two drugs simultaneously. The interactions of the molecules with the cytochrome enzymes predicted by the SwissADME web server are given in Table 3. The majority of the molecules acted as inhibitors of the enzymes, except for the CYP1A2 inhibitor. ZINC 09185092 did not inhibit CYP1A2 and CYP2C19.
| Molecule | Gastrointestinal absorption | BBB permeant | P-gp substrate | CYP1A2 inhibitor | CYP2C19 inhibitor | CYP2C9 inhibitor | CYP2D6 inhibitor | CYP3A4 inhibitor |
|---|---|---|---|---|---|---|---|---|
| A | Low | No | Yes | No | No | Yes | No | Yes |
| B | Low | No | Yes | No | Yes | Yes | Yes | Yes |
| C | High | No | Yes | No | Yes | No | Yes | Yes |
| D | High | No | Yes | No | Yes | Yes | Yes | Yes |
| E | Low | No | Yes | No | Yes | Yes | Yes | Yes |
| F | High | No | Yes | No | Yes | Yes | Yes | Yes |
Table 3: Pharmacokinetics Data of Six Hit Molecules from Zinc Database
A computational ADME investigation established that all molecules do not have the capacity to penetrate the Blood Brain Barrier (BBB), which is an essential criterion for those drug molecules affecting the central nervous system. GI absorption is an important pharmacological property since it determines the effectiveness of drugs. Out of six molecules, three showed low activity (ZINC09185092, ZINC09278965, ZINC2401500) and three displayed high activity (ZINC09014307, ZINC02418731, ZINC9282093).
The Permeability glycoprotein (P-glycoprotein or P-gp) plays a very important role in removing toxic metabolites from the body, and it is found in many organs[24]. If P-gp identifies a drug as a toxin, it will be thrown out by the efflux pump of the protein. This will affect the efficacy of the oral drugs administered. In some cases, similar to CYP450, P-gp may induce the drug's efficacy. In general, the bioavailability of a drug is enhanced if it is inhibited by P-gp. The pharmacokinetics of the molecules predicted by SwissADME showed that all the molecules acted as P-gp substrates.
In conclusion, pharmaceutically important oxadiazole molecule that perturbs the ion channel activity of proteins was selected from the chEMBL database and analyzed for its binding efficacy on the E protein of SARS-CoV-2. The pharmacophore features of the molecule were determined by considering the best pose of the protein-ligand complex. Using the pharmacophore model, lead molecules that possess the same structural features as the oxadiazole were downloaded from the Zinc database. Out of the many molecules in the Zinc database, those with low RMSD were selected for the docking studies. This consists of two triazole and three quinazolinone derivatives. Docking investigations of the molecules were done using the Swissdock web server. Molecules having good binding scores (magnitude >9 kcal/mol) were reported in this work. Pharmacokinetic and ADME investigations were done on the six molecules obtained from the zinc database. To obtain a complete picture of the potency of the drugs, in vitro, in vivo toxicological studies, and clinical trials must be conducted.
Conflict of interests:
The authors declared that there are no conflicts of interest.
References
- Centers for Disease Control and Prevention. Symptoms of COVID-19. Centers for Disease Control and Prevention.
- Coronavirus disease 2019 (COVID-19)-symptoms and causes. Mayo Clinic.
- COVID Live Update: 270247123 Cases and 5320918 deaths from the Coronavirus Worldometer
- Venkatesan P. Repurposing drugs for treatment of COVID-19. Lancet Respir Med 2021;9(7):e63.
[Crossref] [Google Scholar] [PubMed]
- Singh TU, Parida S, Lingaraju MC, Kesavan M, Kumar D, Singh RK. Drug repurposing approach to fight COVID-19. Pharmacol Rep 2020;72:1479-508.
[Crossref] [Google Scholar] [PubMed]
- Mariano G, Farthing RJ, Lale-Farjat SL, Bergeron JR. Structural characterization of SARS-CoV-2: Where we are, and where we need to be. Front Mol Biosci 2020;17:605236.
