- *Corresponding Author:
- Zheng Wu
Department of Respiratory Medicine, The People’s Hospital of Hai’an, Hai’an, Jiangsu 226600, China
E-mail: wuzhen0137@163.com
| Date of Received | 25 January 2022 |
| Date of Revision | 30 October 2022 |
| Date of Acceptance | 10 August 2023 |
| Indian J Pharm Sci 2023;85(4):1178-1183 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The purpose of this research was to examine the effects of sirtuin 6 overexpression on the biological characteristics of A549 cells, which are a form of non-small cell lung cancer. In the laboratory setting, the upregulation of sirtuin 6 expression in A549 cells was accomplished by introducing an overexpression plasmid (plasmid cloning deoxyribonucleic acid 3.1-sirtuin 6). Subsequently, the expression of sirtuin 6 in the transfected A549 cells was assessed through both reverse transcription-polymerase chain reaction and Western blot analysis. Proliferation was evaluated using flow cytometry, while metastatic and invasive potential were examined using the Transwell cultivation system. Additionally, apoptosis was measured utilizing the terminal deoxynucleotidyl transferase dUTP nick end labeling assay. The results revealed a significant increase (p<0.05) in sirtuin 6 expression in the plasmid cloning deoxyribonucleic acid 3.1-sirtuin 6 group compared to the control and plasmid cloning deoxyribonucleic acid 3.1 groups. Moreover, the transfection of A549 cells resulted in suppressed proliferation and invasive ability, without any noticeable impact on cell apoptosis. In conclusion, these findings demonstrate that excessive sirtuin 6 expression impedes the growth and invasive capacity of A549 cells. Targeting sirtuin 6 may represent a promising therapeutic strategy for non-small cell lung cancer treatment.
Keywords
Non-small cell lung cancer, sirtuin 6, A549 cells, adenocarcinoma, Western blot
Lung carcinoma is a highly prevalent malignancy globally, ranking below only breast and prostate carcinoma. It stands as the primary contributor to cancer-related mortality. Typically, males have a higher incidence rate (13 %) compared to females (12 %)[1]. Non-Small Cell Lung Carcinoma (NSCLC) is predominantly comprised of two primary pathological types: adenocarcinoma and squamous cell carcinoma, which together account for approximately 85 % of lung cancer cases. In recent years, there has been a noteworthy rise in the incidence of lung adenocarcinoma, surpassing squamous cell carcinoma to become the predominant form of NSCLC[2]. Currently, the etiology of NSCLC is still not well understood[3,4]. The increasing comprehension of malignant tumors at the cellular, gene and molecular levels has led to a growing interest in the involvement of certain genes and their products in lung cancer[5-7]. The nucleus is where Sirtuin 6 (SIRT6) is mainly found in different types of cells[8]. SIRT6 controls the stability and repair of genomic Deoxyribonucleic Acid (DNA), transcription, aging, and inflammation reduction, as well as metabolism, by performing histone deacetylation[9-13]. Moreover, research findings have suggested that the protein SIRT6 is involved in the regulation of autophagy, a vital process for the survival of cancer cells[13,14]. Analysis of clinical samples from pancreatic cancer[15], colorectal cancer[16], head and neck squamous cell carcinoma has revealed a decrease in the expression of SIRT6[17]. Based on our previous investigation, it has been observed that the expression levels of SIRT6 are reduced in NSCLC. Furthermore, there is a significant correlation between the protein expression level of SIRT6 and the malignancy and prognosis[18]. However, the precise role of SIRT6 in NSCLC is still not fully understood. The objective of this study