- *Corresponding Author:
- Sudha Ramaiah
Medical and Biological Computing Laboratory, School of Biosciences and Technology, VIT University, Vellore-632 014, India
E-mail: sudhaanand@vit.ac.in
| Date of Submission | 12 October 2015 |
| Date of Revision | 19 October 2016 |
| Date of Acceptance | 24 November 2016 |
| Indian J Pharm Sci 2016;78(6):780-788 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Rice (Oryza sativa) is the grain of life. It is one among the main cereal crops and it is the staple food for most of the world’s population. Medicinal rice varieties have defensive and therapeutic properties against many human disorders. Drug resistance has become a major problem in recent years. Escherichia coli have developed resistance to most of the antibiotics including sulfonamides that target dihydropteroate synthase. In the present study, we attempt to identify a novel inhibitor for the dihydropteroate synthase from a medicinal rice variety – “Karungkavuni”. The phytochemical composition of the rice “Karungkavuni” is analyzed through Gas chromatography-mass spectrometry. The isolated compounds with reported antimicrobial activity are subjected to molecular docking procedures to understand the binding behaviour of the ligands with the target. Our analysis reveals that, ethyl iso-allocholate and 9,12,15-octadecatrienoic acid- 2,3-dihydroxypropyl ester as the best binding compounds. Molecular dynamics studies reveal dihydropteroate synthase-ethyl iso-allocholate complex is more stable than other ligands throughout the simulation. Our study demonstrates that ethyl iso-allocholate compound isolated from “Karungkavuni” can serve as a potent inhibitor for dihydropteroate synthase.
Keywords
Dihydropteroate synthase, drug resistance, sulfonamide, binding affinity, folate biosynthesis, molecular docking, molecular simulation
Rice is a monocotyledonous plant, which belongs to the family of poaceae [1]. Several studies suggest that Oryza sativa is domesticated from O. rufipogon, which is an Asian common wild rice [2]. Rice is the main cereal crop of the world and it is the main staple food in most Asian countries. Approximately, 95% of rice is produced in Asia. Rice is consumed mostly in a cooked form for obtaining energy and nutrition. Rice contains nutrients such as carbohydrates, proteins, dietary fiber, vitamins and minerals [3]. Medicinal rice varieties have many defensive and therapeutic properties against human disorders like colon cancer, epilepsy, chronic headache, paralysis, skin diseases, indigestion, rheumatism, arthritis, blood pressure, diabetes, internal rejuvenation of tissues and postnatal weaknesses [4]. Whole grain of rice has some health promoting components. It is suggested that regular intake of whole grains and whole grain products reduces the risk of chronic diseases such as type 2 diabetes and cardiovascular diseases [5]. Antibiotic resistance has become a major concern in recent decades. Pathogenic bacteria have the potential to transmit and acquire resistance to all available therapeutic agents. Development of drug resistance is a major cause that limits the usage of existing antimicrobials that includes sulphonamides, earlier which were quite effective against infections caused by both Gram-positive and Gram-negative bacteria. Hence, sulfonamide resistance is a serious problem in health care settings. Bacterial folate biosynthetic pathway is an ideal target for the antimicrobial therapy [6]. Dihydropteroate synthase (DHPS, EC 2.5.1.15) is a triose-phosphate isomerase enzyme with eight β-strands and eight α-helices. DHPS is the key enzyme in folic acid metabolism as it catalyses the formation of tetrahydrofolate, an important precursor for the synthesis of pyrimidines, purines and amino acids. DHPS plays a vital role in the proliferation of microorganisms and it is encoded by the gene folP [7]. DHPS is involved in the conversion of 6-hydroxymethyl-7,8-dihydropteridine-pyrophosphate and para-amino benzoic acid (PABA) to pyrophosphate and 7,8-dihydropteroate [8]. PABA is synthesized through the upregulation process by the chorismate pathway and supplemented into the folate biosynthesis pathway. DHPS has been identified as the best target for the sulfonamide group of compounds. Generally, sulfonamides inhibit the target by mimicking as a competitive inhibitor and substrate analogs of PABA and leads to “dead end” in the pathway [9]. Resistance to sulfonamides are associated with chromosomal mutations within the folP gene. Drug resistance is the major factor that restricts the current use of sulfa-based inhibitors against pathogenic bacteria including E. coli. Over the past few years, there is an increasing interest in the study of chemical composition of rice varieties. Many researchers have reported the phytochemical composition of the rice and rice by-products [3,10,11]. There are a few reports on the antimicrobial activity of the rice extracts, rice by-products and components isolated from rice [12,13]. However, there are not many reports that describe the mechanism and bioactivity of compounds isolated from medicinal rice varieties.
Thus, in our present study we aim to explore the phytochemical composition in the medicinal rice variety, Karungkavuni through Gas chromatographymass spectrometry (GC-MS) analysis. The compounds with reported antimicrobial activity are selected for the molecular docking study to understand the binding behaviour. Similar to sulpha group of compounds, the selected compounds competitively inhibit the DHPS and might disrupt the folic acid pathway. The stability of the docked complexes is then analysed through molecular dynamics (MD) simulation.
