- *Corresponding Author:
- G. Mustafa
Department of Biochemistry, Government College University, Faisalabad-38060, Pakistan
E-mail: gmustafa_uaf@yahoo.com
| Date of Received | 22 November 2019 |
| Date of Revision | 31 March 2020 |
| Date of Acceptance | 30 April 2020 |
| Indian J Pharm Sci 2020;82(3):465-471 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
In this study, a detailed annotation of human defensin was performed and also found out the best interactors of defensin peptide from human using STRING database. The selected peptide showed best associations with its various isoforms such as neutrophil defensins, α-defensins and β-defensins. A phylogenetic tree was also reconstructed to analyze the evolutionary relationships of human defensin with its selected isoforms. The human neutrophil defensin including all neutrophil defensins appeared in a separate clade. But a great divergence has been observed in case of β-defensins because they all did not fall in a single clade and showing more distantly relationships with α-defensins and with each other as well. In future, this study will provide scientists an ease to explore novel associations of human defensins with its various isoforms and other homologs.
Keywords
Seed storage protein, neutrophil defensins, β-defensins, protein-protein interactions, phylogeny
The vertebrates[1], invertebrates, plants[2], fungi[3-4] and microorganisms[5] tackle a lot of infectious agents. To circumvent these attacks all these organisms have special sorts of survival tricks as defensive mechanisms. These defensive mechanisms include formation of bioactive compounds, which certainly constrain growth of pathogens. Among these compounds, antimicrobial peptides (AMPs) have potential to destroy the cytoplasmic membrane of microbes and these can also bind with targets present inside the cytoplasm[6]. AMPs are natural bioactive compounds and part of innate immunity and these have action of mechanism quite different from antibiotics[7].
Defensins, as their name indicates have their role in defense mechanisms. They belong to antimicrobial peptide super family. Their presence in all mammals as well as in birds, fishes, insects, fungi and plants give evidence about their wide distribution and diversity and also indicate their conserved and primitive expression. Defensin peptides present in mammals have arginine as their primary residues. The peptides are non-glycosylated and cationic in nature. Their molecular masses range from 3.5 to 6 kDa and form 3 intracellular disulfide bridges[8]. Human defensin peptides are divided into 3 categories, α, β and θ-defensins, because of their versatility in amino acid sequence and presence of disulfide bonds between different residues[9]. Both α- and β-defensins consist of a triple-stranded β-sheet with a distinctive defensin fold. It is also evident that mature cyclic θ-defensin peptides are produced by splicing of α-defensin. The θ-defensins are inactivated in humans because of a genetic mutation, which results in premature stop codon[10].
Defensins present in neutrophils are known as human neutrophil peptides. Defensins are also found in natural killer cells in some monocytes and lymphocytes cells. Defensins are also detected in Paneth cells, small intestinal cells, respiratory tract, and in gynecological tracts[11]. Many active β-defensin peptides are identified in mucosal lining, gastrointestinal tract, skin cells, and urogenital lining[12]. Various isoforms of β-defensin (such as DEFB108, DEFB1, DEFB116, DEFB128, DEFB129, DEFB125, DEFB112, DEFB104, DEFB103, DEFB118 and DEFB123) and of α-defensins (such as DEFA1, DEFA2, DEFA3, DEFA4, DEFA5 and DEFA6) have been identified[13].
Defensins have many similar roles in human body but the seminal effect of defensins is their antimicrobial activities which have been evident by in vitro studies[14]. Defensins were reported to have microbicidal effect against Gram-negative and Gram-positive bacteria. It also has fungicidal activity and reported to kill some enveloped viruses[15]. In vivo, the expression of defensins has been checked against microbial infections in mouse, which strongly supported the potential antimicrobial activity of defensins[16] through different modes of actions[17]. From the literature survey it is evident that all types of defensins have many remarkable effects on various activities in mammals and other organisms and also on their metabolic pathways. In this regard, defensins interact with many other peptides and their isoforms in different ways and in different activities. It is needed to perform a detailed study of defensin structural features, significant interactors of defensins and their relationships according to evolutionary perspective. So, in this study, computational biological approaches were used to determine the significant interaction of human defensin protein (neutrophil defensin 3) with its various isoforms. Further, a phylogram was also reconstructed to reveal evolutionary closeness of defensin with its selected isoforms. The study would help researchers to recognize the potential interactors of human defensin, which have their roles in many metabolic pathways and affect different activities and the type of interaction they share with eachother. The phylogram would also assist to study evolutionary perspectives of defensins.