was to assess the impact of SIRT6 on the biological properties of NSCLC A549 cells. The present study obtained approval from the Research Ethics Committee of People's Hospital of Hai'an, ensuring compliance with ethical standards. A549 cells were obtained from the Shanghai Institute of Biochemistry and Cell Biology, which is situated in Shanghai, China. These cells were cultivated in a serum-based medium comprising of Dulbecco's Modified Eagle's Medium (DMEM) containing elevated glucose levels and 10 % fetal bovine serum. Subsequently, the cells were incubated at a temperature of 37° in an incubator with a controlled atmosphere consisting of 95 % air, 5 % Carbon dioxide (CO2), and optimal humidity. Oligobio (China) sold the plasmid cloning Deoxyribonucleic Acid (pcDNA)-3.1 and pcDNA3.1-SIRT6 plasmid. The SIRT 6 plasmid was transfected temporarily following the guidelines provided by Lipofectamine 2000 (Invitrogen, United States of America (USA)). The total Ribonucleic Acid (RNA) was extracted using the UNIQ-10 Spin Column RNA Purification Kit, which was obtained from Sangon in Shanghai, China. The synthesis of the initial complementary DNA (cDNA) strand was carried out using the RevertAidTM kit for first strand cDNA synthesis (Fermentas, Burlington, Canada). Subsequently, the initial cDNA strand was subjected to amplification using the Corbett RG-6000 Polymerase Chain Reaction (PCR) system (QIAGEN, Dusseldorf, Germany) and FastStart Universal SYBR Green Master Mix (Roche, Basel, Switzerland). To optimize the reactions, the annealing temperatures were varied within the range of 50°~55°. The sense and antisense primers used were synthesized as follows: Glyceraldehyde 3-Phosphate Dehydrogenase (GAPDH) 5’-GCAAGTTCAACGGCACAG-3’, 5’-GCCAGTAGACTCCACGACAT-3’; SIRT6 5’-CCCGGATCAACGGCTCTATC-3’ and 5’-GCCTTCACCCTTTTGGGGG-3’. Electrophoresis was performed using a 12 % Sodium Dodecyl Sulfate (SDS)-polyacrylamide gradient gel with 50 µg of total protein loaded per lane. Subsequently, the proteins were transferred onto nitrocellulose membranes (Millipore). The membranes were then washed with rinse buffer at room temperature and blocked with blocking buffer (5 % fat-free milk in rinse buffer) for 30 min. Following that, the membranes were incubated with primary rabbit-anti-SIRT6 antibody (1:100 dilution, Abclonal Biotechnology) for 2 h at room temperature. After rinsing with rinse buffer, the membranes were exposed to Horseradish Peroxidase (HRP)-conjugated secondary antibody (goat-anti-rabbit Immunoglobulin G (IgG), 1:1000 dilution, Santa Cruz) for 2 h at room temperature. The protein bands were visualized using enhanced chemiluminescence reagents (Amersham, Little Chalfont Buckinghamshireand United Kingdom (UK)). Beta (β)-actin was used as the reference protein. Optical densities of the protein bands were analyzed using ImageMaster™ 2D Platinum software (Version 5.0, Amersham Biosciences, Piscataway, New Jersey). The A549 cells, divided into three group’s including the control group consisting of non-transfected A549 cells, the mock group consisting of A549 cells transfected with a mock, and the knockdown group consisting of transfected A549 cells, were cultured in the previously mentioned medium. After 4 d in vitro, the cells were washed once with cold Phosphate Buffer Saline (PBS). Subsequently, the cells were treated with 70 % ethanol, followed by 100 mg/l RNase for 30 min, and stained with 50 mg/l Propidium Iodide (PI) (Sigma) for the same duration. Flow cytometry (Epics XL; Beckman Coulter, Fullerton California, USA) was utilized to analyze the cells. The PI value was calculated using the formula:
PI=(S+G2/M)/(G0/G1+S+G2/M)
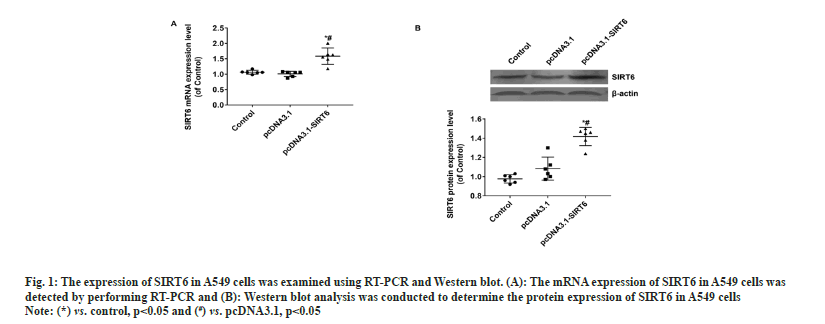
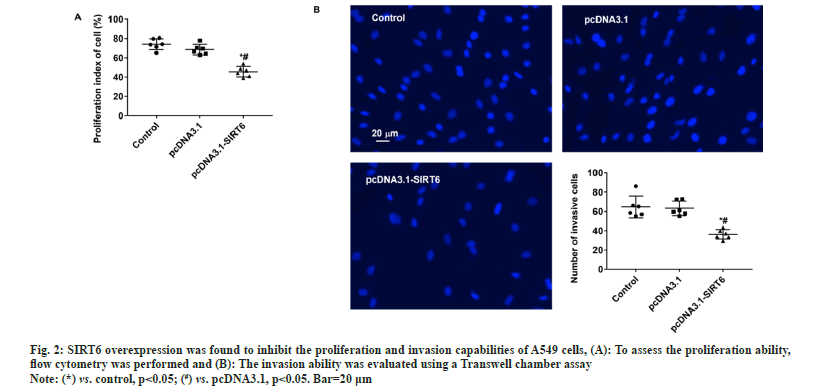
