- *Corresponding Author:
- G. P. Li
Department of Pulmonary and Critical Care Medicine, Chengdu Third People’s Hospital branch of National Clinical Research Center for Respiratory Disease, Affiliated Hospital of ChongQing Medical University, Chengdu 610031,China
E-mail: lzlgp@163.com
| Date of Received | 01 August 2022 |
| Date of Revision | 06 April 2023 |
| Date of Acceptance | 11 September 2023 |
| Indian J Pharm Sci 2023;85(5):1373-1387 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
Abstract
Magnolol is an active polyphenol extracted from the traditional Chinese herb Magnolia officinalis, which can be applied for expectorating phlegm and relieving cough. However, the role and molecular mechanism of Magnolol in the treatment of asthma was not fully explored. To comprehensively analyze the pharmacological targets of Magnolol, ovalbumin-induced asthmatic mice model was established, and subsequently conducted with Magnolol. The pathological changes of lung tissue were observed by Haematoxylin and Eosin, periodic acid-Schiff stain and Masson's trichrome staining. Network pharmacology combined with the transcriptomic analysis was applied to investigate the underlying mechanisms and targets. Potential targets were validated by quantitative reverse transcription polymerase chain reaction and immunofluorescence in vivo. As a result, this study found that the inflammatory infiltrations in Magnolol-treated asthma mice were significantly ameliorated. Using network pharmacology, it was identified 33 asthma-related Magnolol targets, which obviously involved in positive regulation of cytokine production and Th17 cell differentiation pathways. Furthermore, the transcriptome analysis showed that the infiltration score of eosinophils, activated mast cells and M1 macrophages were significantly decreased in Magnolol-treated asthmatic mice. Meanwhile, Hallmark analysis exhibited that the enrichment scores of interferon gamma and alpha responses were remarkably enhanced in Magnolol treated ovalbumin-induced asthmatic mice. Moreover, the potential therapeutic targets, Arginase 1, Phosphodiesterase 4B, Signal transducer and activator of transcription 1 and Matrix metallopeptidase 12 were screened out by using combined analysis of asthma targets, Magnolol targets and differential expressed genes. In particular, gene set enrichment analysis revealed that high Matrix metallopeptidase 12 expression was associated with asthma and cytokine-cytokine receptor interaction pathways. The expression of Matrix metallopeptidase 12 was significantly decreased in asthma group and was restored after Magnolol treatment through quantitative reverse transcription polymerase chain reaction and immunofluorescence validation. Together, this study found that Magnolol might have a therapeutic effect on asthma by regulation of Matrix metallopeptidase 12 through cytokine mediated signaling pathway.
Keywords
Asthma, Magnolol, therapeutic targets, transcriptomics, network pharmacology
Asthma is one of the chronic respiratory diseases that attack children and adults, which accounts for a large proportion of respiratory diseases. According to a study published in The Lancet, it is estimated that there are approximately 45.7 million asthma patients aged 20 y and above in China[1]. Typical asthma symptoms such as coughing, wheezing, tachypnea and chest tightness are usually caused by inflammation and spasm of the small airways, which lead to an estimated 461 000 people worldwide died[2]. These symptoms can fluctuate and could be easily affected by complex genetic and environmental factors. Through genome sequencing analysis, researchers have identified 8 loci with significant genome-wide association with asthma, as well as 16 loci of common risk variant for asthma and 34 loci related with allergic diseases[3]. These loci reveal gene-level alterations in asthma and have certain therapeutic implications. Genetic variation at these loci can influence gene expression, thereby impacting cell function and the pathophysiology of diseases[4,5]. By validating these predicted gene targets, researchers can explore the function of target genes and their role in asthma pathophysiology, leading to the development of drugs targeting these specific genes.
The current recommendation for daily treatment of asthma is to use the lowest effective dose of inhaled corticosteroids[6,7]. But many patients are still unable to get benefits from this treatment. Additionally, there is the risk of systemic side-effects that remains because of cumulative doses, individual differences and the extent of drug absorption in distinct regions of the body[8]. Therefore, extracting therapeutic ingredients from natural plants with better efficacy and fewer side effects has become a growing research focus in asthma management[9]. Network pharmacology is the ideal method to study traditional Chinese medicines, which can construct a complex component-target-disease-molecular pathway model and could systematically explore drug-disease associations[10,11].
For network pharmacology research, initially, public databases[12] and literature, along with bioinformatics technology are used to screen target information of drug components and diseases. Subsequently, a network visualization tool[13] is employed to construct a multi-dimensional biological network model of 'gene-drug-target-disease' by focusing on the network's specific signal nodes. In brief, network pharmacology is crucial in various areas such as drug target discovery, screening of bioactive compounds, toxicity evaluation, mechanism research, and quality control research[14,15]. An et al.[16] conducted a study on the antioxidant effects of ZhiZiDaHuang Decoction (ZZDHT) using a network pharmacology approach, which revealed that ZZDHT has the potential to regulate reactive oxygen species and treat alcoholic liver disease by targeting specific enzymes such as xanthine dehydrogenase, nitric oxide synthase 2, and prostaglandin-endoperoxide synthase 2. Otherwise, transcriptome sequencing technology, widely used in molecular biology research widely, is able to find cellular and molecular pathways in asthma[17,18]. Therefore, drug-disease-target relationships can be more easily analyzed by combining network pharmacology and transcriptome analysis.
Magnolol is a natural lignan extracted from the dried bark of Magnolia officinalis, which has roles of anti-inflammatory, anti-oxidative and even anti-tumor activities, pharmaceutically[19,20]. Studies have shown that magnolol exerts anti-asthmatic effects through the regulation of Janus kinase signal transduction and activation of Notch signaling pathway in Ovalbumin (OVA)-sensitized asthmatic mice[21,22]. However, the gene target of Magnolol to alleviate asthma has not yet been identified. And recently, researchers have posed more attention to the application of network pharmacology and Ribonucleic Acid-sequencing (RNA-seq) in traditional Chinese medicine research. To investigate the molecular mechanism of Magnolol in the treatment of asthma, a mouse model of Magnolol treatment OVA-induced asthma was constructed. Combining network pharmacology and RNA-seq methods, this study may provide new insights into the molecular mechanisms of Magnolol and provide candidate biomarkers for targeted therapy of asthma.
Materials and Methods
Mice and reagents:
Female Bagg Albino/c mice (20-22 g, 6-8 w) were purchased from Chongqing Tengxin Technology Company. Mice were conventionally fed for 1 w under Specific-Pathogen-Free (SPF) rearing environment before animal modeling. During the rearing period, the lights were alternately with 12 h of light/dark cycle and all diets provided ad libitum.
OVA (CAS:9006-59-1) was purchased from Beijing Solarbio Science & Technology Co., Ltd. (Beijing, China). Imject® alum adjuvant (WA313000) was obtained from Thermo Fisher Scientific Inc. (USA). Magnolol (H-004-180116) was purchased from Chengdu Ruifensi Biotechnology Co., Ltd. (Chengdu, China). Ribonucleic Acid (RNA) isolater total RNA extraction reagent (R401-01), HiScript®II Q RT SuperMix (R233-01) and ChamQ Universal SYBR quantitative Polymerase Chain Reaction (qPCR) Master Mix (Q711-02) were purchased from Nanjing Vazyme Biotech Co., Ltd. (Nanjing, China). The goat serum was purchased from Thermo Fisher Scientific Inc. (USA). Anti- Matrix Metallopeptidase 12 (MMP12) (bs-23566R) primary antibodies were purchased from Bioss Inc (Woburn, MA, USA). Goat Anti-Rabbit IgG H&L (Alexa Fluor® 555) were obtained from Abcam (UK). Vector® TrueVIEW™ Autofluorescence Quenching Kit (SP-8400) was acquired from Vector Laboratories (CA, USA).
Model construction of asthma:
Twenty-four mice were evenly divided into Control group (CON), Asthma group (OVA), Magnolol Group (MG) and Magnolol-treated asthma Group (OVA+MG) according to the random number table method. Following the classical method for asthma modeling[17,23], mice were sensitized once by intraperitoneal injection of OVA (20 μg/d) with Al(OH)3 (1 mg/d) on 0th and 7th d. On the 14th d, mice were challenged once with OVA (50 μg/d) by nasal instillation and consecutively instillation for 7 d. Based on the asthma model, the OVA+MG group was given intraperitoneal injection of Magnolol (20 mg/kg/d) starting from the 14th d and continuously injected for 7 d[24-26] before challenge with OVA. The control group was managed with the same volume of PBS for intraperitoneal injection and intranasal instillation. Similarly, the Magnolol group was disposed with PBS, and on the 14th d, equivalent Magnolol was administered intraperitoneally for 7 consecutive d. All animal procedures conform to accordance with the guidelines of the Committee on the Protection and Use of Animals in the Southwest Jiaotong University.