Materials and Methods
Structural annotation of neutrophil defensin 3:
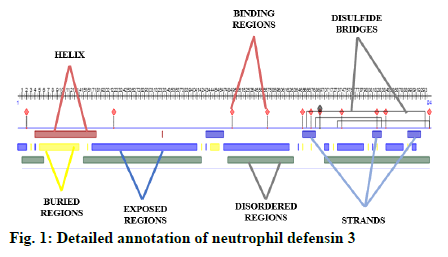
A detailed annotation of neutrophil defensin protein was predicted using an online available software Predict Protein (https://www.predictprotein.org/). This tool predicts the helices, strands, exposed regions, buried regions, disordered regions, protein binding regions and disulfide bridges of protein[1].
Protein-protein interactions of defensin:
The interactions of human defensin was analyzed using online available database STRING[18] that contains predicted and experimentally validated protein-protein interactions. Neutrophil defensin 3 from human was used as a query to get graphical presentation of its interactions with other proteins in the database. Top 10 significant interactors of query protein were explored in the form of direct (physical) and indirect (functional) associations as both types are specific and biologically meaningful.
The type of each interaction between query and its interactors such as gene fusion, coexpression, cooccurrence, homology, neighborhood, literature evidence, experimental evidence or predicted interaction were also studied. Taking in account all these types of interactions we ended up with finding a scored based evaluation of each interactor.
Phylogenetic analysis:
The FASTA sequences of query protein neutrophil defensin 3 from human (GenBank accession No. NP_005208.1) with its 10 significant interactors including neutrophil defensin 4 (accession No. NP_001916.1), neutrophil defensin 1 (accession no. NP_001289194.1), β-defensin 1 (accession no. AAM97293.1), defensin, alpha 1B (accession no. NP_001289194.1), β-defensin 108B (accession no. NP_001002035.1), β-defensin 116 (accession no. NP_001032820.1), β-defensin 128 (accession no. NP_001032821.1), β-defensin 129 (accession no. NP_543021.1), β-defensin 125 (accession no. NP_697020.2) and β-defensin 112 (accession no. NP_001032587.1) were retrieved from protein database of NCBI. All selected isoforms of defensin were derived from Homo sapiens.
The phylogenetic tree of query protein and its selected interactors was reconstructed using MEGA v6[19]. The alignment of retrieved protein sequences was done using ClustalW method in MEGA and phylogram was created by Neighbor-Joining method with 100 bootstrap replications[20].
Statistical analysis:
Overall mean distance was measured to estimate average evolutionary divergence over all sequence pairs using MEGA.
Results and Discussion
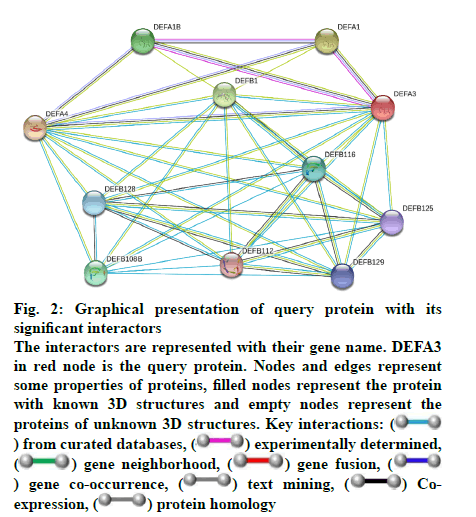
Some important features of defensin protein were also predicted by PredictProtein that is a mega service for sequence analysis. PredictProtein has been used to predict structural and functional features of different proteins since 1992. A detailed annotation and predicted features of neutrophil defensin 3 protein from human are shown in fig. 1 and detail of residues is given in Table 1. Top 10 interactors of human neutrophil defensin 3 predicted through STRING is shown in fig. 2. Edges represent the association between proteins and it does not necessarily mean they have physical associations as some other properties could also be found (Table 2).