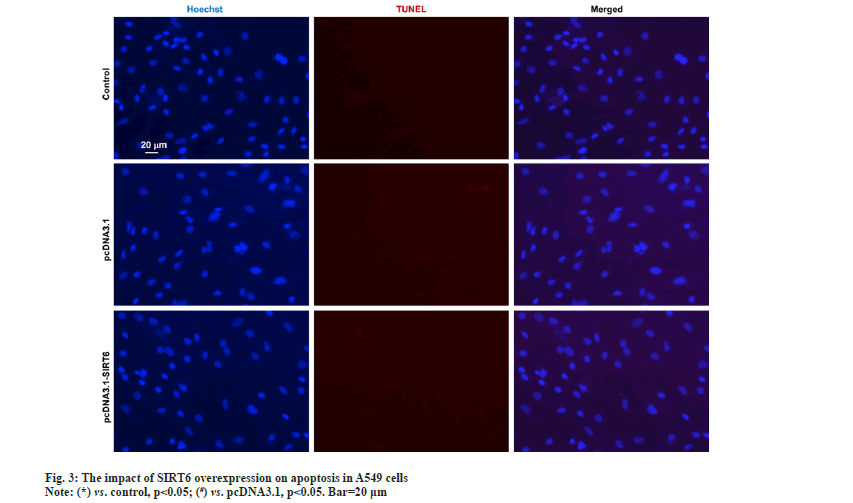
The upper chamber of the transwell cultivation system was filled with A549 cells from three groups at a concentration of 1×105 cells/ml. DMEM/F12 medium filled the lower chamber. Following a 12 h incubation, the cells were treated with 4 % Paraformaldehyde (PFA) for 30 min to fix them, and then stained with Hoechst 33342 at room temperature for another 30 min. Afterwards, the filters were washed completely using distilled water and examined under bright field microscopy to confirm the adherence and migration of the cells. Afterwards, the stationary cells were eliminated from the inner side by using a damp cotton swab. Hoechst 33342 was used to stain the transferred cells. The apoptosis of A549 cells was evaluated using the TUNEL kit (Beyotime Biotechnology, China) according to the manufacturer's instructions. The statistical analysis was performed utilizing Statistical Package for the Social Sciences (SPSS) 22.0, a software program designed for statistical analysis in the field of social sciences. The measurement data of the study were presented as mean±Standard Deviation (SD), and the groups were compared using a statistical method known as one-way Analysis of Variance (ANOVA). Statistical significance was determined for values with a significance level below 0.05. In the pcDNA3.1-SIRT6 group, the mRNA expression of SIRT6 in transfected A549 cells showed a significant increase of approximately 50 % compared to the control and pcDNA3.1 groups (p<0.05). Moreover, there was no statistically significant difference between the control and pcDNA3.1 groups (p>0.05) (fig. 1A). The protein expression of SIRT6 in A549 cells transfected with pcDNA3.1-SIRT6 also exhibited a notable elevation of approximately 40 % compared to the control and pcDNA3.1 groups (p<0.05). Similarly, there was no significant difference observed between the control and pcDNA3.1 groups (p>0.05) as shown in fig. 1B. The A549 cells from three different groups were cultured using the identical conditions described in the materials and methods section. Flow cytometry was used to measure the cell proliferation capacity of three different groups. In the pcDNA3.1-SIRT6 group, the PI of transfected A549 cells was found to be 45.62±5.44, whereas the control and pcDNA3.1 group exhibited PIs of 74.11±5.68 and 68.59±5.44 respectively. The PI of transfected A549 cells in the pcDNA3.1-SIRT6 group exhibited a significant decrease compared to the other two groups (p<0.05) as shown in fig. 2A. In order to examine the involvement of SIRT6 in the invasion of tumor cells, we assessed the capacity of A549 cells to penetrate matrigel-coated filters using a Transwell chamber assay. Following transfection, the number of invasive cells in the pcDNA3.1-SIRT6 group of transfected A549 cells significantly decreased in comparison to the other two groups (p<0.05). Nevertheless, there were no notable distinctions among the remaining two groups as shown in fig. 2B. The TUNEL results showed no presence of apoptotic cells in both the control and pcDNA3.1 groups. Furthermore, the pcDNA3.1-SIRT6 group also did not show any apoptotic cells (fig. 3). Therefore, the overexpression of SIRT6 had no impact on the apoptosis of A549 cells. Multiple initial lines of evidence strongly supported the hypothesis that SIRT6 functions as a tumor suppressor gene[14,19]. Studies on clinical samples obtained from patients with pancreatic cancer[20], colorectal cancer[16], and hepatocellular carcinomahave consistently shown a significant decrease in the expression levels of SIRT6[21]. However, some studies have also reported that SIRTR6 is highly expressed in diseases such as osteosarcoma[22], prostate cancer[23], and papillary thyroid cancer[24]. The