Materials and Methods
Karungkavuni paddy was procured from the organic rice farmers in Tanjavur and Tiruvarur Districts of Tamil Nadu, India.
Preparation of the extract
Fifty grams of Karungkavuni rice was pulverized by using pestle and mortar. Two grams of powdered homogenized rice sample was added in a centrifuge tube containing 20 ml of ethanol. The contents were mixed properly using cyclomixer for 10 min. The resultant mixture was then centrifuged at 10 000 rpm for 10 min. After centrifugation, the separated supernatant was filtered through 0.2 μ filter. Then, the filtrate was stored in a glass vial.
GC-MS analysis
GC-MS analysis of the ethanol extract of Karungkavuni rice was carried out on the Thermo Fisher TSQ Quantum XLS GC-MS/MS using DB5-MS capillary column measuring 30 m×0.25 mm×0.25 μm and composed of 5% phenyl, 95% dimethyl polysiloxane cross bonded liquid phase. Helium of 99.9995% was used as a carrier gas with a flow rate of 1.0 ml/min. Sample vial with 1 ml sample was kept in the autoinjector and 1 μl of sample was injected into the GC column using syringe in split mode. At the time of injection, the system temperature was kept at 290°. The sample was injected with split ratio of 10:1. Initially, the column temperature was programmed at 80° for 2 min, then it was increased to 150° with 40°/min then to 240° with 4°/min then slowly increased to 255° with 2°/min and finally it was increased to 285° with 4°/ min keeping for 3 min. Total GC cycle run time was set to 30 min. The MS scan mode of m/z 50 to 650 was operated and data were evaluated using total ion count. The spectrum of compounds was compared with reference in the National Institute of Standards and Technology (NIST) database.
Statistical analysis
Tests were performed in triplicates (n=3) and the area of percentage observed in GC-MS analysis was expressed as mean±standard deviation.
Dataset
After GC-MS analysis, the compounds reported with antimicrobial activities were retrieved from the National Center for Biotechnology Information (NCBI) PubChem database [14] and the structures were energy minimized. The 3-Dimensional (3-D) structure of the target protein E. coli DHPS (1AJ0) [15] was retrieved from the protein data bank [16]. The water molecules were removed and the hydrogen atoms were added to the protein at the end.
Molecular docking
Molecular docking is a standard procedure in drug discovery process. Surflex-Dock program interfaced with SYBYL X 2.0 (Tripos international, USA) was used for the docking study. The compounds isolated from rice variety with reported antimicrobial activity were docked into the active site of the 3-D structure of DHPS. At first, the optimization of the protein structure was done by detaching the unrelated substructures. Similarly, the other standard parameters such as, protein side chain fixation, addition of hydrogen, removal of water molecules and allocation of unknown atom types were also utilized. The relaxation of bumps was carried out to define the correct configuration and tautomeric states of the protein structure. The protein atoms were assigned with the Kollman-all charges [17]. The whole protein structure was energy minimized by using the AMBER7 F99 force field. Finally, the ligands were energy minimized by using the Powell’s energy minimization algorithm [18]. SYBYL X 2.0 program docks a ligand automatically into the active site of a target protein. It is a protomol-based method and an empirical score is provided. The scoring functions employ properties that include solvation, crash, hydrophobic, polar entropic and repulsive properties. These functional scores explain the interaction between the protein and ligand. The final bioactive conformation was selected based on the highest consensus scoring (C-score) and number of H-bond formation. The C-score also utilizes the other surflex dock scores, crash score, polar score, gold score (G-score), potential mean force (PMF) score, dock score (D-score) and chem-score.
Absorption, distribution, metabolism and excretion (ADME) screening
The compounds retrieved from the NCBI PubChem database were subjected to ADME screening for the prediction of both physiochemical and pharmacological properties. The molinspiration server was used for the prediction of ADME and drug-likeness of the compounds isolated from rice variety. For this, simplified molecule input line entry specification of the compounds was submitted to the molinspiration server, which predicts the number of hydrogen (H)- bond donors (HBD) and number of H-bond acceptors (HBA), log P, molecular volume and rotatable bonds. This online server calculates molecular properties and drug likeness of a given compound rapidly. Based on the predicted properties, the acceptability of the drugs was evaluated by Lipinski’s rule of 5 [19], which was important for the structure-based drug designing.
MD simulation
The MD simulation of free DHPS, DHPS-ethyl isoallocholate and DHPS-9,12,15-octadecatrienoic acid-2,3 dihydroxypropyl ester complexes were performed using Gromacs 4.5.5 package [20]. In the first step, enzyme topology was generated by the default parameters of the Gromacs, whereas the ligand topology was created by the online tool PRODRG server [21]. Gromos96 53a6 force field was used for the generation of topology and interaction parameters. The structures were solvated in the cubic box containing simple-point-charge water molecules. The distance of 1.2 nm was set between the box and protein-ligand complex. Since, proteins were positively charged, the system must be neutralized by the addition of counter ions (Cl- or Na+). The concentration of NaCl was set as 0.145 M. For energy minimization, steepest descent algorithm was applied for first 1000 steps. In the next step, system was equilibrated under restraints position to relax the solvent under constant volume and temperature dynamics (NVT) for 100 ps at 300 K, followed by the production simulations that implemented under constant pressure and temperature dynamics (NPT) for 100 ps. Linear constraint solver algorithm was used for restrain the lengths of all bonds. A coulomb type particle mesh Ewald method is applied for calculating electrostatic interactions. The coulomb and Van der Waals Cut off radii interactions were set to 10.0 Å and 14.0 Å, respectively. Each system was subjected to 50 ns MD simulation. Xm grace tool was used for the generation of graphs [22].
