- *Corresponding Author:
- S. K. Singh
Department of Biotechnology,
Thapar Institute of Engineering and Technology,
Patiala 147004,
Punjab, India
E-mail: sanjaybiosoft@gmail.com
| Date of Submission | 11 January 2020 |
| Date of Revision | 20 June 2020 |
| Date of Acceptance | 05 July 2020 |
| Indian J Pharm Sci 2020;82(4):665-676 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The drug resistance in malarial parasites is increasingly emerging, hence it is essential to discover and develop alternative anti-malarial agents against both new and established drug targets. One of such possible drug targets is thiamine phosphate synthase because of its role and essentialness in the thiamine biosynthesis pathway. The present study aims to model the three-dimensional (3D) structure of thiamine phosphate synthase and to predict the potential inhibitors to derive therapeutic objectives for Plasmodium falciparum. The 3D structure was constructed using SWISS-MODEL and several computer-aided approaches were used for screening of drug-like compounds. In PyRx 0.8, molecular docking was conducted using AutoDock Vina. The absorption, distribution, metabolism and excretion properties were predicted using admetSAR. Post-docking results were analyzed using LigPlot+ program. The 3D model of thiamine phosphate synthase was generated using thiamine phosphate pyrophosphorylase from Pyrococcus furiosus as a template. Out of 156 compounds screened, only those 98 compounds which followed the Lipinski’s rule of five were used for molecular docking. The best 25 docked ligands were further subjected to admetSAR for evaluation of absorption, distribution, metabolism, excretion and toxicity properties. Among these, 3 compounds, 5b (ZINC000003953801), 5m (ZINC000001686969), and 5u (ZINC000002036738) showed good absorption, distribution, metabolism, excretion and toxicity properties. Impact of 14 nsSNPs on the PfThiE protein structure or function was also investigated. The predicted inhibitors in this study may be further oriented to the development of treatment through experimental therapeutic methods to suppress pathogenic action of P. falciparum.
Keywords
Thiamine phosphate synthase, PfThiE protein; malaria, plasmodium falciparum, nsSNP docking, parasitic diseases, protozoan infections, vector borne diseases, drug targets
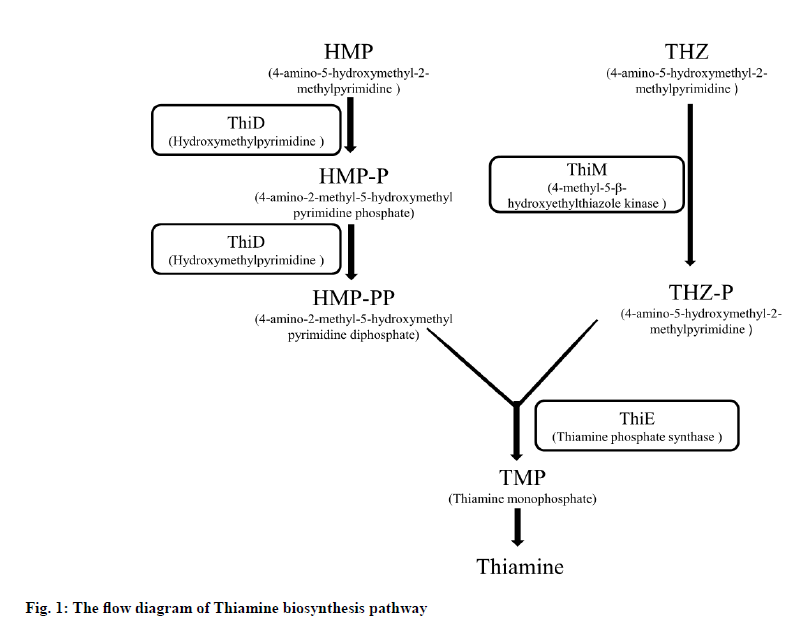
Malaria is caused by the genus Plasmodium parasite and is transmitted by infected female Anopheles mosquitoes through the bites. An estimated 228 million malaria cases and 4,05,000 deaths were reported in 87 nations in 2018 as stated by the World Health Organization in World Malaria Report of 2019[1]. Malaria remains a major public health issue in the world. Drug resistance is increasingly emerging in malaria parasites, so it is important to identify and develop alternative antimalarial agents against both new and existing drug targets. In apicomplexan parasites, thiamine biosynthesis offers a potential and exciting chance to achieve such goals, as the pathway is found only in prokaryotes, fungi, and plants, but is not present in mammals[2,3]. Thiamine pyrophosphate (Thi-PP) is the active form of vitamin B1, which is a co-factor for various enzymes primarily involved in the metabolism of carbohydrates such as 2-oxoglutarate dehydrogenase, transketolase or pyruvate dehydrogenase. For a few days, the culturing of P. falciparum in a thiamine deficient medium showed no adverse effect but a substantial need for 4-amino-5- hydroxymethyl-2-methylpyrimidine (HMP) or thiamine itself for parasite growth was reported after ten days[1]. In thiamine biosynthesis pathway (fig. 1), thiazole THZ-P (5-methyl-4-(beta-hydroxyethyl)thiazole phosphate) and pyrimidine HMP-PP (2-methyl-4-amino-5- hydroxymethylpyrimidine pyrophosphate) moieties are combined to yield thiamine phosphate by PfThiE [3,4]. For several enzymes, thiamine is metabolized as an essential cofactor[5]. So, a novel drug target thiamine phosphate synthase (PfThiE) of P. falciparum which is essential enzyme in thiamine biosynthesis was chosen to screen potent anti-malarial drugs. The human host’s lack of vitamin biosynthesis signifies that inhibition of the parasite pathways can be a way to particularly interfere with the development of parasites[6].