results indicate that SIRT6 may have dual roles in various types of tumors. Studies in cancer cells level at the different cancer also show a similar duality effect. The study by Hang et al,[25] it was discovered that the inhibition of cell proliferation and invasion could be achieved by silencing the SIRT6 gene in Saos-2 and U2OS cell lines. The restoration of SIRT6 levels resulted in the recovery of these cellular biological behaviors in the cell lines. Additionally, their research indicated that SIRT6 could enhance the movement and infiltration of osteosarcoma cells via the ERK1/2/MMP9 pathway[22]. SIRT6 overexpression in the HepG2 cell line suppressed the growth of liver cancer by blocking the ERK1/2 signaling pathway and enhanced cell death by increasing the levels of activated C-caspase-3. The involvement of SIRT6 in cancer is intricate, and it might have diverse functions even within cancer cell lines originating from the identical tumor. In the case of breast cancer, SIRT6 has demonstrated a dual function of both promoting and suppressing tumor growth[26,27]. In their study, Xiong et al. found that the upregulation of SIRT6 can enhance the sensitivity of A549 cells to radiation and suppress tumor growth by reducing the activity of the PI3K/Akt/mTOR signaling pathway[28]. In another study involving NSCLC cell lines, it was observed that SIRT6 showed significant upregulation in NCI-H520, A549 and NCI-H460 cells compared to normal BEAS-2B cells. When SIRT6 was suppressed, cell proliferation decreased, leading to cell cycle arrest and the induction of apoptosis[29]. The findings regarding the relationship between SIRT6 and A549 cells appear to be contradictory. To validate the impact of SIRT6 in A549 cells, we enhanced the expression of SIRT6 in A549 cells using pcDNA3.1-SIRT6. Subsequently, we investigated the proliferation and invasion potential of the transfected A549 cells. Flow cytometry results revealed that the PI of transfected A549 cells in the pcDNA3.1-SIRT6 group exhibited a lower value compared to the other two groups. This observation suggests that overexpression of SIRT6 hindered the cell cycle and suppressed cell proliferation. The Transwell chamber assay results demonstrated a significant decrease in the number of invasive cells in the pcDNA3.1-SIRT6 group compared to the other two groups, indicating that overexpressing SIRT6 inhibited the invasion capability of A549 cells. The findings of Xiong et al.[28] are essentially in line with these results. Nevertheless, the TUNEL outcomes revealed the absence of any apoptotic cells in the 3 groups, suggesting that the overexpression of SIRT6 did not impact the apoptosis of A549 cells. Further verification of these findings and their related mechanisms is required through follow-up studies. Furthermore, the study did not investigate the associated signaling mechanisms, which will be examined in subsequent research. To summarize, our research results suggest that an overexpression of SIRT6 has a negative impact on the growth and invasive capabilities of A549 cells. Therefore, targeting SIRT6 might be a potential therapeutic strategy for the treatment of NSCLC.
Fig. 1: The expression of SIRT6 in A549 cells was examined using RT-PCR and Western blot. (A): The mRNA expression of SIRT6 in A549 cells was
detected by performing RT-PCR and (B): Western blot analysis was conducted to determine the protein expression of SIRT6 in A549 cells.
Note: (*) vs. control, p<0.05 and (#) vs. pcDNA3.1, p<0.05.
Fig. 2: SIRT6 overexpression was found to inhibit the proliferation and invasion capabilities of A549 cells, (A): To assess the proliferation ability,
flow cytometry was performed and (B): The invasion ability was evaluated using a Transwell chamber assay.