Results and Discussion
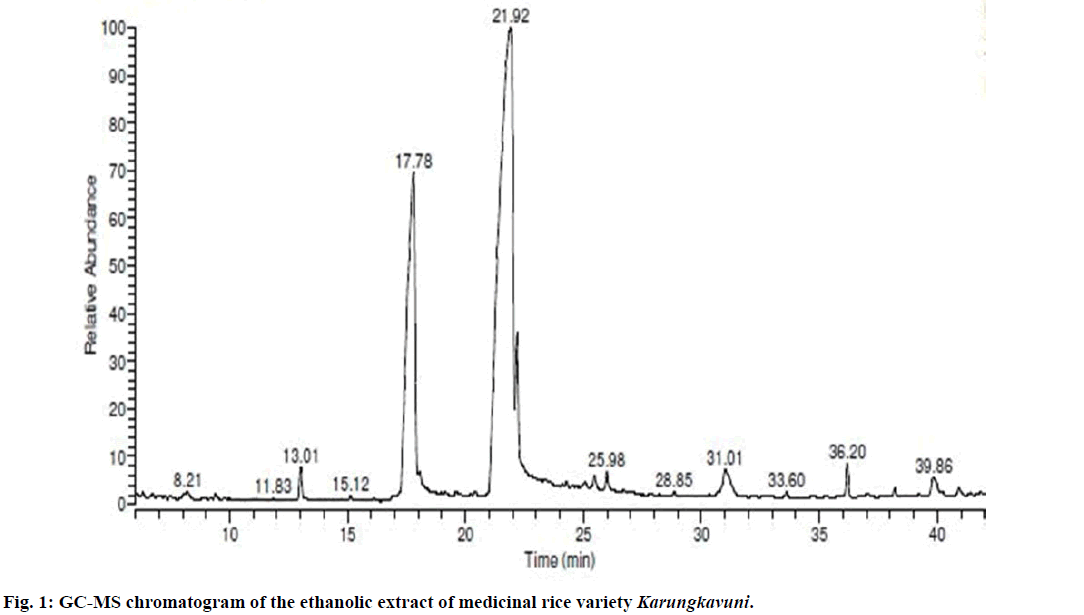
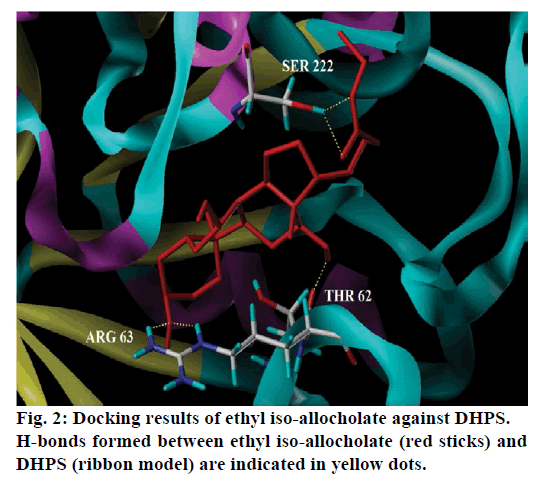
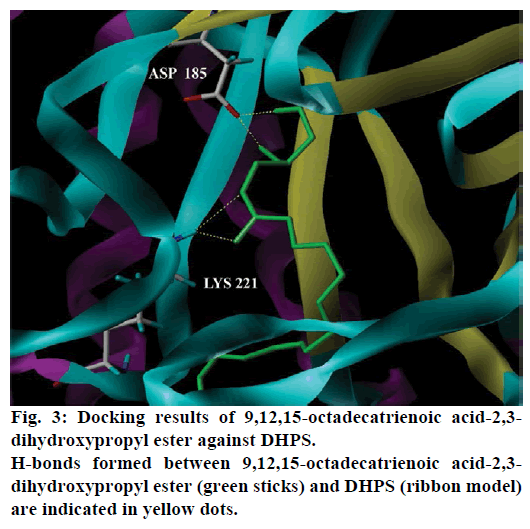
Seventeen different compounds are observed in the GCMS chromatogram of ethanolic extract of the medicinal rice variety Karungkavuni. The chromatogram of the rice is depicted in Figure 1 and the compounds are tabulated in Table 1. These phytochemical compounds are identified using NIST library. Some of the prominent phytochemicals noted in i rice variety are sitosterol, tetradecanoic acid, hexadecanoic acid, cis-vaccenic acid, eicosanoic acid and tocopherol. The identified phytochemical compounds signify the medicinal activity of this rice variety. The biological activities of these compounds are listed in Table 2 [23-34]. Reports suggest that compounds 9,12,15-octadecatrienoic acid-2,3- dihydroxy propyl ester [26], sitosterol [28,29], squalene [30,31] and ethyl iso-allocholate [33] have antimicrobial activity. We selected these four compounds for docking studies to reveal the binding orientation of ligands with the target. The compound structures are retrieved from the NCBI PubChem database and their PubChem IDs are listed in Table 3. These retrieved compounds are docked into the active site of the E. coli DHPS. After docking, the observed results are evaluated based on the C-score and number of H-bonds formed between the target protein and ligand. Out of four compounds screened, ethyl iso-allocholate has the best binding affinity with DHPS, which has C-score value of 4. DHPS-ethyl iso-allocholate interaction displayed totally 5 H-bonds and the residues Arg63, Ser222 and Thr62 of DHPS are involved in the H-bond formation. Ethyl iso-allocholate forms two H-bonds with Arg63 and ser222. The residue Thr62 forms only one H-bond with ethyl iso-allocholate (Figure 2). Similarly, 9,12,15-octadecatrienoic acid-2,3-dihydroxypropyl ester also have binding affinity with DHPS, the C-score value is 3. DHPS-9,12,15-octadecatrienoic acid-2,3- dihydroxypropyl ester interaction has 4 H-bonds formation. The 9,12,15-octadecatrienoic acid-2,3- dihydroxypropyl ester has formed two H-bonds with the residues Asp185 and Lys221 of DHPS as in Figure 3. To understand the interaction of ligands with the protein, different binding energy scores and number of H-bonds formed are tabulated in Table 4.