Pathological and immunofluorescence staining:
Lung tissue from right lower lobe was taken for histopathological and immunofluorescence staining in each group of mice. First, tissues were immersed in 4 % paraformaldehyde for fixation, routinely paraffin-embedded and sliced into 5 μm-thick sections using a microtome at -16°. Hematoxylin-Eosin (HE), Masson and Periodic Acid-Schiff (PAS) staining were performed strictly according to the kit instructions. After the staining process was completed, histopathological changes in immune infiltration, collagen fiber hyperplasia and bronchial mucus secretion were evaluated under a light microscope. In addition, the paraffin sections were dried, conventionally deparaffinized, followed by antigen repairing with sodium citrate.
Then blocked with 10 % goat serum in Phosphate Buffer Solution (PBS), primary antibody MMP12 (Bios, bs-23566R, 1:400) was incubated overnight at 4°, and secondary antibody AlexaFluor® 555 goat anti-rabbit IgG (1:2000) was incubated for 1 h at room temperature. Afterwards sections were washed 3 times with PBS. Following the manufacturer's instructions, the Vector® TrueVIEW™ Autofluorescence Quenching Kit was used to minimize background staining. Nuclei were finally stained using 4′-6-Diamidino-2-Phenylindole dihydrochloride (DAPI) (Biosharp). The images were taken with a laser confocal microscope and analyzed with CellSens Dimension Software (IX73, Olympus, Japan).
Screening and analysis of therapeutic targets of Magnolol and asthma:
PharmMapper database (http://lilab.ecust.edu.cn/pharmmapper/) was used to obtain the drug targets of Magnolol[27]. To begin with, molecular structure of Magnolol was downloaded from Pubchem database in SDF format (PubChem CID 72300). The structure-data format structure file of Magnolol was submitted into the PharmMapper sever to predict the potential targets using the pharmacophore mapping approach, and then gained the activity targets of Magnolol. And the UniProt (https://uniprot.org/) was used for converting target name to gene name. Then, the targets of asthma were downloaded from Therapeutic target database (http://db.idrblab.net/), Online Mendelian Inheritance in Man database (OMIM) (https://www.omim.org/), DisGeNET database (https://www.disgenet.org/) and GeneCards database (https://www.genecards.org). According to the relevance score, 260 asthma target genes were selected for conjoint analysis. Network visualizations were created using Cytoscape (version 3.7.2).
Sample collection and sequencing:
The lung tissues from the 4 groups were collected for transcriptome sequencing. They were rapidly removed from mice in each group, subsequently frozen in liquid nitrogen and stored at -80°. The complete RNA extraction, library construction, and RNA-seq analysis were conducted by Shenzhen Huada Gene Co., Ltd. (Shenzhen, China).
Immune cell infiltration acquisition:
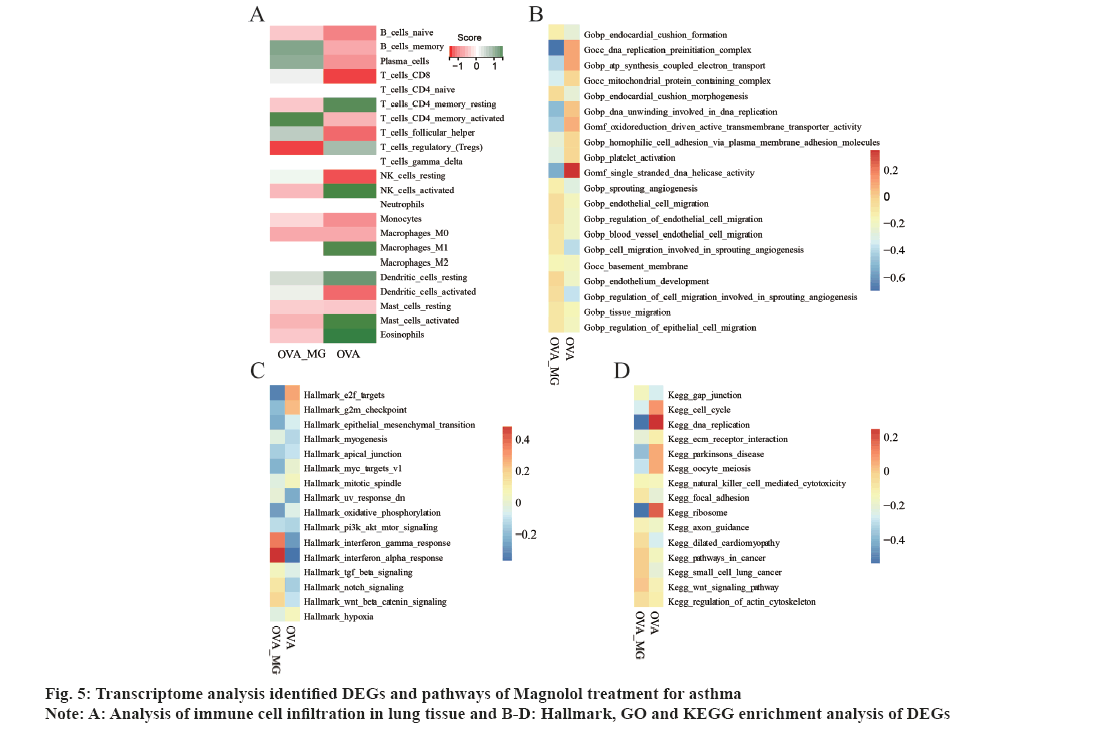
The analysis of immune cell infiltration was based on the gene transcriptomic messenger Ribonucleic Acid (mRNA) expression profile data through mRNA sequencing. The raw transcriptomic mRNA expression data of asthma was preprocessed using the Limma package in the R language. Then, deconvolution analysis was performed by the CIBERSORT method and the LM22 gene signature found on the CIBERSORT website (https://cibersort.stanford.edu/)[28]. The proportions of 22 types of immune cells were calculated that conclude M1 macrophages, M2 macrophages, regulatory T cells, neutrophils, etc. By using Pheatmap package in R, we showed the immune cell expression in the OVA group and the OVA+MG group according to the calculated results.
Combining analysis of Differentially Expressed Genes (DEGs), asthma targets and Magnolol targets:
DESeq2 package in R software (http://bioconductororg/packages/stats/bioc/DESeq2/) was applied to analyze the DEGs based on the Real Counts of gene transcripts (|Log2 fold change|>1, p value<0.05). R package ClusterProfiler (https://bioconductor.org/packages/clusterProfiler/) was used to process the Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), Hallmark analyses. Pathway analysis for asthma target was performed using a hypergeometric distribution across a set of canonical pathways (GO, KEGG, Wiki-pathways and Reactome).
Based on Gene Set Enrichment Analysis (GSEA) analysis of single-gene batch correlation analysis, gene correlation pathway analysis was performed. Using the R package ClusterProfiler, the KEGG and GO pathways were found. Combined analysis with asthma targets and Magnolol targets, the common pathway of these genes was identified.
RNA extraction and real-time Polymerase Chain Reaction (PCR):
Lung tissues were removed from -80° and processed for RNA extraction. RNA was extracted with Trizol and reverse transcribed. SYBER Green method was conducted for the quantitative reverse transcription real-time PCR. HiScript®II Q RT SuperMix and ChamQ Universal SYBR qPCR Master Mix reagents were utilized for reverse transcription and fluorescence quantification by using the Epoch multi-volume spectrophotometer system (biotechnology, USA). Primers were designed using PCR primer design tool (https://eurofinsgenomics.eu/en/ecom/tools/pcr-primer-design/) online website and synthesized by Tsingke Biotechnology Co., Ltd. (Beijing, China). There were at least 3 samples in each group and the detection was repeated 3 times. The 2-ΔΔCT method was used to calculate the relative content of each gene.
Molecular docking:
Docking molecular of Magnolol to the target receptors was explored by Cavity-detection guided Blind Docking (CB-Dock) (http://cao.labshare.cn/cb-dock/)[29]. Among the targets of Magnolol and asthma, along with DEGs, the common genes Arginase 1 (ARG1), Phosphodiesterase 4B (PDE4B), Signal Transducer and Activator of Transcription 1 (STAT1) and MMP12 were selected for docking. The molecular structure of Magnolol was downloaded from the traditional Chinese medicine systems pharmacology database and the protein structures were downloaded from the research collaboration for structural bioinformatics, protein data bank (https://www.rcsb.org/search)[30]. At last, these files were uploaded and submitted to the CB-Dock for visualization.