The present study aimed to investigate the possible effects of nsSNPs in PfThiE and their effects on its structure and function, 3D structure formation and prediction of inhibitors for the modelled structure. Till date no reports are available on the effect of deleterious SNPs and docking studies experimentally or computationally on PfThiE of P. falciparum. Non-synonymous single nucleotide polymorphisms (nsSNPs) lead to variations in the amino acid sequence, as they influence the primary polypeptide directly. These changes are not only associated with their primary sequence modification but can also alter or impair the structure and function of protein in the amino acid sequence. Numerous studies have predicted most deleterious nsSNPs among recorded polymorphisms and understood their effect on protein function, structure, and stability[7,8]. Many researchers have been studied SNPs of P. falciparum[9,10]. Functional analysis, stability analysis and conservation analysis were performed for PfThiE protein. The 3D structure of PfThiE was developed and validated. Several potent ligand molecules were identified by virtual screening method and evaluated through ADMET (Absorption, Distribution, Metabolism, Excretion and Toxicity) properties. The interactions between proteins and ligands were studied in this study using molecular docking. In the lack of the molecular structure, the proposed 3D model will be useful in providing a novel target against malaria for structure-based drug design.
Materials and Methods
Identification of nonsynonymous SNPs:
The protein sequence of PfThiE protein was retrieved from the NCBI Gene Bank (XP_966127.2) and UniProtKB (C6KSY2) databases. The Polymorphism information for the PfThiE (PF3D7_0614000) was retrieved from PlasmoDB database (https://plasmodb. org/plasmo/). In this study, only nsSNPs were used for analysis because non-synonymous mutations can change the protein sequences, which ultimately change structure and function of protein.
Functional analysis of nonsynonymous SNPs:
SIFT - Sorting Intolerant from Tolerant [11] (http:// sift.bii.a-star.edu.sg/www/SIFT_seq_submit2.html), PROVEAN - Protein Variation Effect Analyzer [12] (http:// provean.jcvi.org/), PredictSNP - Consensus classifiers for prediction of disease-related mutations [13] (https:// loschmidt.chemi.muni.cz/predictsnp/) and SNAP2 - Screening for Non-acceptable Polymorphisms [14] (https://rostlab.org/services/snap2web/) web-based tools were used to predict whether a substitution of amino acid affects the biological function of a protein. SIFT is a multi-step algorithm that uses homology sequences to distinguish amino acid substitutions. SIFT expects deleterious substitutions with values < 0.05. PROVEAN is a sequence-based method that utilizes clustering of BLAST hits. For each supporting sequence, a delta alignment score is calculated, and then combined in and through clusters for PROVEAN score calculation. PROVEAN scores below -2.5 are known to be having a deleterious effect for individual SNPs. PredictSNP was specifically designed to combine the predicted outcomes of a number of tools to create a consensus forecast. By using various biophysical features, evolutionary knowledge and several features of structure, SNAP2 determines whether or not an SNP is likely to alter the function of proteins. It provides prediction results in the form of effect or neutral and a score ranging from -100 to 100.
Protein stability analysis:
I-Mutant 2.0[15] (http://folding.biofold.org/i-mutant/imutant2.0. html) has been used for the analysis of protein stability and alterations by taking into account the SNPs. Protein sequence, temperature (25oC), pH (7.0), and detailed SNP data are the input parameters for this tool. It provides prediction in the form of either Reliability Index (RI) or Free Energy change value (DDG).
ConSurf server:
Using a Bayesian algorithm ConSurf [16] (consurf.tau. ac.il/) was used to evaluate the evolutionary stability of amino acid positions in the protein. Conserved regions were predicted using conservation scores and a colour scheme and further divided into different nine-degree scales. The score of conservation is 1 - 4 for variable, 5 - 6 for intermediate and 7 - 9 for conserved regions.
Prediction of 3D structure and validation of modelled protein:
The Protein Data Bank (https://www.rcsb.org/) lacks the 3D structure of thiamine phosphate synthase (PfThiE). Therefore, SWISS-MODEL (https://swissmodel. expasy.org/) [17] was used to build 3D structure of protein by submitting FASTA protein sequence. SWISSMODEL is a fully automated server that uses the crystal structure of similar protein as a template to predict 3D protein structures. Depending on global model quality estimation (GMQE) and qualitative model energy analysis (QMEAN) values, the most reliable 3D structure has been selected. Further, Verify3D, ERRAT and PROCHECK tools available on SAVES v5.0 (https://servicesn.mbi.ucla.edu/SAVES/) were used to validate the predicted 3D model of protein.
Alignment of model and the template structure:
The Dali [18] (http://ekhidna2.biocenter.helsinki.fi/dali/) web server was used for comparing 3D structure of proteins. For structure comparisons, this offers four choices, PDB search, PDB25, Pairwise and All against all. Pairwise structure comparison was used to compare template structure and modelled structure. It also provides secondary amino acid structure of a protein by means of DSSP.
Screening of compounds:
Several thiamine phosphate synthase (EC 2.5.1.3) inhibitors and their analogs were taken from BRENDA (BRaunschweig ENzyme DAtabase), PubChem (https://pubchem.ncbi.nlm.nih.gov/), ZINC [19] (https:// zinc.docking.org/) and DrugBank (https://www. drugbank.ca/) compound databases and also 3D structure of protein modelled by SWISS-MODEL was uploaded to MTiOpenScreen [20] (http://bioserv.rpbs. univ-paris-diderot.fr/services/MTiOpenScreen/) for screening of drug-like compounds. MTiOpenScreen conducts automatic virtual ligand screening, based on AutoDock Vina docking. This enables a curated library of small compounds to be screened in order to identify compounds that are likely to bind to a given protein receptor. This comprises five in-house prepared libraries, containing drug-like molecules. There are many compound library filters available for screening customization.
Molecular Docking:
AutoDock Vina 1.1.2 [21] in PyRx 0.8 [22] was used to do molecular docking. ZINC database was used for retrieval of compounds in Structure Data File (SDF) format. Open Babel (http://openbabel.org) tool was used to convert various file formats. PyRx was initially used to minimize compounds energy and convert all molecules to AutoDock Ligand (PDBQT) format. The compounds without any predefined binding sites were docked against the entire surface of protein. The outcomes of docking results were reported in the form of binding energy. LigPlot+[23] program was used for the analysis of post-docking results. Using PyMOL, the docked complexes which showing lowest binding affinity values were further analyzed hydrogen and hydrophobic bond interaction analysis.