| RTa | Name of the compound | Similarity index (%) | MWb | Area (%) |
|---|---|---|---|---|
| 5.69 | D-ribo-hexose, 2,6-dideoxy-3-o-methyl- | 85 | 176 | 0.88±0.03 |
| 6.31 | Ascaridole epoxide | 82 | 184 | 0.13±0.02 |
| 8.2 | D-glucopyranoside | 79 | 504 | 0.86±0.07 |
| 9.39 | 3-tret-butyl-4-hydroxyanisole | 84 | 180 | 0.28±0.03 |
| 13.01 | Tetradecanoic acid | 86 | 228 | 0.86±0.08 |
| 17.76 | Hexadecanoic acid | 89 | 256 | 26.76±0.92 |
| 21.89 | Cis-vaccenic acid | 92 | 282 | 64.40±0.77 |
| 25.06 | 9,12,15-octadecatrienoic Acid-2,3-dihydroxy propyl ester | 86 | 352 | 0.32±0.06 |
| 25.47 | Cis-13-eicosenoic acid | 81 | 310 | 0.48±0.05 |
| 25.98 | Eicosanoic acid | 88 | 312 | 0.49±0.05 |
| 31.03 | Sitosterol | 86 | 414 | 2.13±0.10 |
| 36.2 | Squalene | 85 | 410 | 0.86±0.07 |
| 38.21 | Heptacosane | 82 | 380 | 0.21±0.25 |
| 39.86 | 9,19-cyclolanostan-3-ol,24-methylene-(3a) | 89 | 440 | 0.97±0.02 |
| 40.94 | Tocopherol | 91 | 416 | 0.47±0.03 |
| 42.13 | Ethyl iso-allocholate | 93 | 436 | 0.25±0.03 |
| 43.25 | Olean-13-(18)-ene | 86 | 410 | 0.29±0.04 |
aRT-retention time; bMW-molecular weight. Analysis was done in triplicates; data presented as mean ± standard deviation
Table 1: Phytochemicals identified in ethanolic extract of medicinal rice variety Karungkavuni using gc-ms peak
| Name of the compound | Compound nature | Bioactivity |
|---|---|---|
| D-ribo-hexose, 2,6-dideoxy-3-o-methyl- | Heterocyclic | Sugar moiety of cardiac glycosides |
| Ascaridole epoxide | Heterocyclic | Anthelminthic |
| D-glucopyranoside | Heterocyclic | Preservative |
| 3-tret-butyl-4-hydroxyanisole | Phenolic | Antioxidant, antimutagenic activity |
| Tetradecanoic acid | Acid | Antioxidant, anticancer |
| Hexadecanoic acid | Acid | Antioxidant, hypocholesterolemic |
| Cis-vaccenic acid | Acid | Anticancer, anti-inflammatory |
| 9,12,15-octadecatrienoic acid-2,3-dihydroxy propyl ester | Alpha linolenic acid | Antimicrobial activity, anti-inflammatory, anticancer |
| Eicosanoic Acid | Acid | Anticancer, antioxidant, anti-inflammatory, cardioprotective |
| Sitosterol | Phytosterol | Antimicrobial activity, reduces blood levels of cholesterol |
| Squalene | Tritrepene | Antibacterial, antioxidant, antitumor, cancer preventive |
| Heptacosane | Heterocyclic | Insect pheromone constituent |
| 9,19-cyclolanostan-3-ol, 24-methylene-(3a) | Tritrepene alcohol |
Used as an active ingredient of a drug |
| Tocopherol | Alcohol | Antioxidant, antiageing |