Note: (*) vs. control, p<0.05; (#) vs. pcDNA3.1, p<0.05. Bar=20 μm.
Funding:
This work was supported by Nantong Science Plan guiding project (JCZ21091).
Ethical approval:
This study was approved by the Research Ethics Committee of People’s Hospital of Hai’an.
Author’s contributions:
Zheng Wu was responsible for the experimental design, while Bojin Zhu and Zheng Wu conducted the experiments. The data analysis was carried out by Bojin Zhu, and the paper was authored by Zheng Wu. All authors have reviewed and approved the final draft of the manuscript to ensure its accuracy.
Conflict of interests:
The authors declared no conflict of interests.
References
- Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin 2020;70(3):145-64.
[Crossref] [Google Scholar] [PubMed]
- Skrickova J. Lung cancer. Cas Lek Cesk 157(5):226-36.
- Yin X, Li Y, Wang H, Jia T, Wang E, Luo Y, et al. Small cell lung cancer transformation: From pathogenesis to treatment. Semin Cancer Biol 2022;86(2):595-606.
[Crossref] [Google Scholar] [PubMed]
- Musial C, Zaucha R, Kuban-Jankowska A, Konieczna L, Belka M, Marino Gammazza A, et al. Plausible role of estrogens in pathogenesis, progression and therapy of lung cancer. Int J Env Res Public Health 2021;18(2):648.
[Crossref] [Google Scholar] [PubMed]
- Black RC, Khurshid H. NSCLC: An update of driver mutations, their role in pathogenesis and clinical significance. Rhode Island Med J 2015;98(10):25.
[Google Scholar] [PubMed]
- Lin S, Zhen Y, Guan Y, Yi H. Roles of Wnt/β-catenin signaling pathway regulatory long non-coding RNAs in the pathogenesis of non-small cell lung cancer. Cancer Manag Res 2020;12:4181-91.
[Crossref] [Google Scholar] [PubMed]
- Doghish AS, Ismail A, Elrebehy MA, Elbadry AM, Mahmoud HH, Farouk SM, et al. A study of miRNAs as cornerstone in lung cancer pathogenesis and therapeutic resistance: A focus on signaling pathways interplay. Pathol Res Pract 2022;237:154053.
[Crossref] [Google Scholar] [PubMed]
- He Y, Xiao Y, Yang X, Li Y, Wang B, Yao F, et al. SIRT6 inhibits TNF-α-induced inflammation of vascular adventitial fibroblasts through ROS and Akt signaling pathway. Exp Cell Res 2017;357(1):88-97.
[Crossref] [Google Scholar] [PubMed]
- Yang Y, Zhu M, Liang J, Wang H, Sun D, Li H, et al. SIRT6 mediates multidimensional modulation to maintain organism homeostasis. J Cell Physiol 2022;237(8):3205-21.
[Crossref] [Google Scholar] [PubMed]
- Yuan Z, Zeng Y, Tian Y, Wang S, Hong B, Yang M. SIRT6 serves as a polyhedron in glycolytic metabolism and ageing-related diseases. Exp Gerontol 2022;162:111765.
[Crossref] [Google Scholar] [PubMed]
- Jing XD, Tang Q, He JH. The role of SIRT6 in nonalcoholic steatohepatitis. Sheng Li Xue Bao 2021;73(5):745-54.
[Google Scholar] [PubMed]
- Fiorentino F, Carafa V, Favale G, Altucci L, Mai A, Rotili D. The two-faced role of SIRT6 in cancer. Cancers 2021;13(5):1156.
[Crossref] [Google Scholar] [PubMed]
- Akter R, Afrose A, Rahman MR, Chowdhury R, Nirzhor SS, Khan RI, et al. A comprehensive analysis into the therapeutic application of natural products as SIRT6 modulators in Alzheimer’s disease, aging, cancer, inflammation, and diabetes. Int J Mol Sci 2021;22(8):4180.
[Crossref] [Google Scholar] [PubMed]
- Li Y, Jin J, Wang Y. SIRT6 widely regulates aging, immunity, and cancer. Front Oncol 2022;12:861334.
[Crossref] [Google Scholar] [PubMed]
- Gong S, Xiong L, Luo Z, Yin Q, Huang M, Zhou Y, et al. SIRT6 promotes ferroptosis and attenuates glycolysis in pancreatic cancer through regulation of the NF‑κB pathway. Exp Ther Med 2022;24(2):502.