Molecular features analyses:
The ADME and drug-likeness predictions of compounds were carried out using SwissADME [24] (http://www. swissadme.ch/). The SMILES of compounds have been used in SwissADME web tool as input. Further, ADMET and the pharmacokinetic properties were evaluated using admetSAR [25] (http://lmmd.ecust.edu. cn/admetsar2) web server to ensure the druggability potential of compounds.
Results
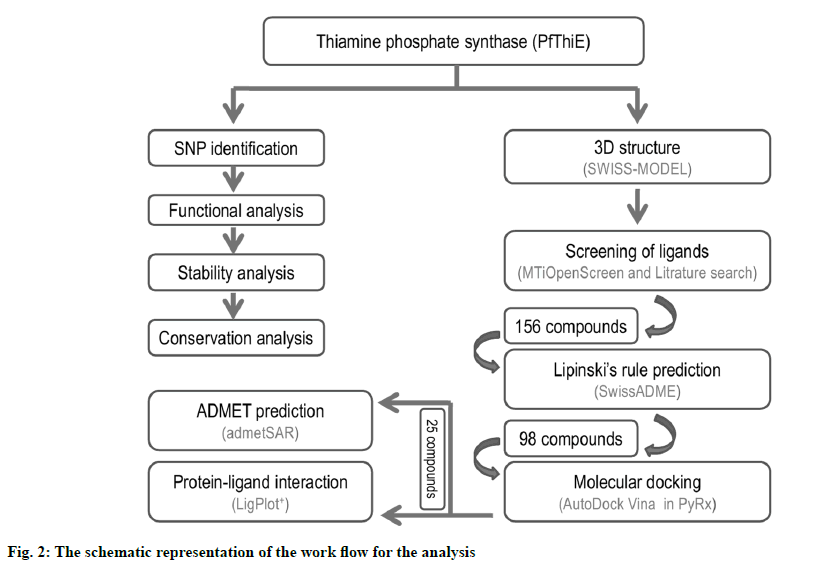
There are two key parts of this study: sequence and structure. Sequence part used the protein sequence to identify SNP, functional analysis, stability analysis, conservation analysis, and 3D structure modelling. The P. falciparum thiamine phosphate synthase (PfThiE) consists of 1617 bp, with a total of 538 amino acids in its protein. The Thiamin phosphate pyrophosphorylase gene studied had a total of 14 nsSNPs in PlasmoDB database. For further study, all these non-synonymous coding SNPs have been chosen. Second part starts with the modelled 3D structure of protein. The 3D structure of protein was used for the screening of compounds, ADMET analysis and molecular docking studies. The schematic representation of the work flow for the analysis is depicted in fig. 2.
Functional analysis of nsSNPs:
To examine whether these SNPs have any impact on protein structure or function of PfThiE, we subjected all nsSNPs to five separate damaging prediction tools. The protein amino acid sequence in FASTA format and list of mutation positions and mutations were submitted to prediction tools for predicting harmful effects. The SIFT sequence tool predicted all of 14 variants that had an effect on protein function in PfThiE. Two nsSNPs (G165D and H190P) with a score of 0.01 and one nsSNP (I220V) with a score of 0.02 were deleterious, while the remaining 11 nsSNPs exhibited a deleterious score of 0.00. Two nsSNPs (A145V and G165D) were considered to be deleterious with the PROVEAN method, with a PROVEAN score below -2.5, and the remaining nsSNPs (12) were recognized as neutral. The PROVEAN method utilizes -2.5 as a cut-off value for all predictions. According to PredictSNP, in PF3D7_0614000, 5 nsSNPs (G165D, S330C, G401E, S427W and C456F) were predicted to be deleterious while 9 were found to be in neutral. The results from the SNAP2 server anticipated that two variants (G165D and S427W) would be effective, while the remaining 12 nsSNPs were intended to be neutral. Only one nsSNP (G165D) was estimated to affect protein function by all prediction tools (SIFT, PROVEAN, PredictSNP and SNAP2) (Table 1).
| Amino acid change | SIFT | PROVEAN | PredictSNP | SNAP2 |
|---|---|---|---|---|
| A145V | Affect protein function | Deleterious | Neutral | Neutral |
| G165D | Affect protein function | Deleterious | Deleterious | Effect |
| H190P | Affect protein function | Neutral | Neutral | Neutral |
| I220V | Affect protein function | Neutral | Neutral | Neutral |
| N239H | Affect protein function | Neutral | Neutral | Neutral |
| L310V | Affect protein function | Neutral | Neutral | Neutral |
| D311E | Affect protein function | Neutral | Neutral | Neutral |
| S330C | Affect protein function | Neutral | Deleterious | Neutral |
| N355S | Affect protein function | Neutral | Neutral | Neutral |
| G401E | Affect protein function | Neutral | Deleterious | Neutral |
| S427W | Affect protein function | Neutral | Deleterious | Effect |
| I433L | Affect protein function | Neutral | Neutral | Neutral |
| C456F | Affect protein function | Neutral | Deleterious | Neutral |
| D494E | Affect protein function | Neutral | Neutral | Neutral |
Table 1:The Nssnps That Predicted To Affect Protein Function By Sift, Provean, Predictsnp And Snap2 Tools In Pfthie
Analysis of mutation effects on the protein stability:
I-Mutant further evaluated all 14 SNPs for their effect on the stability of proteins. Reliability index (RI) was predicted for each mutation. Among the 14 SNPs proposed to predict stability, 13 predicted a decrease in the stability of the protein while one was found to increase the stability. With the exception of D494E, all nsSNPs decreased the stability of proteins with a range in the RI of 0 - 9 after mutation. Analysis of mutation effects on the protein stability of 14 nsSNPs is provided in Table 2. This result suggests that these mutations of PfThiE may directly or indirectly destabilize the amino acid interactions, leading to functional protein deviations.