Data processing and statistical analysis:
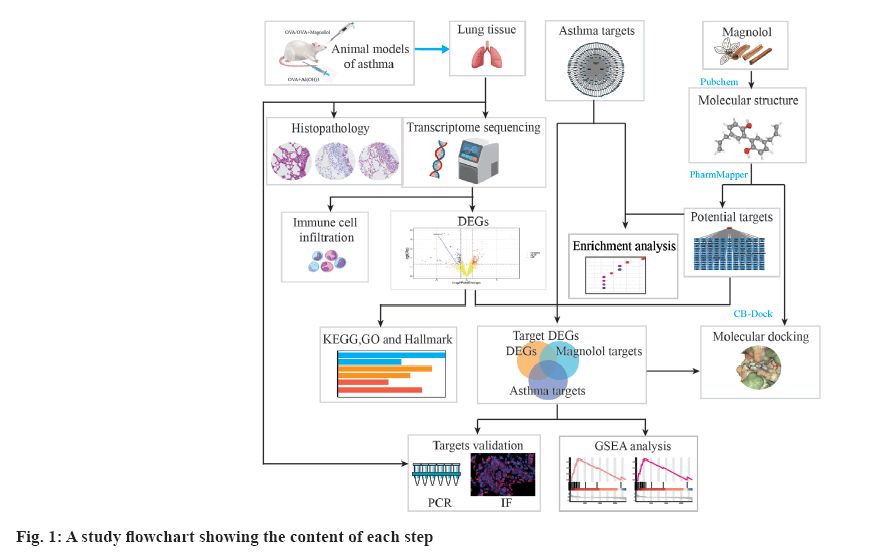
R language 4.1.0 software was used for data analysis, and Statistical Package for Social Sciences 18.0 software was used for data statistics. Measurement data were expressed as mean±standard error of mean, and the homogeneity of variance was compared by least significant difference-t test and the comparison among multiple groups was analyzed by one-way analysis of variance. Inspection level α=0.05. The R packages were used to filter and plot the data (fig. 1).
Results and Discussion
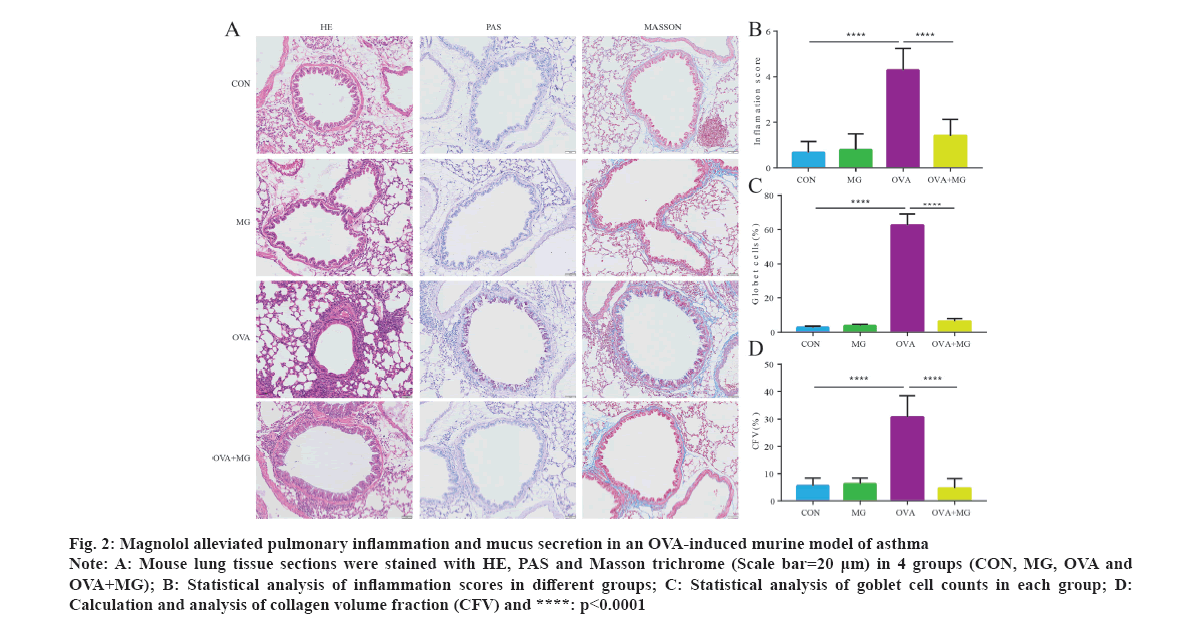
HE staining of the mice lung tissue shows the pathological conditions of bronchial mucosal epithelium in different groups (fig. 2A). In the CON and MG groups, the bronchial epithelial structure remains intact, with minimal infiltration of inflammatory cells. However, in the OVA group, there is significant deformation and detachment of the bronchial mucosal epithelium, accompanied by a large infiltration of inflammatory cells, mainly eosinophils. Additionally, congestion of the alveolar walls and a reduction in alveolar spaces are observed. The inflammatory cells in the OVA+MG group were significantly attenuated, leading to a significant decrease in airway inflammation compared to the OVA group. To evaluate the extent of pulmonary fibrosis, Masson staining was conducted to visualize collagen deposition. In the transverse section of mice lung, a substantial amount of blue collagen was observed around the large-sized bronchi in the OVA group. But collagen deposition was considerably less pronounced after treatment with Magnolol. PAS staining was primarily used to detect glycogen deposition, which indicates excessive mucus secretion. After PAS staining, glycogen in lung epithelial cells appeared as purple-red and was located in the cytoplasm. The number of PAS-positive cells was determined through staining, revealing a strong presence of intracytoplasmic mucus in the OVA group. The differences between the OVA group and OVA+MG group are significant (p<0.05) (fig. 2B-fig. 2D).
Fig 2: Magnolol alleviated pulmonary inflammation and mucus secretion in an OVA-induced murine model of asthma
Note: A: Mouse lung tissue sections were stained with HE, PAS and Masson trichrome (Scale bar=20 μm) in 4 groups (CON, MG, OVA and OVA+MG); B: Statistical analysis of inflammation scores in different groups; C: Statistical analysis of goblet cell counts in each group; D: Calculation and analysis of collagen volume fraction (CFV) and ****: p<0.0001
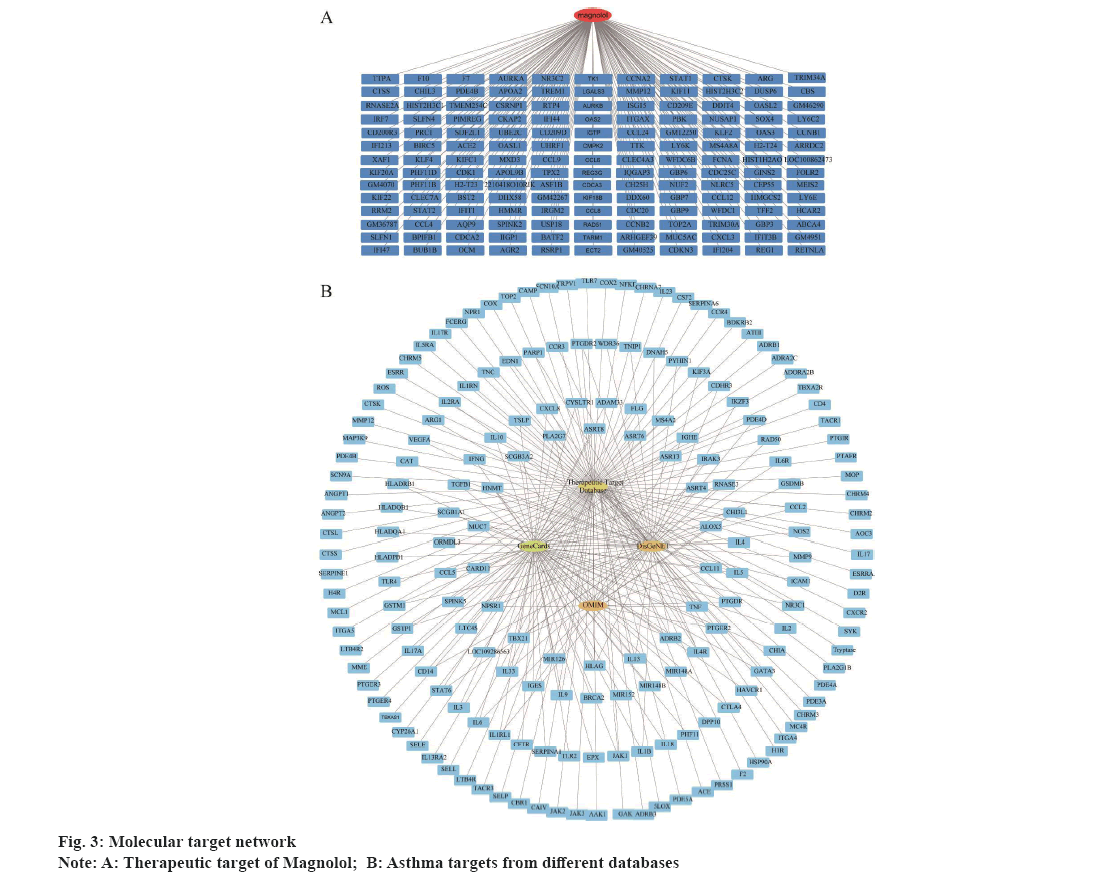
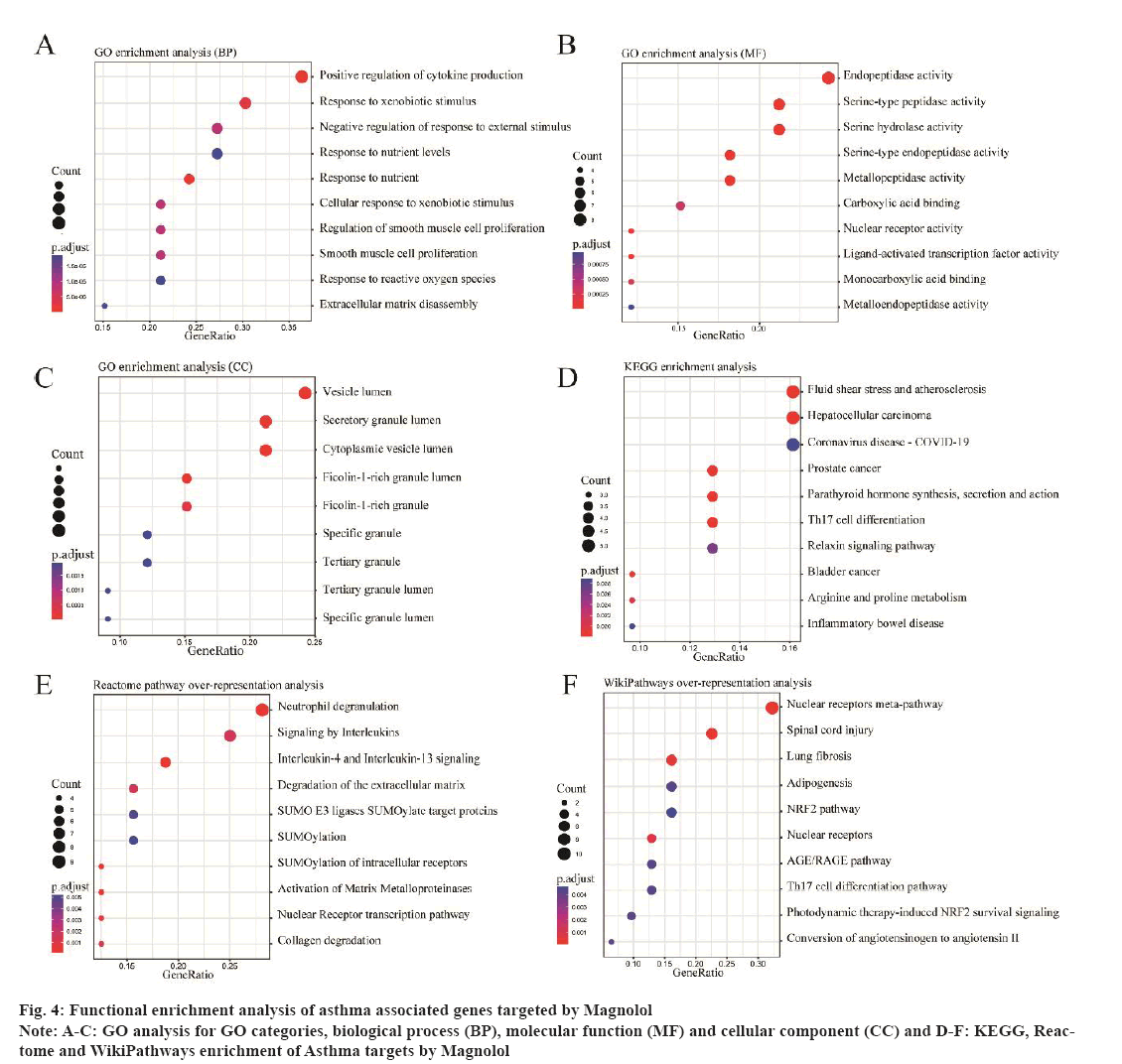