Fig. 2: Graphical presentation of query protein with its
significant interactors
The interactors are represented with their gene name. DEFA3
in red node is the query protein. Nodes and edges represent
some properties of proteins, filled nodes represent the protein
with known 3D structures and empty nodes represent the
proteins of unknown 3D structures. Key interactions: ( ) from curated databases, (
) from curated databases, (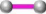 ) experimentally determined,
(
) experimentally determined,
( ) gene neighborhood, (
) gene neighborhood, ( ) gene fusion, (
) gene fusion, ( ) gene co-occurrence, (
) gene co-occurrence, ( ) text mining, (
) text mining, ( ) Coexpression,
(
) Coexpression,
( ) protein homology
) protein homology
| Description | Residues |
|---|---|
| Protein binding regions | 2, 22, 49-50, 57-63, 65, 67, 69-72, 74-80, 82, 84-92, 94 |
| Strands | 43-47, 65-68, 81-83, 89-92 |
| Helix | 4-18, 33 |
| Exposed region | 0-2, 17-41, 47-62, 64, 70-71, 73-79, 81, 84-88, 90-91 |
| Buried region | 3, 5-14, 16-42, 44-46, 65, 67, 72, 80, 82, 89 |
| Disulfide bond | 66-93, 68-83, 73-94 |
| Disordered region | 1-6, 15-42, 48-63, 70-94 |
Table 1: Description of Residues of Neutrophil Defensin 3
| Gene name | Protein name |
|---|---|
| DEF 3 | Neutrophil defensin 3(Query) |
| DEFA4 | Neutrophil defensin 4 |
| DEFA1 | Neutrophil defensin 1 |
| DEFB1 | β-defensin 1 |
| DEFA1B | Defensin, α 1B |
| DEF B108B | β-defensin 108B |
| DEFB116 | β-defensin 116 |
| DEFB128 | β-defensin 128 |
| DEFB129 | β-defensin 129 |
| DEFB125 | β-defensin 125 |
| DEFB112 | β-defensin 112 |
Table 2: Genes with Their Proteins of Human Neutrophil Defensin 3 and its Interactors
The STRING database has few evidence channels such as cooccurrence, coexpression, gene fusion, and homology. On the basis of these evidence channels the score-based interaction evaluation was also revealed. The coexpression channel represents that pair of query protein and its interactor has similar expression pattern under fluctuating conditions. Neighborhood channel indicates the proteins which are consistently observed in neighborhood of each other in the genome, e.g. genes present in an operon. Gene fusion channel determines the pair of proteins where their respective homologous genes have been fused to form single protein coding gene. Cooccurrence channel represents the similarity of two proteins in phylogenetic distribution of their orthologs. The homology channel represents the same ancestry of protein pair[18]. Each interactor was found with different scores based on above evidence channels. Scores of protein-protein interactions are given in Table 3.
| Protein name | Coexpression | Homology | Gene fusion | Neighborhood | Score |
|---|---|---|---|---|---|
| Neutrophil defensin 4 | yes | Yes | No | No | 0.977 |
| Neutrophil defensin 1 | yes | Yes | No | No | 0.969 |
| β-defensin 1 | No | No | No | No | 0.966 |
| Defensin, α 1B | yes | Yes | No | No | 0.952 |
| β-defensin 108B | No | No | No | No | 0.943 |
| β-defensin 116 | No | No | No | No | 0.940 |
| β-defensin 128 | No | No | No | No | 0.933 |
| β-defensin 129 | No | No | No | No | 0.932 |
| β-defensin 125 | No | No | No | No | 0.930 |
| β-defensin 112 | No | No | No | No | 0.930 |
Table 3: Scores of Evidence Channels Of Human Neutrophil Defensin 3 With Its Interactors
A phylogram of human neutrophil defensin 3 and its selected interactors was reconstructed (fig. 3). From the phylogram it was revealed that human neutrophil defensin 3 has strong evolutionary relationships with other neutrophil defensins as all neutrophil defensins (α-defensins) appeared as a monophyletic group. A wide divergence in β-defensins was found because they did not fall in a clade. The α-neutrophil defensins had a stronger evolutionary relationship with β-defensin 108B as compared to other β-defensins. The β-defensins were found to be distantly related. Taking into account the wide degree of divergence among β-defensins depicted from the phylogram, it could be hypothesized that there would be a horizontal gene transfer among β-defensins.