| I-Mutant 2.0 | ||||
|---|---|---|---|---|
| Position | WT | NEW | Stability | RI |
| 145 | A | V | Decrease | 1 |
| 165 | G | D | Decrease | 6 |
| 190 | H | P | Decrease | 5 |
| 220 | I | V | Decrease | 8 |
| 239 | N | H | Decrease | 9 |
| 310 | L | V | Decrease | 9 |
| 311 | D | E | Decrease | 2 |
| 330 | S | C | Decrease | 2 |
| 355 | N | S | Decrease | 7 |
| 401 | G | E | Decrease | 0 |
| 427 | S | W | Decrease | 0 |
| 433 | I | L | Decrease | 2 |
| 456 | C | F | Decrease | 5 |
| 494 | D | E | Increase | 4 |
Table 2: I-Mutant 2.0 Outcomes For 14 Nssnps In The Protein Pfthie
Conservation analysis of deleterious nsSNPs:
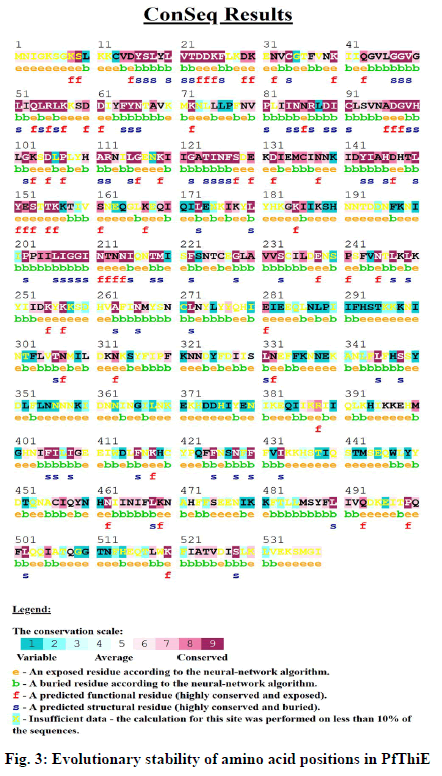
To further explore the possible effects of deleterious nsSNPs, the evolutionary conservation of amino acid residues of PfThiE protein was calculated using ConSurf web server. The ConSurf tool predictions consist of a structural protein representation which includes the colorimetric conservation score (fig. 3). ConSurf predicted I433, A145 and C456 with conservative score 9, 8 and 7 respectively. Conservation score 6 was projected for G165, L310 and D311 while score 4 was for N239, S330 and D494. However, the remaining 5 amino acids (H190, I220, N355, G401 and S427) with conservative score 1 were predicted in variable region. Positions I433, A145 and C456 were expected in highly conserved regions so mutation in these amino acids suggests more possibility of altering the protein structure. The residues that are highly conserved are sometimes important for biological function. The ConSurf findings are presented in Table 3.
| Position | Amino Acid | Conservation Score | ConSurf Prediction |
|---|---|---|---|
| 145 | A145 | 8 | Buried |
| 165 | G165 | 6 | Exposed |
| 190 | H190 | 1 | Exposed |
| 220 | I220 | 1 | Exposed |
| 239 | N239 | 4 | Exposed |
| 310 | L310 | 6 | Buried |
| 311 | D311 | 6 | Exposed |
| 330 | S330 | 4 | Exposed |
| 355 | N355 | 1 | Exposed |
| 401 | G401 | 1 | Exposed |
| 427 | S427 | 1 | Buried |
| 433 | I433 | 9 | Highly conserved and buried (s) |
| 456 | C456 | 7 | Buried |
| 494 | D494 | 4 | Exposed |
Table 3: Conservation Profile Of Amino Acids In Pfthie
Protein 3D modeling and structural analysis:
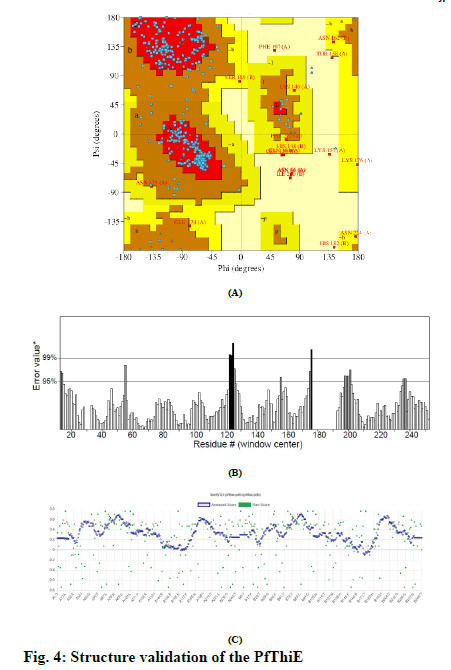
Total three 3D structure of PfThiE were generated by SWISS-MODEL. The best model with GMQE scores 0.22 and QMEAN score −4.71 was generated using thiamine phosphate pyrophosphorylase from P. furiosus (pdb ID: 1xi3.1.A) as a template for this purpose. The structure for PfThiE was predicted as homo-dimer. Ramachandran plot of the 3D model showed 82.7% of its residues in the core while 13.3% in allowed, 2.0% in generously allowed and 2.0% in disallowed regions (fig. 4A). Overall ERRAT quality score of 80.8 suggested that the structure could be regarded as a good model (fig. 4B). Verify 3D result passed the model with 81.25% of the residues have averaged 3D-1D score > = 0.2 (fig. 4C).