To predict protein targets of Magnolol, the three-Dimensional (3D) structure of Magnolol was obtained from PubChem database and uploaded to PharmMapper database. Drug target identification is an essential initial stage in the process of drug discovery. It involves the utilization of various algorithms to identify genes and proteins. By uploading the molecular information of the 3D structure and specifying optional parameters, the website generates a list of results that match the target pharmacophore based on the input molecule and the specified filter parameters[31]. As a result, a total of 286 protein targets were obtained (fig. 3A). The top 10 most fitting targets for Magnolol were shown in Table 1. Genes associated with asthma were screened in GeneCards, Therapeutic Target Database, DisGeNET and OMIM databases, resulting in 260 asthma-related genes (fig. 3B). By intersecting the targets of Magnolol with all the asthma related genes, 33 co-targets were identified. In addition, GO enrichment analysis of the 33 genes suggested these genes were significantly association with Positive regulation of cytokine production, Endopeptidase activity and Serine-type peptidase activity (fig. 4A-fig. 4C). In the meantime, pathway analysis demonstrated that these co-target genes are closely linked to Th17 cell differentiation, neutrophil degranulation and signaling by interleukins pathways (fig. 4D-fig. 4F).
| Target Name | Symbol | Pharma Model | Norm Fit |
|---|---|---|---|
| Aldo-keto reductase family 1 member C2 | AKR1C2 | 1j96_v | 1 |
| Steryl-sulfatase | STS | 1p49_v | 0.9998 |
| Apolipoprotein A-II | APOA2 | 1l6l_v | 0.9984 |
| Peptidyl-prolyl cis-trans isomerase A | PPIA | 1w8l_v | 0.9982 |
| Carbonic anhydrase 2 | CA2 | 1if4_v | 0.996 |
| Proto-oncogene serine/threonine-protein kinase Pim-1 | PIM1 | 3bgp_v | 0.9937 |
| Serum albumin | ALB | 1e7e_v | 0.9929 |
| Glutathione S-transferase P | GSTP1 | 1lbk_v | 0.9926 |
| Androgen receptor | AR | 2pir_v | 0.9924 |
| Carbonic anhydrase 1 | CA1 | 1czm_v | 0.9919 |
Table 1: Top 10 Magnolol Targets From Phammapper
To further explore the relationship between asthma and Magnolol targets, lung tissue from CON group, MG group, OVA group and OVA+MG group were applied for RNA-seq. And to systematically evaluate the transcriptomic alterations in the asthmatic mice treated with Magnolol, we applied differential expression analysis to identify the aberrantly expressed genes between OVA and OVA+MG group (|Log2 fold change|>1, p value<0.05). A total of 403 DEGs were identified, with 217 up-regulated genes and 186 down-regulated genes. Additionally, immune cells infiltration analysis was performed (fig. 5A). The score of eosinophils, activated mast cells resting dendritic cells and M1 macrophages was significantly lower in OVA+MG group compared to the OVA group. In contrast, the levels of memory B cells, Plasma cells and activated CD4 T cells were higher in OVA+MG group than those in OVA group. Furthermore, GSEA analysis was performed (fig. 5B-fig. 5D), which revealed that the enrichment scores of Interferon (IFN) gamma response and IFN alpha response in OVA+MG group were remarkably enhanced compared to the OVA group. Conversely, the enrichment scores of cell cycle, DNA replication, ribosome and G2M checkpoint in OVA group were significantly higher than those in OVA+MG group.