[Crossref] [Google Scholar] [PubMed]
- Qi J, Cui C, Deng Q, Wang L, Chen R, Zhai D, et al. Downregulated SIRT6 and upregulated NMNAT2 are associated with the presence, depth and stage of colorectal cancer. Oncol Lett 2018;16(5):5829-37.
[Crossref] [Google Scholar] [PubMed]
- Lu CT, Hsu CM, Lin PM, Lai CC, Lin HC, Yang CH, et al. The potential of SIRT6 and SIRT7 as circulating markers for head and neck squamous cell carcinoma. Anticancer Res 2014;34(12):7137-43.
[Google Scholar] [PubMed]
- Zhu B, Yan Y, Shao B, Tian L, Zhou W. Downregulation of SIRT6 is associated with poor prognosis in patients with non-small cell lung cancer. J Int Med Res 2018;46(4):1517-27.
[Crossref] [Google Scholar] [PubMed]
- de Céu Teixeira M, Sanchez-Lopez E, Espina M, Garcia ML, Durazzo A, Lucarini M, et al. Sirtuins and SIRT6 in carcinogenesis and in diet. Int J Mol Sci 2019;20(19):4945.
[Crossref] [Google Scholar] [PubMed]
- Demir IE, Ceyhan GO, Friess H. Epigenomic therapies: The potential of targeting SIRT6 for the treatment of pancreatic cancer. Exp Opin Ther Targets 2017;21(1):1-3.
[Crossref] [Google Scholar] [PubMed]
- Liu J, Yu Z, Xiao Y, Meng Q, Wang Y, Chang W. Coordination of FOXA2 and SIRT6 suppresses the hepatocellular carcinoma progression through ZEB2 inhibition. Cancer Manag Res 2018;10:391-402.
[Crossref] [Google Scholar] [PubMed]
- Lin H, Hao Y, Zhao Z, Tong Y. Retracted: Sirtuin 6 contributes to migration and invasion of osteosarcoma cells via the ERK1/2/MMP9 pathway. FEBS Open Bio 2017;7(9):1291-301.
[Crossref] [Google Scholar] [PubMed]
- Liu Y, Xie QR, Wang B, Shao J, Zhang T, Liu T, et al. Inhibition of SIRT6 in prostate cancer reduces cell viability and increases sensitivity to chemotherapeutics. Protein Cell 2013;4(9): 702–10.
[Crossref] [Google Scholar] [PubMed]
- Qu N, Hu JQ, Liu L, Zhang TT, Sun GH, Shi RL, et al. SIRT6 is upregulated and associated with cancer aggressiveness in papillary thyroid cancer via BRAF/ERK/Mcl‑1 pathway. Int J Oncol 2017;50(5):1683-92.
[Crossref] [Google Scholar] [PubMed]
- Zhang ZG, Qin CY. Sirt6 suppresses hepatocellular carcinoma cell growth via inhibiting the extracellular signal‑regulated kinase signaling pathway. Mol Med Rep 2014;9(3):882-8.
[Crossref] [Google Scholar] [PubMed]
- Choe M, Brusgard JL, Chumsri S, Bhandary L, Zhao XF, Lu S, et al. The RUNX2 transcription factor negatively regulates SIRT6 expression to alter glucose metabolism in breast cancer cells. J Cell Biochem 2015;116(10):2210-26.
[Crossref] [Google Scholar] [PubMed]
- Thirumurthi U, Shen J, Xia W, LaBaff AM, Wei Y, Li CW, et al. MDM2-mediated degradation of SIRT6 phosphorylated by AKT1 promotes tumorigenesis and trastuzumab resistance in breast cancer. Sci Signal 2014;7(336):ra71.
[Crossref] [Google Scholar] [PubMed]
- Xiong L, Tan B, Lei X, Zhang B, Li W, Liu D, et al. SIRT6 through PI3K/Akt/mTOR signaling pathway to enhance radiosensitivity of non‐Small cell lung cancer and inhibit tumor progression. IUBMB Life. 2021;73(9):1092-102.
[Crossref] [Google Scholar] [PubMed]
- Krishnamoorthy V, Vilwanathan R. Silencing Sirtuin 6 induces cell cycle arrest and apoptosis in non-small cell lung cancer cell lines. Genomics 2020;112(5):3703-12.
[Crossref] [Google Scholar] [PubMed]