Target and template alignment:
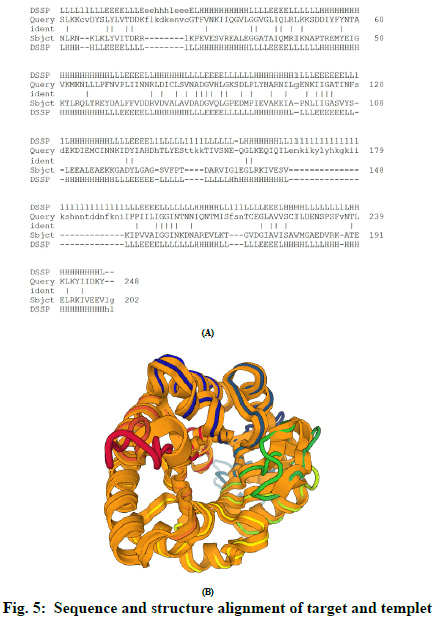
The Dali web server was used for alignment of target and template 3D structure of proteins. Alignment of template and modelled structure and sequence was performed in Dali (fig. 5A & fig 5B). Alignment score was predicted in the form of Z-score 32.9 and 32.4 for chain A and chain B, respectively. The identical amino acids are labelled with vertical bars. It also provides secondary structure of amino acid of protein by DSSP.
Screening of compounds:
Several compound databases were searched for thiamine phosphate synthase inhibitors, and 12 from BRENDA and 6 from Drug Bank were taken. Also 39 top Inhibitors against Mycobacterium tuberculosis thiamin phosphate synthase from Khare et al.[24] study was retrieved from PubChem database. MTiOpenScreen screened top 100 drug-like compounds from 10,000 compound libraries that may be inhibitors for thiamine phosphate synthase. 156 compounds were screened from all searches. These 156 drug-like compounds were retrieved in SDF format.
Molecular properties analysis:
The ADME and drug-likeness predictions of 156 compounds were carried out using SwissADME. Out of 156 compounds, 98 follows the Lipinski’s rule of five is provided as supplementary information (Table S1).
Molecular Docking and interaction analysis:
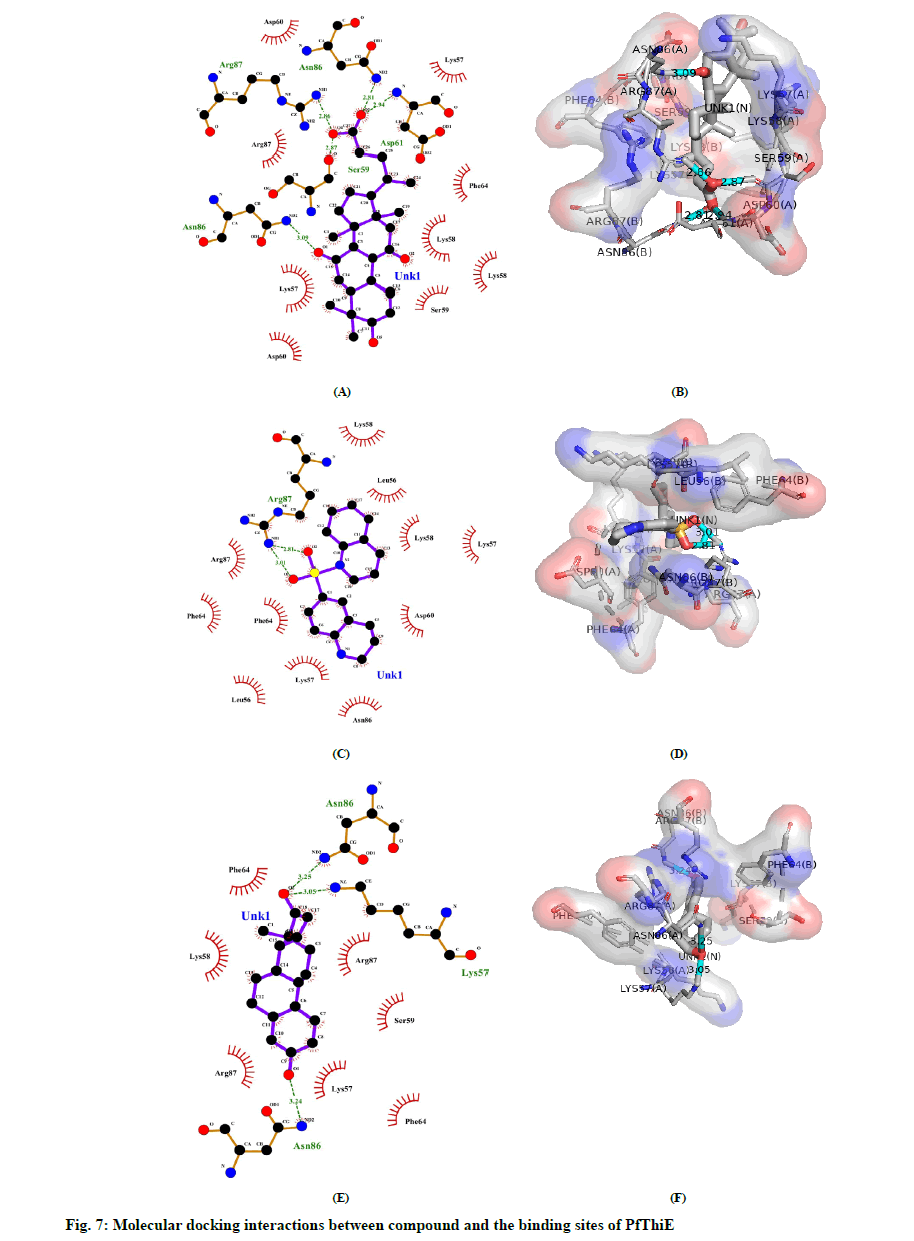
Molecular Docking was performed by using AutoDock Vina in PyRx 0.8 with 98 compounds which followed the Lipinski’s rule. PyRx was initially used to minimize compounds energy and convert all molecules to AutoDock Ligand (PDBQT) format. The value of the grid box was set to center_x = 17.7657, center_y = 21.3541, center_z = 69.9140 while size_x = 65.1899, size_y = 58.2979, and size_z = 83.6737. The default exhaustiveness value was 8. All 98 compounds without any specified binding sites were docked against whole surface of the protein. Table 4 lists the outcomes of docking results that were shown as binding affinity less than -8.0 in at least one pose. The graphical representation of the 25 best screened compounds is depicted in the fig 6. LigPlot+ software was used to predict all residues that interact with the cofactors of protein. Compound 5b had hydrogen bonds with amino acids SER59, ASP61, ASN86 AND ARG87, whereas hydrophobic interaction with LYS57, LYS58, SER59, ASP60, PHE64 and ARG87 residues (fig 7A and 7B). Compound 5m had only one hydrogen bond with amino acid ARG87, while LEU56, LYS57, LYS58, ASP60, PHE64, ARG86 and ARG87 residues had hydrophobic interaction (fig 7C and 7D). Amino acids LYS57 and ASN86 were connected to compound 5u with hydrogen bonds, while residues LYS58, SER59, PHE64 and ARG87 had hydrophobic interactions (fig 7E and 7F).