Fig. 3: Evolutionary relationship of human neutrophil defensin 3 with its selected interactors
The optimal tree with the sum of branch length = 3.46403302 is shown. The percentage of replicate trees in which the associated
taxa clustered together in the bootstrap test (100 replicates) is given above the branches. The tree is drawn to scale, with branch
lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were
computed using the p-distance method and are in the units of the number of amino acid differences per site. The analysis involved
11 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 53 positions in the
final dataset. Overall mean distance (d) was found to be 0.782
Defensins are considered as ubiquitous, present in all living organisms. For this reason, this microbial arsenal interacts with many other peptides to fulfill many crucial biological defensive and metabolic pathways[6]. They play their roles in inflammation, proliferation, homeostasis, and in wound healing in diabetic cells[21]. Anticancer activity of defensins has also been reported[22]. The proteins recognize pathogens and this quality makes them more efficient antibiotic and antiviral drugs[23]. Defensins act as antimicrobial compounds. The small cationic α-defensin, β-defensins and θ-defensins are part of innate immunity and modulators of adaptive immunity. The α-defensins mostly express in neutrophil and β-defensins express in skin cells and mucosae[13]. θ-defensins are mostly found in granules of neutrophils, monocytes and leukocyte of rhesus macaque monkey[24]. Defensin peptides have unavoidable effects on human body as their presence is needed for male fertility[25]. Remarkably, knock down of Defb15 leads to a reduction in fertility and embryonic development failure. In addition, the recombinant Defb15 has antimicrobial activity in a dose-dependent fashion. These results suggest that Defb15 plays a dual role in both sperm maturation and pathogen defense[26].
This recent exceptional study integrates a deep mechanistic understanding about annotation of protein, potential interactors and sort of interaction of human defensin supported by their evolutionary concepts. Human neutrophil defensin 3 has showed β strands, and a short helix. The study by Dhople et al.[27] showed 3 β strands and a short helix at N-terminal region of human defensin. The current findings showed 3 disulfide bonds, these predicted results are also corelated to work of Dhople et al.[27], who has also determined three disulfide bonds in human neutrophil defensin 3, which are cause of protein stability. The exposed, buried regions and protein binding regions of defensin predicted in current study, showed similarity with results of Bauer et al.[28]. The predicted disordered regions are similar to the results of Mattar et al.[29], who experimentally proved that disordered regions play important role in antimicrobial properties of defensin.
The human neutrophil defensin 3 showed significant interactions with 10 proteins, which are isoforms of defensin[30]. Neutrophil defensin 3 showed most significant interactions with neutrophil defensin 4 having score of 0.977, which is maximum value of association. In the phylogenetic analysis they showed close relationships and had same ancestor. Coexpression and homology have been observed in both neutrophil defensins and both have inhibitory effects towards Gram-negative bacteria[15,31]. Priyadarshini et al.[32] also studied and showed combined effects of α-defensin 1 and α-defensin 4. The count of both isoforms was found to be increased in nasal aspirates from children with naturally occurring adenovirus infection.
Neutrophil defensin 1 was found as the second most meaningful interactor of neutrophil defensin 3. It also showed coexpression and homology with query protein with score of 0.969. They also have inhibitory activities against Gram-positive and Gram-negative bacteria. In phylogenetic analysis both defensins also evolved from the same ancestor. Zhang et al.[33] also showed similar results regarding collective effect of α-defensin 1 and α-defensin 3 as they collectively influence the antiHIV activity of CD8. The third interactor was β-defensin 1 with a score of 0.966. It shared distant ancestry with query protein in phylogenetic analysis. It also did not show coexpression and homology in our study but the relevance of both α-defensin 3 and β-defensins 1 has been proved in the pathogenesis of diabetes and diabetic complications[34].