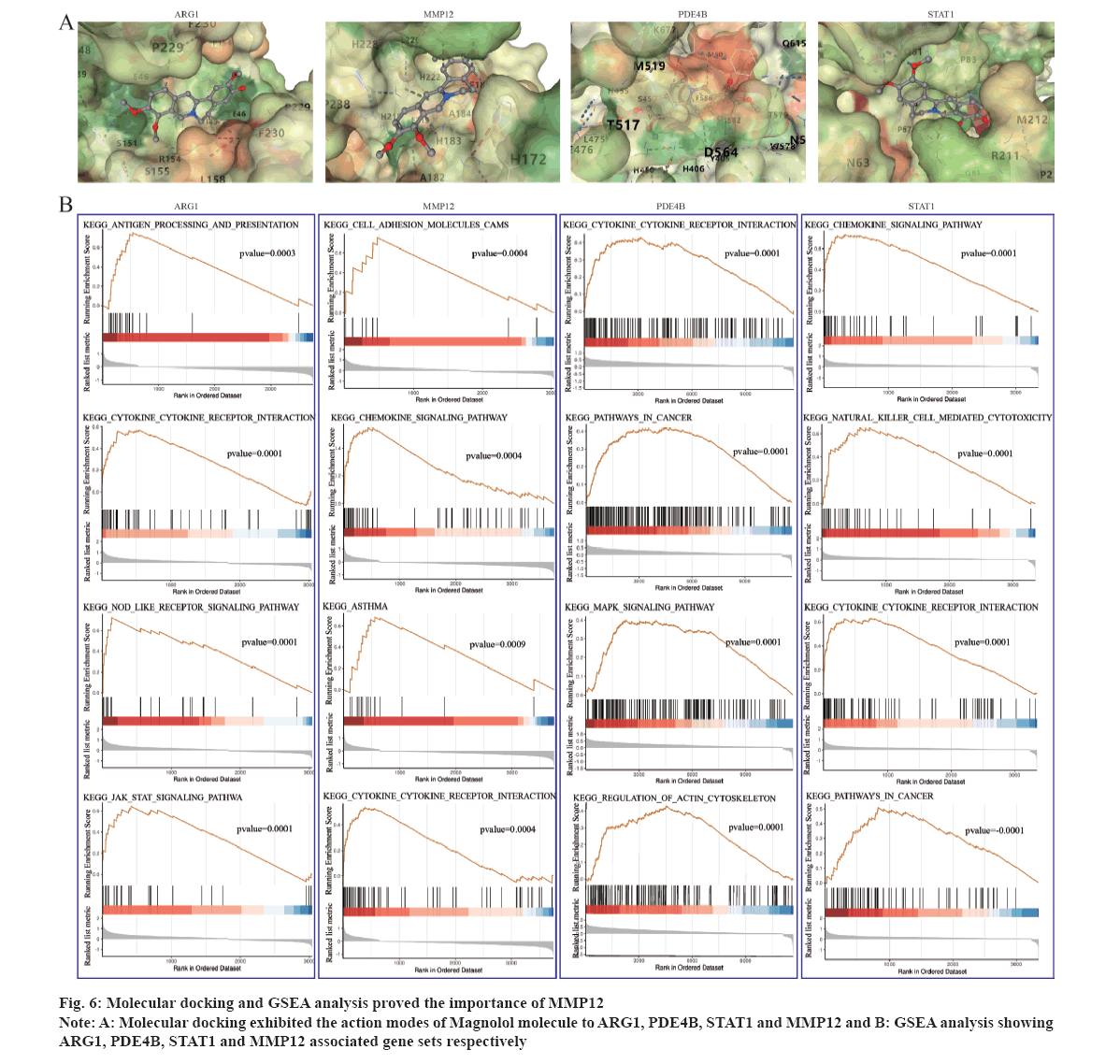
Previous sections have identified the therapeutic targets of Magnolol and asthma, as well as the DEGs through transcriptome sequencing. To investigate the potential of Magnolol in asthma treatment, combined analysis was conducted to identify the common genes among Magnolol target genes, asthma target genes and DEGs. As a result, ARG1, MMP12, STAT1 and PDE4B were identified as potential therapeutic targets of asthma by Magnolol. Molecular docking, a commonly used molecular modeling tool, was employed to explore the interactions between Magnolol and these core target genes. Specifically, ARG1, MMP12, STAT1 and PDE4B were selected for molecular docking verification. As shown in fig. 6A, the binding of Magnolol to these key targets can be seen through the analysis of hydrogen bonds and their binding sites. The logarithms of the free binding energy of Magnolol-ARG1, Magnolol-PDE4B, Magnolol-STAT1 and Magnolol-MMP12 calculated by CB-Dock were -6, -7, -6.4 and -7.1 kcal/ mol (Table 2). Therefore, this work found that MMP12 binds most closely to Magnolol molecule.
| Gene symbol | Protein name | PDP | Ligand | Average binding score |
|---|---|---|---|---|
| ARG1 | Arginase 1 | 4d9o | Magnolol | -6 |
| PDE4B | Phosphodiesterase 4B | 4myq | Magnolol | -7 |
| MMP12 | Matrix metallopeptidase 12 | 1os9 | Magnolol | -7.1 |
| STAT1 | Signal transducer and activator of transcription 1 | 4u2x | Magnolol | -6.4 |
Table 2: Molecular Docking Score
Then, a further GSEA pathway enrichment analysis was performed to understand the function of these four potential therapeutic targets using 59 normal lung samples from the Cancer Genome Atlas (TCGA) (fig. 6B). The result showed that high expression of ARG1 was correlated with cytokine-cytokine receptor interaction, Janus kinase (JAK)/STAT signaling pathway, nucleotide-binding and oligomerization domain like receptor signaling pathway and antigen processing and presentation. For MMP12, high expression was found to be involved in cell adhesion molecules, asthma, cytokine-cytokine receptor interaction and chemokine signaling pathway. As to STAT1, it was associated with natural killer cell mediated cytotoxicity, pathways in cancer, cytokine-cytokine receptor interaction and chemokine signaling pathway. PDE4B displayed correlation with Mitogen-Activated Protein Kinase (MAPK) signaling pathway, pathways in cancer, regulation of actin cytoskeleton and cytokine-cytokine receptor interaction pathway. All of these four genes were found to be related to immune responses and cytokine response, suggesting their important position as the therapeutic targets in asthma. And MMP12 was directly related to Asthma pathway, indicating its potential as an important target gene for Magnolol treatment of asthma.
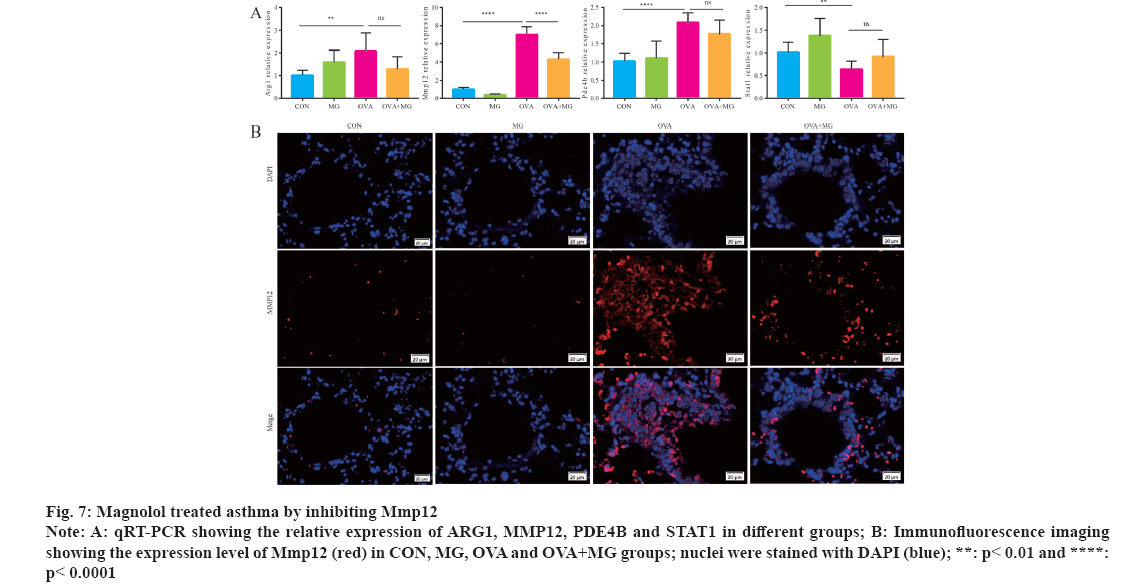
Next, the transcript and protein levels of the four potential targets in mice were validated by using qRT-PCR and immunofluorescence. For transcript levels, the results demonstrated that MMP12 expression significantly increased in OVA group, but decreased with Magnolol treatment (fig. 7A). Further analysis, tissue immunofluorescence exhibited that the expressions of MMP12 in the OVA group were higher than those in the CON group. In contrast, a predominant decrease in MMP12 protein was observed in the OVA+MG group (fig. 7B). These findings suggest that MMP12 plays a crucial role in the treatment of asthma with Magnolol.