| Compounds | Binding affinity (kcal/mol) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
|
Pose 1 | Pose 2 | Pose 3 | Pose 4 | Pose 5 | Pose 6 | Pose 7 | Pose 8 | Pose 9 |
| CID 272364 | -11.2 | -11.2 | -11.1 | -10.8 | -10.7 | -10.7 | -10.7 | -10.6 | -10.4 |
| CID 219835 | -9.6 | -8.1 | -7.9 | -7.7 | -7.6 | -7.6 | -7.6 | -7.5 | -7.3 |
| ZINC000022910880 | -9.3 | -9.1 | -9 | -8.9 | -8.9 | -8.8 | -8.8 | -8.8 | -8.3 |
| ZINC000001493878 | -9 | -8.7 | -8.7 | -8.5 | -8.4 | -8.4 | -8.3 | -8 | -8 |
| CID 63114 | -9 | -8.6 | -8.4 | -8.3 | -8.3 | -7.4 | -7.3 | -7.3 | -7.3 |
| ZINC000001578333 | -8.9 | -8.3 | -8.3 | -8.2 | -8 | -7.9 | -7.8 | -7.7 | -7.7 |
| ZINC000115619865 | -8.8 | -8.5 | -8.3 | -8.2 | -8 | -7.9 | -7.8 | -7.7 | -7.7 |
| CID 291365 | -8.8 | -8.4 | -8.3 | -8.2 | -7.9 | -7.8 | -7.8 | -7.5 | -7.2 |
| ZINC000004720969 | -8.7 | -8.5 | -8.4 | -8.1 | -8.1 | -7.8 | -7.6 | -7.4 | -7.3 |
| ZINC000004214702 | -8.6 | -8.5 | -8.2 | -8.2 | -8.1 | -8.1 | -8 | -7.8 | -7.6 |
| ZINC000004215333 | -8.6 | -8.5 | -8.4 | -8.3 | -8.2 | -7.6 | -7.5 | -7.4 | -7.4 |
| ZINC000100030989 | -8.6 | -8 | -7.9 | -7.6 | -7.4 | -7.3 | -7.3 | -7.3 | -7.1 |
| CID 221226 | -8.6 | -8.2 | -8.1 | -8 | -7.6 | -7.5 | -7.4 | -6.8 | -6.8 |
| ZINC000000615883 | -8.6 | -8.5 | -8.2 | -7.9 | -7.7 | -7.6 | -7.6 | -7.5 | -7.4 |
| CID 338190 | -8.5 | -7.8 | -7.8 | -7.8 | -7.8 | -7.7 | -7.3 | -7.3 | -7.3 |
| ZINC000019804668 | -8.4 | -7.4 | -7.3 | -7.3 | -7.2 | -7.2 | -7 | -7 | -7 |
| ZINC000040863182 | -8.4 | -8.2 | -8.1 | -7.8 | -7.6 | -7.6 | -7.6 | -7.6 | -7.3 |
| CID 222169 | -8.4 | -8.1 | -7.8 | -7.5 | -7.4 | -7.4 | -7.2 | -7.1 | -7.1 |
| ZINC000000607731 | -8.3 | -7.7 | -7.6 | -7.5 | -7.3 | -7 | -6.9 | -6.9 | -6.9 |
| ZINC000002008866 | -8.2 | -7.9 | -7.8 | -7.8 | -7.6 | -7.3 | -7.3 | -7.1 | -7.1 |
| ZINC000002036738 | -8.2 | -7.7 | -7.7 | -7.3 | -7.2 | -7.2 | -7 | -6.9 | -6.7 |
| CID 227464 | -8.2 | -8.2 | -7.7 | -7.7 | -7.6 | -7.5 | -7.3 | -7.2 | -7.2 |
| CID 304907 | -8.2 | -8.2 | -8.1 | -7.9 | -7.7 | -7.7 | -7.6 | -7.5 | -7.5 |
| ZINC000034035805 | -8.1 | -8 | -7.9 | -7.7 | -7.7 | -7.7 | -7.6 | -7.6 | -7.6 |
| CID 135451590 | -8.1 | -7.3 | -7 | -6.9 | -6.7 | -6.6 | -6.5 | -6.5 | -6.5 |
Table 4: Docking Results Of Different Poses Showing Binding Affinity With Less Than -8.0 In At Least One Pose.