[Crossref] [Google Scholar] [PubMed]
- Satarker S, Nampoothiri M. Structural proteins in severe acute respiratory syndrome coronavirus-2. Arch Med Res 2020;51(6):482-91.
[Crossref] [Google Scholar] [PubMed]
- Yadav R, Chaudhary JK, Jain N, Chaudhary PK, Khanra S, Dhamija P, et al. Role of structural and non-structural proteins and therapeutic targets of SARS-CoV-2 for COVID-19. Cells 2021;10(4):821.
[Crossref] [Google Scholar] [PubMed]
- Schoeman D, Fielding BC. Coronavirus envelope protein: Current knowledge. Virol J 2019;16(1):1-22.
[Crossref] [Google Scholar] [PubMed]
- Mandala VS, McKay MJ, Shcherbakov AA, Dregni AJ, Kolocouris A, Hong M. Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers. Nat Struct Mol Biol 2020;27(12):1202-8.
[Crossref] [Google Scholar] [PubMed]
- Chai J, Cai Y, Pang C, Wang L, McSweeney S, Shanklin J, Liu Q. Structural basis for SARS-CoV-2 envelope protein recognition of human cell junction protein PALS1. Nat Commun 2021;12(1):3433.
[Crossref] [Google Scholar] [PubMed]
- Bianchi M, Benvenuto D, Giovanetti M, Angeletti S, Ciccozzi M, Pascarella S. Sars-CoV-2 envelope and membrane proteins: Structural differences linked to virus characteristics? BioMed Res Int 2020;2020.
[Crossref] [Google Scholar] [PubMed]
- BIOVIA, Dassault Systèmes, BIOVIA Workbook, BIOVIA Pipeline Pilot, Release 2020.
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 2004;25(13):1605-12.
[Crossref] [Google Scholar] [PubMed]
- Grosdidier A, Zoete V, Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res 2011;39(suppl_2):W270-7.
[Crossref] [Google Scholar] [PubMed]
- Daina A, Michielin O, Zoete V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 2017;7(1):42717.
[Crossref] [Google Scholar] [PubMed]
- Schneidman-Duhovny D, Dror O, Inbar Y, Nussinov R, Wolfson HJ. Deterministic pharmacophore detection via multiple flexible alignment of drug-like molecules. J Comput Biol 2008;15(7):737-54.
[Crossref] [Google Scholar] [PubMed]
- Schneidman-Duhovny D, Dror O, Inbar Y, Nussinov R, Wolfson HJ. PharmaGist: A webserver for ligand-based pharmacophore detection. Nucleic Acids Res 2008;36(suppl_2):W223-8.
[Crossref] [Google Scholar] [PubMed]
- Koes DR, Camacho CJ. ZINCPharmer: Pharmacophore search of the ZINC database. Nucleic Acids Res 2012;40(W1):W409-14.
[Crossref] [Google Scholar] [PubMed]
- Loureiro DR, Soares JX, Costa JC, Magalhães ÁF, Azevedo CM, Pinto MM, et al. Structures, activities and drug-likeness of anti-infective xanthone derivatives isolated from the marine environment: A review. Molecules 2019;24(2):243.
[Crossref] [Google Scholar] [PubMed]
- Arnott JA, Planey SL. The influence of lipophilicity in drug discovery and design. Expert Opin Drug Discov 2012;7(10):863-75.
[Crossref] [Google Scholar] [PubMed]
- Ghose AK, Viswanadhan VN, Wendoloski JJ. Prediction of hydrophobic (lipophilic) properties of small organic molecules using fragmental methods: An analysis of ALOGP and CLOGP methods. J Phys Chem A 1998;102(21):3762-72.
- Lynch T, Price AM. The effect of cytochrome P450 metabolism on drug response, interactions, and adverse effects. Am Fam Physician 2007;76(3):391-6.
[Google Scholar] [PubMed]
- Shirasaka Y, Sakane T, Yamashita S. Effect of P‐glycoprotein expression levels on the concentration‐dependent permeability of drugs to the cell membrane. J Pharm Sci 2008;97(1):553-65.
[Crossref] [Google Scholar] [PubMed]