| Ethyl iso-allocholate | Steroid derivative | Antimicrobial activity |
| Olean-13-(18)-ene | Triterpenoids | Antitumor activity |
Table 2: Biological activities of phytochemicals present in the ethanolic extracts of medicinal rice variety Karungkavuni
| Pubchem ID | Compound name | Structure |
|---|---|---|
| 6452096 | Ethyl iso-allocholate | 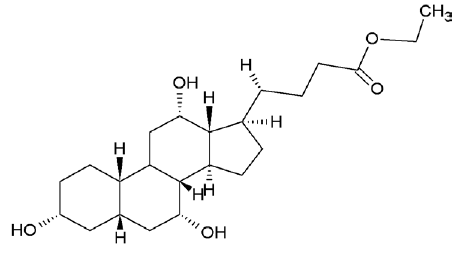 |
| 5367328 | 9,12,15-octadecatrienoic acid-2,3-dihydroxypropyl ester | 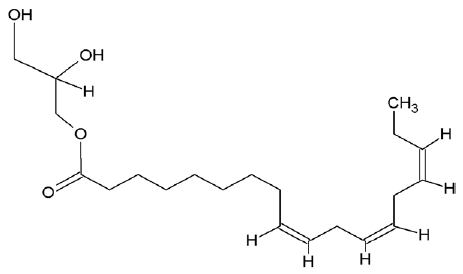 |
| 1105 | Squalene | 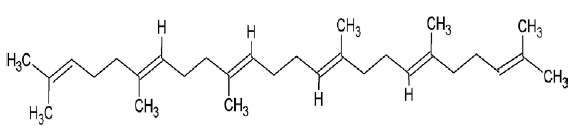 |
| 222284 | Sitosterol | 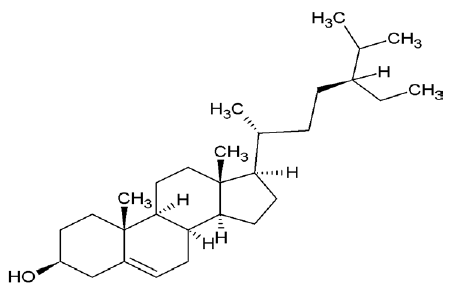 |
Table 3: Compounds reported with antimicrobial activity and their pubchem ids
| Pubchem ID |
Compound name |
Crash scorea |
Polar scoreb |
D scorec | PMF scored | G scoree |
Chem scoref |
C scoreg |
H bond |
|---|---|---|---|---|---|---|---|---|---|
| 6452096 | Ethyl iso-allocholate | -10.21 | 1.18 | -143.39 | -25.77 | -328.52 | -25.78 | 4 | 4 |
| 5367328 | 9,12,15-octadecatrienoic acid- 2,3-dihydroxy propyl ester | -1.73 | 3.29 | -86.65 | -35.92 | -168.63 | -16.74 | 3 | 4 |
| 1105 | Squalene | -3.67 | 0.00 | -169.21 | -18.13 | -346.84 | -36.97 | 3 | - |
| 222284 | Sitosterol | -5.85 | 0.29 | -114.79 | -29.34 | -275.419 | -25.235 | 2 | 1 |
Table 4: Surflex dock scores of compounds isolated from medicinal rice variety Karungkavuni
Rice contains a consortium of many bioactive components. The compounds isolated from rice varieties are subjected to physicochemical and pharmaceutical property prediction by the Lipinski’s rule of 5. It accepts compounds with one violation. This rule explains about the importance of physiochemical property prediction in the pharmacokinetics of a drug in the human body and also with ADME prediction. This rule restricts compounds with less than 5 HBD, molecular weight below 500 Da, less than 5 log P and 10 HBA. The Molinspiration prediction reveals that the 4 compounds have shown possible drug likeness characteristics as listed in Table 5. The compounds squalene and sitosterol has log P value greater than 5. Also, the compounds squalene and 9,12,15-octadecatrienoic acid-2,3-dihydroxypropyl ester has more than 10 rotatable bond counts. All the 4 compounds are having the drug likeness properties. The rotatable bonds are used as a filter to distinguish the drug molecule. Increase in the rotatable bond count may leads to the decreased oral bioavailability [35].