Evaluation of ADMET and pharmacokinetic properties:
Twenty-five best docked ligands were further subjected to admetSAR for evaluation of ADMET properties (Table 5). Out of 25 compounds, 14 compounds not exhibited toxicity to AMES. Blood Brain Barrier penetration was shown by all except 5b, 5s and 5u. There was no hepatotoxicity shown with only three compounds. The water solubility of all docked compounds is greater than -3.05. Compounds 5a, 5b, 5m, and 5v demonstrated Caco2 permeability. Intestinal absorption (human) was observed in all compounds. The acute oral toxicity of the maximum compounds was estimated as class III while 5d and 5s were indicated as II and IV, respectively. Eye corrosion and Eye irritation was not observed in any compounds.
| Model Name | 5b | 5m | 5u |
|---|---|---|---|
| Ames mutagenesis | - | - | - |
| Acute Oral Toxicity (c) | III | III | III |
| Androgen receptor binding | + | + | + |
| Aromatase binding | + | - | + |
| Avian toxicity | - | - | - |
| Blood Brain Barrier | - | + | - |
| BRCP inhibitior | - | - | + |
| Biodegradation | - | - | - |
| BSEP inhibitior | + | - | - |
| Caco-2 | + | + | + |
| Carcinogenicity (binary) | - | - | - |
| Carcinogenicity (trinary) | Non-required | Non-required | Danger |
| crustacea aquatic toxicity | - | - | + |
| CYP1A2 inhibition | - | - | + |
| CYP2C19 inhibition | - | + | - |
| CYP2C9 inhibition | - | - | - |
| CYP2C9 substrate | - | - | + |
| CYP2D6 inhibition | - | - | - |
| CYP2D6 substrate | - | + | + |
| CYP3A4 inhibition | - | + | - |
| CYP3A4 substrate | + | + | + |
| CYP inhibitory promiscuity | - | + | - |
| Eye corrosion | - | - | - |
| Eye irritation | - | - | - |
| Estrogen receptor binding | + | - | + |
| Fish aquatic toxicity | + | + | + |
| Glucocorticoid receptor binding | + | - | + |
| Honey bee toxicity | + | - | + |
| Hepatotoxicity | - | - | - |
| Human either-a-go-go inhibition | + | - | + |
| Human Intestinal Absorption | + | + | + |
| Human oral bioavailability | - | + | - |
| MATE1 inhibitior | - | - | - |
| micronuclear | - | + | - |
| Acute Oral Toxicity | 3.02 | 3.50 | 2.26 |
| OATP1B1 inhibitior | + | + | + |
| OATP1B3 inhibitior | + | + | + |
| OATP2B1 inhibitior | - | - | - |
| OCT1 inhibitior | + | - | + |
| OCT2 inhibitior | - | - | - |
| P-glycoprotein inhibitior | - | - | - |
| P-glycoprotein substrate | - | - | - |
| PPAR gamma | + | - | - |
| Plasma protein binding | 0.84 | 1.19 | 0.95 |
| Subcellular localzation | Mitochondria | Lysosomes | Mitochondria |
| Tetrahymena pyriformis | 0.65 | 2.17 | 0.73 |
| Thyroid receptor binding | + | + | + |
| UGT catelyzed | - | - | + |
| Water solubility | -3.98 | -3.70 | -4.78 |
Table 5: Admet And Pharmacokinetic Properties Of 3 Best Compounds
Discussion
The pathway of thiamine biosynthesis is present only in prokaryotes, fungi, and plants, but is not found in mammals[2]. THZ-P and HMP-PP moieties are combined in the thiamine biosynthesis pathway to produce thiamine phosphate by thiamine phosphate synthase (PfThiE)[3]. PfThiE is an important enzyme in the thiamine biosynthesis [5]. Thiamine phosphate synthase inhibitors have been studied in many species including M. tuberculosis [26], Pyrobaculum calidifontis[27], Escherichia coli [28] and Zea mays [29] but comprehensive research has not been done in P. falciparum. PfThiE is an essential enzyme in thiamine biosynthesis [30]. The thiamine biosynthesis pathway of the parasite has been proposed as a novel and indispensable antimalarial target [4]. So, current research attempted to explore the possible effects of nsSNPs in PfThiE and their effects on its structure and function, 3D structure formation and prediction of inhibitors for the modelled structure.
Rapid adaptation to changes in the environment due to high mutation rate in P. falciparum may result in drug resistance to standard medicines [2]. Thus, new drug targets are needed to develop potential inhibitors against the disease. In the present study, the impact of nsSNPs of PfThiE was investigated using various bioinformatics tools. All of the 14 nsSNPs obtained were submitted to functional analysis. SIFT tool predicted that all 14 variants had an effect on protein function. All four tools predicted G165D to affect protein function. 5 nsSNPs (G165D, S330C, G401E, S427W and C456F) by PredictSNP, 2 nsSNPs (A145V and G165D) by PROVEAN and also 2 nsSNPs (G165D and S427W) by SNAP2 considered deleterious. Furthermore, after mutation as a consequence of these nsSNPs, I-Mutant 2.0 displayed a decrease in stability except for D494E, indicating to some degree that the protein is directly or indirectly destabilized. Phylogenetic analysis using ConSurf predicted that only two nsSNPs were found in highly conserved regions. This result shows that the majority of highly conserved residues are stable.
The pharmaceutical industry increasingly utilizes computational techniques to minimize time and financial costs in the drug discovery and development process. In this study, a computational approach was used to systematically evaluate the nsSNPs to predict deleterious mutations and after that 3D model of P. falciparum thiamine phosphate synthase was developed using the X-ray crystal structure of P. furiosus thiamine phosphate pyrophosphorylase as the crystal structure of PfThiE was not available. Various validation approaches found the overall structure to be an excellent model
Among the 156 potential inhibitor compounds screened by different computational tools, 98 compounds followed Lipinski’s rule of five and these were chosen for molecular docking studies. Top 25 compounds exhibited dock score values between -8.1 and -11.2 kcal/ mol. 14 of the 25 compounds did not exhibit AMES toxicity. Only three compounds (5b, 5 m, and 5u) had all the good ADMET properties. Compounds (5a, 5b, 5e, 5h, 5i, 5 m, 5n, 5o, 5r, 5v, 5w, and 5y) investigated by Khare et al [24] as inhibitors against M. tuberculosis thiamine phosphate synthase had demonstrated strong binding affinity to PfThiE. All the three compounds (5b, 5 m, and 5u) had hydrogen bonds and hydrophobic interaction with amino acids. Further, these compounds have good ADMET properties.