The fourth interactor is α-defensin 1B. Neutrophil defensin 3 and α-defensin 1B have same antibacterial, fungicide and antiviral activities. It has antimicrobial activity against Gram-negative and Gram-positive bacteria it has score value 0.952 and in our phylogeny analysis and shared same ancestor with query protein. An association study of α-defensin 3 and α-defensin 1B has been reported by Wallace et al.[35] who found that both proteins cumulatively affect the lung functions in smokers. The fifth interactor of neutrophil α-defensin 3 is β-defensin 108B, its association score is 0.943. This interactor also shared antimicrobial activities with query protein. But it does not share coexpression, homology, and occurrence with query protein. It has been reported that neutrophil defensin 3 Chertov[36] and DEFB-108[37] are induced by interleukin.
β-Defensin 116 was the sixth interactor of query protein and its association score was 0.940. It showed an association with query protein but did not show close evolutionary relationship. The seventh interactor was β-defensin 128 with and association score of 0.933. It showed distant evolutionary relationship with query protein. The 8th, 9th and 10th interactors were β-defensins 129, 125 and 112, respectively. All these β-defensins have been reported for their antimicrobial activities. Their association scores were 0.932, 0.930, and 0.930, respectively. They all showed no coexpression, cooccurrence and homology with the query protein and showed distant evolutionary relationships. The β-defensin isoforms (i.e. 112, 116, 125, 128 and 129) have been reported for their antimicrobial activities similar to α-defensins[38-39]. All defensin isoforms indicate that they all share common ancestor. The study of their adjacent chromosomal locations and same precursor genes indicated that all vertebrate defensins have arisen from common ancestor[10].
Protein-protein interactions or associations are very significant to determine the functions of respective proteins and effects of each interaction on one another. Defensins are one of the rising AMPs and because of their different potentials they would be used as new antimicrobial drugs in future. It is increasingly becoming apparent that microbes are developing resistance against previously discovered drugs. Taking in account, such potential abilities of bioactive compounds, these have such fire inside to get the title of next generation antimicrobial drugs. There are many such bioactive compounds need to be more investigated in different aspects to get fully aware of their potentials.
AMPs are the oldest components of innate immunity as these are the part of all living organisms. The diverse evolutionary history of defensins explains their varied antimicrobial activities and its worthy existence. The current study was about detailed annotation of residues of defensin protein, associations of human defensin with other proteins and types of their interactions. The study also explained those common features of interactions among defensins, which they share with each other. Because such association studies are need of the hour to determine functions, conditions and properties of defensins. The evolutionary study is the backbone of any biological process. The phylogeny of human defensin indicated its evolutionary relatedness with selected interactors and how these interactors work in coordination and how the presence or absence of any interactor could affect the processes it is involved in. The study also disclosed the novelty of defensins and some therapeutic defensins could be screened and selected in future on the basis of their antimicrobial potentials to make accessible the synthesis of these next generation antimicrobial drugs at commercial levels.
Conflict of interest:
The authors declare that they have no conflict of interest.
References
- Mustafa G, Iqbal MJ, Hassan M, Jamil A. Bioinformatics characterization of growth differentiation factor 11 of Oryctolagus cuniculus. J Chem Soc Pak 2017a;39:1089-95.
- Mustafa G, Arif R, Atta A, Sharif S, Jamil A. Bioactive compounds from medicinal plants and their importance in drug discovery in Pakistan. Mat Sci Pharm 2017b;1:17-26.
- Mustafa G, Arif R, Bukhari SA, Ali M, Sharif S, Atta A. Structural and functional annotation of citrate synthase from Aspergillus niger ANJ-120. Pak J Pharm Sci 2018;31:709-17.
- Sharif S, Atta A, Huma T, Shah AA, Afzal G, Rashid S, et al. Anticancer, antithrombotic, antityrosinase, and anti-α-glucosidase activities of selected wild and commercial mushrooms from Pakistan. Food Sci Nutr 2018;6:1-7.
- Mustafa G, Jamil A. Comparative analyses of gene clusters and Ksα genes involved in the biosynthesis of Chromomycin A3 and Mithramycin. Indian J Pharm Sci 2017;79:707-14.