Fig 7: Magnolol treated asthma by inhibiting Mmp12
Note: A: qRT-PCR showing the relative expression of ARG1, MMP12, PDE4B and STAT1 in different groups; B: Immunofluorescence imaging showing the expression level of Mmp12 (red) in CON, MG, OVA and OVA+MG groups; nuclei were stained with DAPI (blue); **: p< 0.01 and ****: p< 0.0001
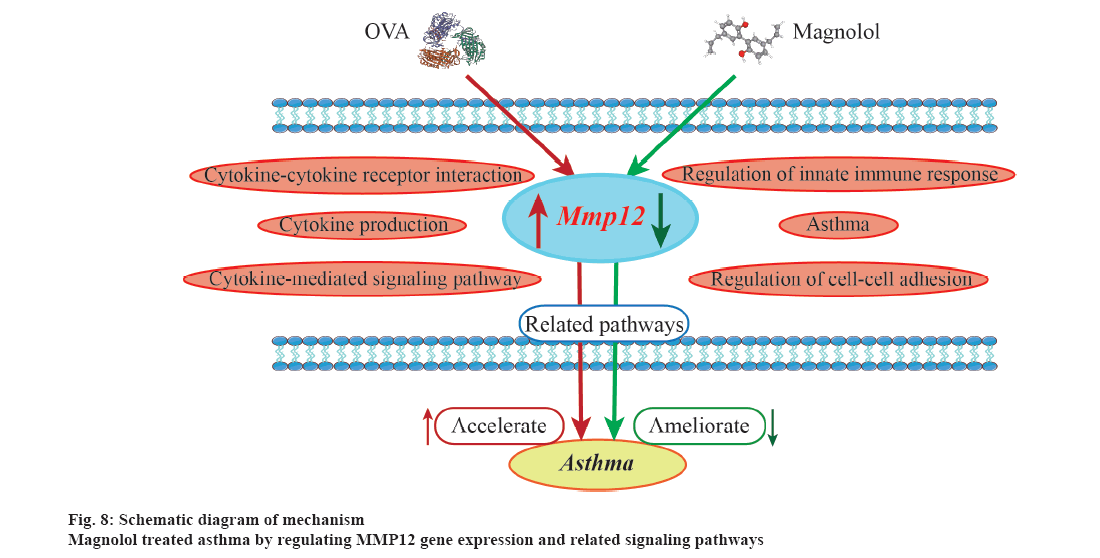
Asthma management has become a major public health concern worldwide due to its high prevalence and poor control[32]. At present, there are certain shortcomings in the available treatment options of asthma. New therapies to improve airway inflammation deserve further investigation. The active ingredients of traditional Chinese medicine are natural products with low toxicity and diverse sources, processed unique anti-inflammatory properties[33]. Previous animal models and in vitro studies have showed that Magnolol significantly ameliorate airway inflammation in asthma[21,22]. But, the exact gene targets of the anti-inflammatory effects of Magnolol have not been reported. Therefore, it is of great significance to establish a mouse model of Magnolol-treated OVA-induced asthma to study the targets of Magnolol and lay the foundation for subsequent drug research and development (fig. 8).
Network pharmacology provides an effective way to studying the complex mechanisms between drugs, targets and diseases from the perspective of biomolecule networks. This approach has several advantages compared to traditional drug discovery methods in the success rate, efficiency, and predictive ability[34,35]. In brief, Network pharmacology can comprehensively understand the pathogenesis of diseases by considering the multi-target characteristics of diseases; reduce costs and side effects associated with new drug development; optimize drug design and delivery strategies. Through the analysis of transcriptome gene sequencing and network pharmacology, novel strategies for discovering gene targets of Magnolol in the treatment of asthma can be realized.
Consequently, Magnolol treatment obviously attenuates airway inflammation, mucus production and subepithelial fibrillar collagen deposition in OVA-induced asthmatic mice. The improvement of airway inflammation was consistent with the results of other study[22]. Additionally, this study identified changes in gene expression and diverse pathways. The analysis of therapeutic targets of Magnolol on asthma revealed a strong association with Neutrophil degranulation and Cytokine positive regulation pathways. Besides, Hallmark analysis of DEGs showed that the IFN-α and IFN-γ response pathways were down-regulated in the OVA group. This down-regulation could activate the JAK-STAT signaling pathway after binding to the receptor, playing a crucial role in anti-virus and immune regulation[36]. It is worth noting that viral infection, particularly rhinovirus, are known to be one of the main triggers of acute asthma exacerbations, attributed to about 90 % of exacerbations in both adults and children[37]. Hence, the down-regulation of IFN-α and IFN-γ response pathways suggests that asthma patients themselves might be at risk of exacerbation.
Furthermore, four genes, ARG1, MMP12, STAT1 and PDE4B have been identified as potential therapeutic targets for asthma by Magnolol treatment. Understanding the function and mechanism of these genes may provide a rationale for using specific inhibitors in the treatment of asthma. ARG1 primarily response for metabolizing the amino acid L-arginine to produce urea and ornithine in the liver[38-40]. However, during infection or tissue injury caused by inflammation, immune cells play a critical role in ARG1 enzyme activity outside the liver[41-43]. ARG1 is a key feature of alternatively activated macrophages, which can recruit fibroblasts to synthesize collagen and promotes tissue fibrosis at the wound site[44-46]. Research has also shown that Arg1 regulates the metabolism of group 2 innate lymphoid cells and the development of type 2 inflammation[47]. These findings suggest a link between Arg1 and the inflammation and fibrosis in asthma.
Phosphodiesterase 4B (PDE4B) is a member of the phosphodiesterase protein family, which plays an important role in the breakdown of intracellular cyclic Adenosine Monophosphate (cAMP)[48]. In recent years, several PDE4B inhibitors have been developed as therapeutic agents due to their demonstrated anti-inflammatory and anti-fibrotic properties[49,50]. In a placebo-controlled trial, researchers investigated the effectiveness and safety of BI1015550, an oral preferential inhibitor of the PDE4B subtype, in patients with idiopathic pulmonary fibrosis. The results showed that treatment with BI1015550 prevented a decline in lung function among patients with idiopathic pulmonary fibrosis (NCT04419506)+. Similarly, a transcription factor, STAT1 is primarily activated by IFNs, growth factors (such as epidermal growth factor and platelet-derived growth factor), or oxidative stress[52-54]. STAT1 is inducted as a protective response to injury and is responsible for activating transcription of key genes involved in cell viability, growth arrest, apoptosis and differentiation[55,56]. In a study, OVA-sensitized STAT1-/- mice that were subsequently exposed to Multi-Walled Carbon Nanotubes (MWCNTs) via oropharyngeal aspiration developed a significant higher degree of airway fibrosis than compared to STAT1+/+ mice. This suggests that STAT1 plays a critical role in directing the immune response to MWCNTs and allergens[57]. Although the difference was not statistically significant, treatment with Magnolol resulted in increased expression of STAT1 and decreased level of ARG1, PDE4B in asthmatic mice. This suggests a potential relationship between Magnolol and these gene targets.
In this study, MMP12 was identified, whose expression was significantly decreased in asthma model and was restored by Magnolol treatment. MMP12 can be targeted for the treatment of asthma, as it is capable of degrading fibrillar collagen[58]. MMP12 encodes the gene macrophage elastase, which is involved in the breaking down of the extracellular matrix and associated with several inflammatory lung diseases including asthma, chronic obstructive pulmonary disease and idiopathic fibrosis[59,60]. A variant MMP12 has been found to be associated with disease severity in a preclinical study involving children and young adults. Animal models of allergic asthma have shown that inhibiting of MMP12 can reduce early and late phase airway responses to inhaled allergens[59]. In a recent animal study, the MMP12 inhibitor FP-025 significantly reduced inflammatory infiltrates in HDM-induced asthmatic mice in a dose-dependent manner[61]. Although the current phase IIα clinical trial (NCT03858686) of MMP12 inhibitors in asthma did not demonstrate clinical efficacy[62]. Targeting MMP12 still has huge potential in the treatment of lung diseases and may provide further insights into the translation of MMP12 into clinical outcomes.
In this study, bio-information, network pharmacology and animal experiments were applied to explore the potential targets of Magnolol in the treatment of asthma. Magnolol is found to significantly ameliorate airway inflammation by reducing the number of inflammatory cells, the grades of fibrosis and the precipitation of glycogen. By intersecting targets of asthma, targets of Magnolol and DEGs, four common genes ARG1, MMP12, STAT1 and PDE4B were identified potential therapeutic targets of Magnolol in the treatment of asthma. Further molecular docking and GSEA pathway analysis provided additional confirmation of MMP12 as a gene of greater significance, which was also supported by tissue qRT-PCR and immunofluorescence analysis. In short, this study demonstrated that Magnolol seem to be a promising candidate for therapeutic treatment of asthma and MMP12 is vital target for further research.