P. falciparum thiamine phosphate synthase (PfThiE) is an essential enzyme in the thiamine biosynthesis which is not present in humans that can be considered as potential drug target to combat malaria menace.
It is one of the few untouched targets for developing anti-malaria drugs. The PfThiE molecular model was developed in the present study by using crystal structure of P. furiosus thiamine phosphate pyrophosphorylase as a template. Potential ligands were tried to be identified through docking-based virtual screening with drug-likeness and ADMET analysis. In this analysis, various bioinformatics approaches were also used to examine the effect of non-synonymous SNPs of PfThiE. ConSurf prediction suggests that no mutation was present in the binding site of this protein. Among the compounds being screened, 5b (ZINC000003953801), 5m (ZINC000001686969), and 5u (ZINC000002036738) had the high binding affinity and good ADMET properties. Also, for further confirmation of the protein target and potential ligands, experimental characterization is also required.
Acknowledgments
Authors thankful to Department of Biotechnology, Thapar Institute of Engineering & Technology, Patiala, India, for providing required facilities to carry out this research work.
References
- WHO (2020) World malaria report 2019. World Health Organization.
- Wrenger C, Eschbach ML, Müller IB, Laun NP, Begley TP, Walter RD. Vitamin B1 de novo synthesis in the human malaria parasite Plasmodium falciparum depends on external provision of 4-amino-5-hydroxymethyl-2-methylpyrimidine. Biol Chem 2006;387:41-51.
- Wrenger C, Knöckel J, Walter RD, Müller IB. Vitamin B1 and B6 in the malaria parasite: requisite or dispensable? Braz J Med Biol Res 2008;41:82-8.
- Zhang Y, Taylor SV, Chiu HJ, Begley TP. Characterization of the Bacillus subtilis thiC operon involved in thiamine biosynthesis. J Bacteriol 1997;179:3030-5.
- Chan XW, Wrenger C, Stahl K, Bergmann B, Winterberg M, Müller IB, et al. Chemical and genetic validation of thiamine utilization as an antimalarial drug target. Nat Commun 2013;4:2060.
- Müller S, Kappes B. Vitamin and cofactor biosynthesis pathways in Plasmodium and other apicomplexan parasites. Trends Parasitol 2007;23(3):112-21.
- Desai M, Chauhan JB (2017) Computational analysis for the determination of deleterious nsSNPs in human MTHFD1 gene. Comput Biol Chem 70:7-14.
- Solayman M, Saleh MA, Paul S, Khalil MI, Gan SH. In silico analysis of nonsynonymous single nucleotide polymorphisms of the human adiponectin receptor 2 (ADIPOR2) gene. Comput Biol Chem 2017;68:175-85.
- Subudhi AK, Boopathi PA, Pandey I, Kaur R, Middha S, Acharya J, et al. Disease specific modules and hub genes for intervention strategies: A co-expression network based approach for Plasmodium falciparum clinical isolates. Infect Genet Evol 2015;35:96-108.
- Singh SK, Reddy SM. Investigation of hub genes and their nonsynonymous single nucleotide polymorphism analysis in Plasmodium falciparum for designing therapeutic methodologies using next‑generation sequencing approach. Indian J Pharmacol 2019;51:389-99.
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009;4:1073-81.
- Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015;31:2745-7.
- Bendl J, Stourac J, Salanda O, Pavelka A, Wieben ED, Zendulka J et al. PredictSNP: robust and accurate consensus classifier for prediction of disease-related mutations. PLoS Comput Biol 2014;10: e1003440.
- Hecht M, Bromberg Y, Rost B. Better prediction of functional effects for sequence variants. BMC Genomics 2015;16:S1.
- Capriotti E, Fariselli P, Casadio R. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res 2005;33:W306-10.
- Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res 2016;44:W344-50.
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 2018;46:W296-W303.
- Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res 2010;38:W545-9.
- Sterling T, Irwin JJ. ZINC 15--Ligand Discovery for Everyone. J Chem Inf Model 2015; 55:2324-37.
- Labbé CM, Rey J, Lagorce D, Vavruša M, Becot J, Sperandio O et al. MTiOpenScreen: a web server for structure-based virtual screening. Nucleic Acids Res 2015;43:W448-54.
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010;31:455-61.
- Dallakyan S, Olson AJ. Small-molecule library screening by docking with PyRx. Methods Mol Biol 2015;1263:243-50.
- Laskowski RA, Swindells MB. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model 2011;51:2778-86.
- Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 2017;7:42717.
- Yang H, Lou C, Sun L, Li J, Cai Y, Wang Z et al. admetSAR 2.0: web-service for prediction and optimization of chemical ADMET properties. Bioinformatics 2019;35:1067-9.
- Khare G, Kar R, Tyagi AK. Identification of inhibitors against Mycobacterium tuberculosis thiamin phosphate synthase, an important target for the development of anti-TB drugs. PLoS One 2011;6:e22441.
- Hayashi M, Kobayashi K, Esaki H, Konno H, Akaji K, Tazuya K et al. Enzymatic and structural characterization of an archaeal thiamin phosphate synthase. Biochim Biophys Acta2014;1844:803-9.
- Kawasaki T. Thiamine phosphate pyrophosphorylase. Methods Enzymol 1979;62:69-73.
- Rapala-Kozik M, Olczak M, Ostrowska K, Starosta A, Kozik A. Molecular characterization of the thi3 gene involved in thiamine biosynthesis in Zea mays: cDNA sequence and enzymatic and structural properties of the recombinant bifunctional protein with 4-amino-5-hydroxymethyl-2-methylpyrimidine (phosphate) kinase and thiamine monophosphate synthase activities. Biochem J 2007;408:149-59.
- Liu X, Wang Y, Liang J, Wang L, Qin N, Zhao Y, et al. In-depth comparative analysis of malaria parasite genomes reveals protein-coding genes linked to human disease in Plasmodium falciparum genome. BMC Genomics 2018;19:312.