| Pubchem ID | Compound name | LogP (<5) |
Molecular weight (<500 Da) |
HBA count (<10) |
HBD count (<5) |
Rotatable bond count (<10) |
|---|---|---|---|---|---|---|
| 222284 | Sitosterol | 8.62 | 414.72 | 1 | 1 | 6 |
| 5367328 | 9,12,15-octadecatrienoic acid- 2,3-dihydroxypropyl ester | 4.87 | 352.51 | 4 | 2 | 17 |
| 1105 | Squalene | 9.62 | 410.73 | 0 | 0 | 15 |
| 6452096 | Ethyl iso-allocholate | 4.01 | 436.63 | 5 | 3 | 6 |
Table 5: Molinspiration predicted molecular properties of the compounds isolated from medicinal rice variety Karungkavuni
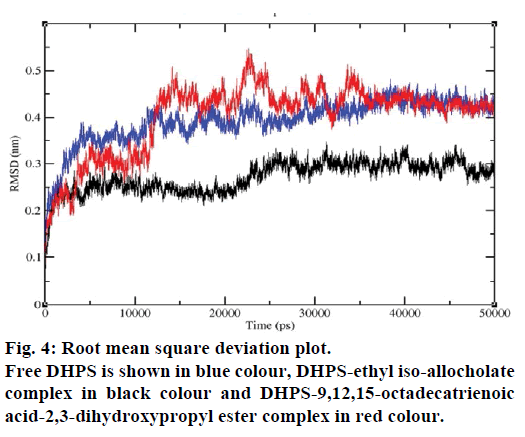
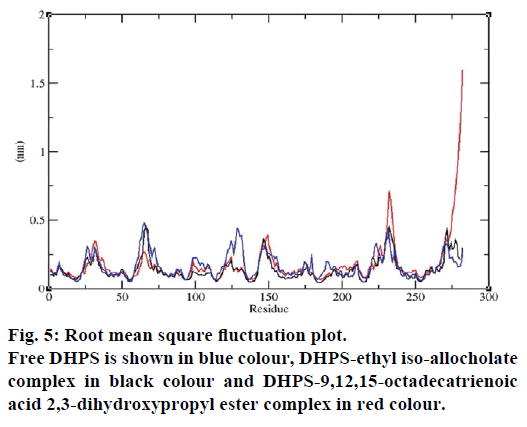
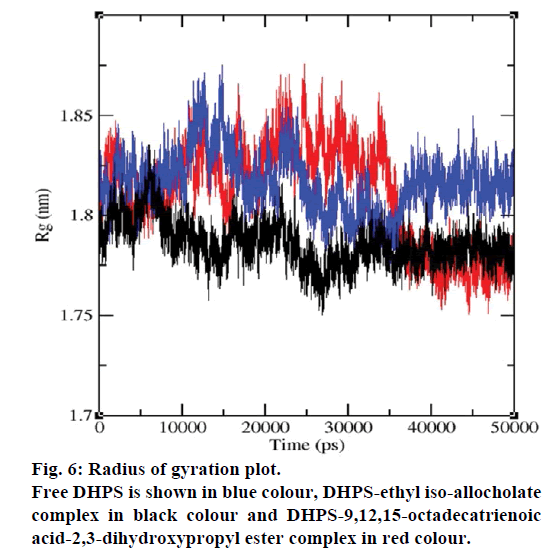
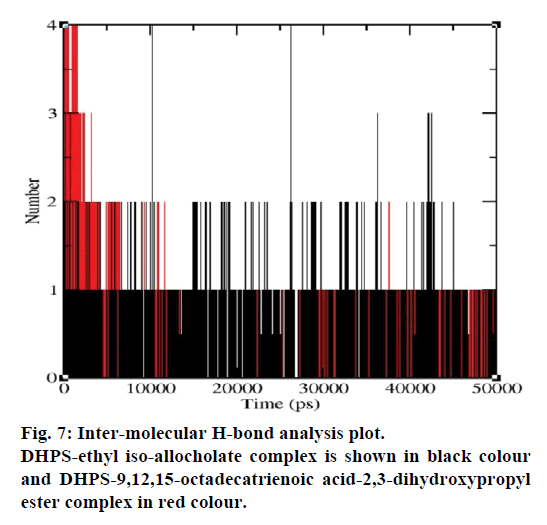
MD simulations of free DHPS, DHPS-ethyl isoallocholate and DHPS-9,12,15-octadecatrienoic acid- 2,3 dihydroxypropyl ester complexes are conducted to understand the overall stability of the complexes on a nanosecond time scale. The complex structures are subjected to 50 ns MD simulations. To investigate the stability of conformations in MD simulations, we analyse root-mean-square deviation (RMSD), root mean-square fluctuations (RMSF), Rg and number of intermolecular H-bonds formed between the protein and ligand. RMSD is used to calculate the average displacement of atoms for a particular frame with respect to the reference frame. RMSD backbone values of free DHPS, DHPS-ethyl iso-allocholate and DHPS- 9,12,15-octadecatrienoic acid-2,3-dihydroxypropyl ester complexes are calculated against the simulation time scale between 0 and 50 000 ps (Figure 4). This plot reveals that three trajectories have the RMSD values between 0.1 nm and 0.54 nm throughout the simulation. For free DHPS system, the RMSD value is increased to 0.45 nm at 38 000 ps and there after the value gets decreased to 0.42 nm. For DHPS- 9,12,15-octadecatrienoic acid-2,3-dihydroxypropyl ester complex, initially the RMSD value increase to 0.45 nm till 15 000 ps and then, there is a slight fluctuations in the value. After 35 000 ps, the values get decreased and reach 0.42 nm. In case of DHPS-ethyl iso-allocholate complex, the RMSD value increased up to 0.3 nm at 25 000 ps. Then, there is no further increase in the RMSD value and the DHPS-ethyl isoallocholate system attains equilibrium. Comparing the values of the DHPS-ethyl iso-allocholate and DHPS- 9,12,15-octadecatrienoic acid-2,3-dihydroxypropyl ester systems with the free DHPS system, it reveals that DHPS-ethyl iso-allocholate system shows a lower RMSD value. The decrease in the RMSD value of the DHPS-ethyl iso-allocholate system indicates increased stability and rigidity of DHPS upon binding with the ethyl iso-allocholate. RMSF is calculated to analyse the flexibility of residues on protein structure and the ligand-binding regions. The fluctuation scores reveal the behaviour of the residues throughout the simulation. The flexibility observed in the residues of free DHPS, DHPS-ethyl iso-allocholate and DHPS- 9,12,15-octadecatrienoic acid-2,3-dihydroxypropyl ester complexes are shown in Figure 5. The free DHPS residues GLY31, GLY65, SER98 and ALA128 have highest fluctuations of 0.29, 0.46, 0.21 and 0.43 nm, respectively. In case of DHPS-ethyl iso-allocholate complex, it shows more fluctuations at 0.26, 0.42, 0.12 and 0.13 nm, respectively. For DHPS-9,12,15- octadecatrienoic acid-2,3-dihydroxypropyl ester complex, the fluctuations are seen at 0.34, 0.25, 0.18 and 0.15 nm, respectively. When comparing the fluctuation scores of free DHPS system with other two complexes, it reveals that the DHPS-ethyl isoallocholate complex has the lesser fluctuation. These results interpret that the DHPS system shows more