Acknowledgements:
Data could be obtained upon request. This work was supported by the National Natural Science Foundation of China (81970026, 82000029, 82200079), Natural Science Foundation of Sichuan (2022NSFCS1324), Chengdu High-level Key Clinical Specialty Construction Project (ZX20201202020), Chengdu Science and Technology Bureau (2021-YF09-00102-SN, 2020-YF05-00003-SN). The authors declare that they have no conflict of interest.
Ethics statement:
The animal experimental procedures were reviewed and approved by the Animal Ethic Committee at Southwest Jiaotong University.
Author’s contribution:
L. Luo, J. Y. Wang and A. Y. Xiong contributed equally to this work.
Conflict of interest:
The authors declared no conflict of interests.
References
- Huang K, Yang T, Xu J, Yang L, Zhao J, Zhang X, et al. Prevalence, risk factors, and management of asthma in China: A national cross-sectional study. Lancet 2019;394(10196):407-18.
[Crossref] [Google Scholar] [PubMed]
- Vos T, Lim SS, Abbafati C, Abbas KM, Abbasi M, Abbasifard M, et al. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2020;396(10258):1204-22.
[Crossref] [Google Scholar] [PubMed]
- Chang D, Hunkapiller J, Bhangale T, Reeder J, Mukhyala K, Tom J, et al. A whole genome sequencing study of moderate to severe asthma identifies a lung function locus associated with asthma risk. Sci Rep 2022;12(1):5574.
- Vicente CT, Revez JA, Ferreira MA. Lessons from ten years of genome?wide association studies of asthma. Clin Transl Immunol 2017;6(12):e165.
[Crossref] [Google Scholar] [PubMed]
- Demenais F, Margaritte-Jeannin P, Barnes KC, Cookson WO, Altmüller J, Ang W, et al. Multiancestry association study identifies new asthma risk loci that colocalize with immune-cell enhancer marks. Nat Genet 2018;50(1):42-53.
[Crossref] [Google Scholar] [PubMed]
- Woods CP, Argese N, Chapman M, Boot C, Webster R, Dabhi V, et al. Adrenal suppression in patients taking inhaled glucocorticoids is highly prevalent and management can be guided by morning cortisol. Eur J Endocrinol 2015;173(5):633-42.
[Crossref] [Google Scholar] [PubMed]
- Schwartz RH, Neacsu O, Ascher DP, Alpan O. Moderate dose inhaled corticosteroid-induced symptomatic adrenal suppression: Case report and review of the literature. Clin Pediatr 2012;51(12):1184-90.
[Crossref] [Google Scholar] [PubMed]
- Ye Q, He XO, D’Urzo A. A review on the safety and efficacy of inhaled corticosteroids in the management of asthma. Pulm Ther 2017;3:1-8.
- Rahman MM, Bibi S, Rahaman MS, Rahman F, Islam F, Khan MS, et al. Natural therapeutics and nutraceuticals for lung diseases: Traditional significance, phytochemistry, and pharmacology. Biomed Pharmacother 2022;150:113041.
[Crossref] [Google Scholar] [PubMed]
- Xin WA, Zi-Yi WA, Zheng JH, Shao LI. TCM network pharmacology: A new trend towards combining computational, experimental and clinical approaches. Chin J Nat Med 2021;19(1):1-11.
[Crossref] [Google Scholar] [PubMed]
- Zhang R, Zhu X, Bai H, Ning K. Network pharmacology databases for traditional Chinese medicine: Review and assessment. Front Pharmacol 2019;10:123.
[Crossref] [Google Scholar] [PubMed]
- Shao LI, Zhang B. Traditional Chinese medicine network pharmacology: Theory, methodology and application. Chin J Nat Med 2013;11(2):110-20.
[Crossref] [Google Scholar] [PubMed]
- Csermely P, Korcsmáros T, Kiss HJ, London G, Nussinov R. Structure and dynamics of molecular networks: A novel paradigm of drug discovery: A comprehensive review. Pharmacol Ther 2013;138(3):333-408.
[Crossref] [Google Scholar] [PubMed]
- Xin WA, Zi-Yi WA, Zheng JH, Shao LI. TCM network pharmacology: A new trend towards combining computational, experimental and clinical approaches. Chin J Nat Med 2021;19(1):1-11.
[Crossref] [Google Scholar] [PubMed]
- Luo TT, Lu Y, Yan SK, Xiao X, Rong XL, Guo J. Network pharmacology in research of Chinese medicine formula: Methodology, application and prospective. Chin J Integr Med 2020;26(1):72-80.
[Crossref] [Google Scholar] [PubMed]
- An L, Feng F. Network pharmacology-based antioxidant effect study of Zhi-Zi-Da-Huang decoction for alcoholic liver disease. Evid Based Complement Alternat Med 2015;2015:492470.
[Crossref] [Google Scholar] [PubMed]
- Zhang L, He X, Xiong Y, Ran Q, Xiong A, Wang J, et al. Transcriptome-wide profiling discover: PM2. 5 aggravates airway dysfunction through epithelial barrier damage regulated by Stanniocalcin 2 in an OVA-induced model. Ecotoxicol Environ Saf 2021;220:112408.
[Crossref] [Google Scholar] [PubMed]
- Akula N, Marenco S, Johnson K, Feng N, Zhu K, Schulmann A, et al. Deep transcriptome sequencing of subgenual anterior cingulate cortex reveals cross-diagnostic and diagnosis-specific RNA expression changes in major psychiatric disorders. Neuropsychopharmacol 2021;46(7):1364-72.
- Akula N, Marenco S, Johnson K, Feng N, Zhu K, Schulmann A, et al. Deep transcriptome sequencing of subgenual anterior cingulate cortex reveals cross-diagnostic and diagnosis-specific RNA expression changes in major psychiatric disorders. Neuropsychopharmacol 2021;46(7):1364-72.
- Tang Y, Wang L, Yi T, Xu J, Wang J, Qin JJ, et al. Synergistic effects of autophagy/mitophagy inhibitors and magnolol promote apoptosis and antitumor efficacy. Acta Pharm Sin B 2021;11(12):3966-82.
[Crossref] [Google Scholar] [PubMed]
- Huang Q, Han L, Lv R, Ling L. Magnolol exerts anti-asthmatic effects by regulating Janus kinase-signal transduction and activation of transcription and Notch signaling pathways and modulating Th1/Th2/Th17 cytokines in ovalbumin-sensitized asthmatic mice. Korean J Physiol Pharmacol 2019;23(4):251-61.
[Crossref] [Google Scholar] [PubMed]
- Wang J, Xian M, Cao H, Wu L, Zhou L, Ma Y, et al. Prophylactic and therapeutic potential of magnolol-loaded PLGA-PEG nanoparticles in a chronic murine model of allergic asthma. Front Bioeng Biotechnol 2023;11:1182080.
[Crossref] [Google Scholar] [PubMed]
- He X, Zhang L, Xiong A, Ran Q, Wang J, Wu D, et al. PM2. 5 aggravates NQO1-induced mucus hyper-secretion through release of neutrophil extracellular traps in an asthma model. Ecotoxicol Environ Saf 2021;218:112272.
[Crossref] [Google Scholar] [PubMed]
- Santos JH, Quimque MT, Macabeo AP, Corpuz MJ, Wang YM, Lu TT, et al. Enhanced oral bioavailability of the pharmacologically active lignin magnolol via Zr-based metal organic framework impregnation. Pharmaceutics 2020;12(5):437.
[Crossref] [Google Scholar] [PubMed]
- Hattori M, Endo Y, Takebe S, Kobashi K, Fukasaku N, Namba T. Metabolism of Magnolol from magnoliae cortex. II.: Absorption, metabolism and excretion of [ring-14C] Magnolol in Rats. Chem Pharm Bull 1986;34(1):158-67.
[Crossref] [Google Scholar] [PubMed]
- Yunhe F, Bo L, Xiaosheng F, Fengyang L, Dejie L, Zhicheng L, et al. The effect of magnolol on the toll-like receptor 4/nuclear factor kappa B signaling pathway in lipopolysaccharide-induced acute lung injury in mice. Eur J Pharmacol 2012;689(1-3):255-61.
[Crossref] [Google Scholar] [PubMed]
- Wang X, Pan C, Gong J, Liu X, Li H. Enhancing the enrichment of pharmacophore-based target prediction for the polypharmacological profiles of drugs. J Chem Inf Model 2016;56(6):1175-83.
[Crossref] [Google Scholar] [PubMed]
- Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol 2018;1711:243-59.
[Crossref] [Google Scholar] [PubMed]
- Liu Y, Grimm M, Dai WT, Hou MC, Xiao ZX, Cao Y. CB-Dock: A web server for cavity detection-guided protein–ligand blind docking. Acta Pharmacol Sin 2020;41(1):138-44.