fluctuation without ethyl iso-allocholate. Hence, the DHPS becomes more rigid as a result of binding of ethyl iso-allocholate, when compared to the binding of 9,12,15-octadecatrienoic acid-2,3-dihydroxypropyl ester. Rg is plotted to identify the level of compaction in the structure of free DHPS, DHPS-ethyl iso-allocholate complex and DHPS-9,12,15-octadecatrienoic acid-2,3- dihydroxypropyl ester complex. The Rg of Cα atoms of free DHPS, DHPS-ethyl iso-allocholate and DHPS- 9,12,15-octadecatrienoic acid-2,3-dihydroxypropyl ester complexes versus time at 300 K are plotted in Figure 6. The Rg value of DHPS structure has shown to increase up to 1.86 nm, later the value get decreased to 1.78 nm and after 35 000 ps, the value again increases and attains equilibrium at 1.82 nm. For DHPS-ethyl isoallocholate complex, the Rg plot shows fluctuation at the beginning and after 35 000 ps, it attains constant value of 1.78 nm. Further, not much variation is observed in Rg plot. In case of DHPS-9,12,15-octadecatrienoic acid-2,3-dihydroxypropyl ester, the Rg value get increased and reach 1.86 nm at 25 000 ps and started decreasing and reach 1.75 nm after 40 000 ps. It shows decreased value for DHPS-9,12,15-octadecatrienoic acid-2,3-dihydroxypropyl ester complex. Finally the intermolecular H-bond analysis reveals that a total of 4 H-bonds are formed between DHPS-ethyl isoallocholate complex and on average of 2 H-bonds are found throughout the simulation (Figure 7). For DHPS- 9,12,15-octadecatrienoic acid-2,3-dihydroxypropyl ester, 4 H-bonds are formed and on average of 1 one H-bond is found throughout the simulation. The number of H-bonds formed between the complexes is involved in the stabilization of the complex. Overall, our results indicate that ethyl iso-allocholate isolated from Karungkavuni rice variety could serve as a potent inhibitor for E. coli DHPS. We believe our results will be a good starting point for further experimental validation on ethyl iso-allocholate to be developed as a potent antibacterial agent.
Acknowledgements
RS and AA gratefully acknowledge the Indian Council of Medical Research (ICMR), Government of India Agency for the research Grant (IRIS ID: 2014-0099). The authors would like to thank Mr. Krishnanunni and R. G. Swetha for their help in the initial part of our study. The authors would also like to thank the management of VIT University for providing the necessary facilities to carry out this research project.
Conflict of interest
The authors declare that there is no conflict of interest.
Financial support and sponsorship
Nil.
References
- Esa NM, Ling TB, Peng LS. By-products of rice processing: An overview of health benefits and applications.J Rice Res 2013;1:107.
- Kovach MJ, Sweeney MT, McCouch SR. New insights into the history of rice domestication. Trends Genet 2007;23:578-87.
- Ham H, Oh SK, Lee JS, Choi IS, Jeong HS, Kim IH, et al. Antioxidant activities and contents of phytochemicals in methanolic. Extracts of specialty rice cultivars in Korea. Food Sci Biotechnol 2013;22:631-7.
- Kirtikar KR, Basu BD. Indian medicinal plants. 2nd ed. New Delhi, India: Bishen Singh Mahendra Pal Singh; 1933.
- Gani A, Wani SM, Masoodi FA, Hameed G. Whole-grain cereal bioactive compounds and their health benefits: A review. J Food Process Technol 2012;3:146.
- Swarbrick J, Iliades P, Simpson JS, Macreadie I. Folate biosynthesis-reappraisal of old and novel targets in the search for new antimicrobials. Open Enzym Inhib J 2008;1:12-33.
- Babaoglu K, Qi J, Lee RE, White SW. Crystal structure of 7, 8-dihydropteroate synthase from Bacillus anthracis: mechanism and novel inhibitor design. Structure 2004;12:1705-17.