- Burley SK, Berman HM, Christie C, Duarte JM, Feng Z, Westbrook J, et al. RCSB Protein Data Bank: Sustaining a living digital data resource that enables breakthroughs in scientific research and biomedical education. Protein Sci 2018;27(1):316-30.
[Crossref] [Google Scholar] [PubMed]
- Wang X, Shen Y, Wang S, Li S, Zhang W, Liu X, et al. PharmMapper 2017 update: A web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res 2017;45(W1):W356-60.
[Crossref] [Google Scholar] [PubMed]
- Masoli M, Fabian D, Holt S, Beasley R, Global Initiative for Asthma (GINA) Program. The global burden of asthma: Executive summary of the GINA Dissemination Committee report. Allergy 2004;59(5):469-78.
[Crossref] [Google Scholar] [PubMed]
- Wang W, Yao Q, Teng F, Cui J, Dong J, Wei Y. Active ingredients from Chinese medicine plants as therapeutic strategies for asthma: Overview and challenges. Biomed Pharmacother 2021;137:111383.
[Crossref] [Google Scholar] [PubMed]
- Wu X, Jiang R, Zhang MQ, Li S. Network?based global inference of human disease genes. Mol Syst Biol 2008;4(1):189.
[Crossref] [Google Scholar] [PubMed]
- Zhang GB, Li QY, Chen QL, Su SB. Network pharmacology: A new approach for Chinese herbal medicine research. Evid Based Complement Alternat Med 2013;2013:621423.
[Crossref] [Google Scholar] [PubMed]
- Hosmillo M, Chaudhry Y, Nayak K, Sorgeloos F, Koo BK, Merenda A, et al. Norovirus replication in human intestinal epithelial cells is restricted by the interferon-induced JAK/STAT signaling pathway and RNA polymerase II-mediated transcriptional responses. MBio 2020;11(2):10-128.
- Mikhail I, Grayson MH. Asthma and viral infections: An intricate relationship. Ann Allergy Asthma Immunol 2019;123(4):352-8.
[Crossref] [Google Scholar] [PubMed]
- Rath M, Müller I, Kropf P, Closs EI, Munder M. Metabolism via arginase or nitric oxide synthase: Two competing arginine pathways in macrophages. Front Immunol 2014;5:532.
[Crossref] [Google Scholar] [PubMed]
- Thomas AC, Mattila JT. “Of mice and men”: Arginine metabolism in macrophages. Front Immunol 2014;5:479.
[Crossref] [Google Scholar] [PubMed]
- Murray PJ. Amino acid auxotrophy as a system of immunological control nodes. Nat Immunol 2016;17(2):132-9.
[Crossref] [Google Scholar] [PubMed]
- Raber P, Ochoa AC, Rodríguez PC. Metabolism of L-arginine by myeloid-derived suppressor cells in cancer: Mechanisms of T cell suppression and therapeutic perspectives. Immunol Invest 2012;41(6-7):614-34.
[Crossref] [Google Scholar] [PubMed]
- Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature 2013;496(7446):445-55.
[Crossref] [Google Scholar] [PubMed]
- Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol 2011;11(11):723-37.
[Crossref] [Google Scholar] [PubMed]
- Gibbons MA, MacKinnon AC, Ramachandran P, Dhaliwal K, Duffin R, Phythian-Adams AT, et al. Ly6Chi monocytes direct alternatively activated profibrotic macrophage regulation of lung fibrosis. Am J Respir Crit Care Med 2011;184(5):569-81.
[Crossref] [Google Scholar] [PubMed]
- Rodriguez PC, Quiceno DG, Ochoa AC. L-arginine availability regulates T-lymphocyte cell-cycle progression. Blood 2007;109(4):1568-73.
[Crossref] [Google Scholar] [PubMed]
- Pesce JT, Ramalingam TR, Mentink-Kane MM, Wilson MS, El Kasmi KC, Smith AM, et al. Arginase-1–expressing macrophages suppress Th2 cytokine–driven inflammation and fibrosis. PLoS Pathog 2009;5(4):e1000371.
[Crossref] [Google Scholar] [PubMed]
- Monticelli LA, Buck MD, Flamar AL, Saenz SA, Tait Wojno ED, Yudanin NA, et al. Arginase 1 is an innate lymphoid-cell-intrinsic metabolic checkpoint controlling type 2 inflammation. Nat Immunol 2016;17(6):656-65.
[Crossref] [Google Scholar] [PubMed]
- Azam MA, Tripuraneni NS. Selective phosphodiesterase 4B inhibitors: A review. Sci Pharm 2014;82(3):453-82.
[Crossref] [Google Scholar] [PubMed]
- Phillips JE. Inhaled phosphodiesterase 4 (PDE4) inhibitors for inflammatory respiratory diseases. Front Pharmacol 2020;11:259.
[Crossref] [Google Scholar] [PubMed]
- Matsuhira T, Nishiyama O, Tabata Y, Kaji C, Kubota-Ishida N, Chiba Y, et al. A novel phosphodiesterase 4 inhibitor, AA6216, reduces macrophage activity and fibrosis in the lung. Eur J Pharmacol 2020;885:173508.
[Crossref] [Google Scholar] [PubMed]
- Richeldi L, Azuma A, Cottin V, Hesslinger C, Stowasser S, Valenzuela C, et al. Trial of a preferential phosphodiesterase 4B inhibitor for idiopathic pulmonary fibrosis. N Engl J Med 2022;386(23):2178-87.
[Crossref] [Google Scholar] [PubMed]
- Aaronson DS, Horvath CM. A road map for those who don't know JAK-STAT. Science 2002;296(5573):1653-5.
[Crossref] [Google Scholar] [PubMed]
- Simon AR, Takahashi S, Severgnini M, Fanburg BL, Cochran BH. Role of the JAK-STAT pathway in PDGF-stimulated proliferation of human airway smooth muscle cells. Am J Physiol Lung Cell Mol Physiol 2002;282(6):L1296-304.
[Crossref] [Google Scholar] [PubMed]
- Gorina R, Sanfeliu C, Galitó A, Messeguer À, Planas AM. Exposure of glia to pro?oxidant agents revealed selective Stat1 activation by H2O2 and Jak2?independent antioxidant features of the Jak2 inhibitor AG490. Glia 2007;55(13):1313-24.
[Crossref] [Google Scholar] [PubMed]
- Ramana CV, Gil MP, Han Y, Ransohoff RM, Schreiber RD, Stark GR. Stat1-independent regulation of gene expression in response to IFN-γ. Proc Natl Acad Sci USA 2001;98(12):6674-9.
[Crossref] [Google Scholar] [PubMed]
- Bonner JC. Mesenchymal cell survival in airway and interstitial pulmonary fibrosis. Fibrogenesis Tissue Repair 2010;3:1-4.
[Crossref] [Google Scholar] [PubMed]
- Thompson EA, Sayers BC, Glista-Baker EE, Shipkowski KA, Ihrie MD, Duke KS, et al. Role of signal transducer and activator of transcription 1 in murine allergen–induced airway remodeling and exacerbation by carbon nanotubes. Am J Respir Cell Mol Biol 2015;53(5):625-36.
[Crossref] [Google Scholar] [PubMed]
- Sprangers S, Everts V. Molecular pathways of cell-mediated degradation of fibrillar collagen. Matrix Biol 2019;75:190-200.
[Crossref] [Google Scholar] [PubMed]
- Mukhopadhyay S, Sypek J, Tavendale R, Gartner U, Winter J, Li W, et al. Matrix metalloproteinase-12 is a therapeutic target for asthma in children and young adults. J Allergy Clin Immunol 2010;126(1):70-6.
[Crossref] [Google Scholar] [PubMed]
- Dancer RC, Wood AM, Thickett DR. Metalloproteinases in idiopathic pulmonary fibrosis. Eur Respir J 2011;38(6):1461-7.
[Crossref] [Google Scholar] [PubMed]
- Ravanetti L, Dekker T, Guo L, Dijkhuis A, Dierdorp BS, Diamant Z, et al. Efficacy of FP-025: A novel matrix metalloproteinase-12 (MMP-12) inhibitor in murine allergic asthma. Allergy 2023;78(2):559-62.
[Crossref] [Google Scholar] [PubMed]
- Abd-Elaziz K, Jesenak M, Vasakova M, Diamant Z. Revisiting matrix metalloproteinase 12: Its role in pathophysiology of asthma and related pulmonary diseases. Curr Opin Pulm Med 2021;27(1):54-60.
[Crossref] [Google Scholar] [PubMed]