- Shiota T, Baugh CM, Jackson R, Dillard R. The enzymatic synthesis of hydroxymethyl dihydropteridine pyrophosphate and dihydrofolate.Biochem 1969;8:5022-8.
- Levy C, Minnis D, Derrick JP. Dihydropteroate synthase from Streptococcus pneumoniae: structure, ligand recognition and mechanism of sulphonamide resistance. Biochem J 2008;412:379-88.
- Kaneda I, Kubo F, Sakurai H. Antioxidative compounds in the extracts of black rice brans. J Health Sci 2006;52:495-511.
- Krishnanunni K, Senthilvel P, Ramaiah S, Anbarasu A. Study of chemical composition and volatile compounds along with in vitro assay of antioxidant activity of two medicinal rice varieties: Karungkuravai and mappilai samba. J Food Sci Technol 2015;52:2572-84.
- Park HL, Yoo Y, Hahn TR, Bhoo SH, Lee SW, Cho MH. Antimicrobial activity of uv-induced phenylamides from rice leaves. Molecules 2014;19:18139-51.
- Zhai C, Lan J, Wang H, Li L, Cheng X, Liu G. Rice dehydrin k-segments have in vitro antibacterial activity. Biochemistry (Mosc) 2011;76:645-50.
- Li Q, Cheng T, Wang Y, Bryant SH. PubChem as a public resource for drug discovery. Drug Discov Today 2010;15:1052-7.
- Achari A, Somers DO, Champness JN, Bryant PK, Rosemond J, Stammers DK. Crystal structure of the anti-bacterial sulfonamide drug target dihydropteroate synthase. Nat Struct Biol 1997;4:490-7.
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The protein data bank. Nucleic Acids Res 2000;28:235-42.
- Xiong B, Gui CS, Xu XY, Luo C, Chen J, Luo HB, et al. A 3D model of SARS_CoV 3CL proteinase and its inhibitors design by virtual screening. Acta Pharmacol Sin 2003;24:497-504.
- Fletcher R, Powell MJD. A rapidly convergent descent method for minimization. Comput J 1963;6:163-8.
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 2001;46:3-26.
- Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced and scalable molecular simulation. J Chem Theory Comput 2008;4:435-47.
- Schuttelkopf AW, Van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes.Acta Crystallogr D Biol Crystallogr 2004;60:1355-63.
- Turner PJ. XMGRACE, Version 5.1.19, Center for Coastal and Land-Margin Research, Oregon Graduate Institute of Science and Technology, Beaverton, ORE, USA, 2005.
- Lemke TL, Williams DA. Foye's Principles Of Medicinal Chemistry. 7th ed. Philadelphia: Lippincott Williams Wilkins; 2012.
- http://www.ars-grin.gov/duke/
- Sakai A, Miyata N, Takahashi A. Promoting activity of 3-tert-butyl-4-hydroxyanisole (BHA) in BALB/3T3 cell transformation. Cancer Lett 1997;115:213-20.
- Musa AM, Ibrahim MA, Aliyu AB, Abdullahi MS, Tajuddeen N, Ibrahim H, et al. Chemical composition and antimicrobial activity of hexane leaf extract of Anisopus mannii (Asclepiadaceae). J Intercult Ethnopharmacol 2015;4:129-33.
- Pandey S, Satpathy G, Gupta RK. Evaluation of nutritional and phytochemical, antioxidant and antibacterial activity of exotic fruit Limonia acidissima. J Pharmacogn Phytochem 2014;3:81-8.
- Fernandez ML, Vega-Lopez S. Efficacy and safety of sitosterol in the management of blood cholesterol levels. Cardiovasc Drug Rev 2005;23:57-70.
- Woldeyes S, Adane L, Tariku Y, Muleta D, Begashaw T. Evaluation of antibacterial activities of compounds isolated from Sida rhombifolia Linn. (Malvaceae). Nat Prod Chem Res 2012;1:101.
- Gopinath S, Sakthidevi G, Muthukumaraswamya S, Mohan VR. GC-MS analysis of bioactive constituents of Hypericum mysorense (Hypericaceae). J Curr Chem Pharm Sci 2013;3:6-15.
- Prabhadevi V, Sahaya SS, Johnson M, Venkatramani B, Janakiraman N. Phytochemical studies on Allamanda cathartica L. using GC-MS. Asian Pac J Trop Biomed 2012;2:550-4.
- Hmimid F, Lahlou FA, Loutfi M, Bourhim N. Phytochemical screening chemical composition and toxicity of Euphorbia regis-jubae (Webb and Berth). J Toxicol Environ Health Sci2012;4:130-9.
- Muthulakshmi A, Margret JR, Mohan VR. GC-MS Analysis of bioactive components of Feronia elephantum Correa (Rutaceae). J App Pharm Sci 2012;2:69-74.
- Nishino H, Nishino A, Takayasu J, Hasegawa T, Iwashima A, Hirabayashi K, et al. Inhibition of the tumor-promoting action of 12-O-tetradecanoylphorbol-13-acetate by some oleanane-type triterpenoid compounds. Cancer Res 1988;48:5210-5.
- Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem 2002;45:2615-23.