- Meneguetti BT, Machado LDS, Oshiro KG, Nogueira ML, Carvalho CM, Franco OL. Antimicrobial peptides from fruits and their potential use as biotechnological tools-a review and outlook. Front Microbiol 2017;7:2136.
- Fox JL. Antimicrobial peptides stage a comeback. Nat Biotechnol2013;31:379-82.
- Ganz T. Defensins and other antimicrobial peptides: a historical perspective and an update. Comb Chem High Throughput Screen 2005;8:209-17.
- Raschig J, Mailänder-Sánchez D, Berscheid A, Berger J, Strömstedt AA, Courth LF, et al. Ubiquitously expressed Human Beta Defensin 1 (hBD1) forms bacteria-entrapping nets in a redox dependent mode of action. PLOS Pathog 2017;13:1006261.
- Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol 2003;3:710.
- Spencer JD, Hains DS, Porter E, Bevins CL, DiRosario J, Becknell B, et al. Human alpha defensin 5 expression in the human kidney and urinary tract. PloS One 2012;7:31712.
- Klotman ME, Chang TL. Defensins in innate antiviral immunity. Nat Rev Immunol 2006;6:447-56.
- Suarez-Carmona M, Hubert P, Delvenne P, Herfs M. Defensins: “Simple” antimicrobial peptides or broad-spectrum molecules. Cytokine Growth Factor Rev 2015;26:361-70.
- Tran D, Tran PA, Tang YQ, Yuan J, Cole T, Selsted ME. Homodimeric θ-defensins from rhesus macaque leukocytes: isolation, synthesis, antimicrobial activities, and bacterial binding properties of the cyclic peptides. J Biol Chem 2002;277:3079-84.
- Schneider JJ, Unholzer A, Schaller M, Schäfer-Korting M, Korting HC. Human defensins. J Mol Med 2005;83:587-95.
- Wilson CL, Ouellette AJ, Satchell DP, Ayabe T, López-Boado YS, Stratman JL, et al. Regulation of intestinal α-defensin activation by the metalloproteinase matrilysin in innate host defense. Science 1999;286:113-17.
- Yudin AI, Tollner TL, Li MW, Treece CA, Overstreet JW, Cherr GN. ESP13.2, a member of the β-defensin family, is a macaque sperm surface-coating protein involved in the capacitation process. Biol Reproduct 2003;69:1118-28.
- Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res 2016;21:937.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013;30:2725-29.
- Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol 1987;4:406-25.
- Bals R. Epithelial antimicrobial peptides in host defense against infection. Respir Res2000;1:141150.
- Hanaoka Y, Yamaguchi Y, Yamamoto H, Ishii M, Nagase T, Kurihara H, et al. In vitro and in vivo anticancer activity of β-defensin-3 and its mouse homolog. Anticancer Res 2016;36:5999-04.
- Hirsch T, Spielmann M, Zuhaili B, Fossum M, Metzig M, Koehler T, et al. Human beta‐defensin‐3 promotes wound healing in infected diabetic wounds. J Gene Med 2009;11:220-28.
- Wohlford-Lenane CL, Meyerholz DK, Perlman S, Zhou H, Tran D, Selsted ME, et al. Rhesus theta-defensin prevents death in a mouse model of severe acute respiratory syndrome coronavirus pulmonary disease. J Virol2009;83:11385-90.
- Tollner TL, Venners SA, Hollox EJ, Yudin AI, Liu X, Tang G, et al. A common mutation in the defensin DEFB126 causes impaired sperm function and subfertility. Sci Trans Med 2011;3:9265.
- Zhao Y, Diao H, Ni Z, Hu S, Yu H, Zhang Y. The epididymis-specific antimicrobial peptide β-defensin 15 is required for sperm motility and male fertility in the rat (Rattus norvegicus). Cell Mol Life Sci 2011;68:697-08.
- Dhople V, Krukemeyer A, Ramamoorthy, A. The human beta-defensin-3, an antibacterial peptide with multiple biological functions. Biochimica et Biophysica Acta 2006;1758:1499-12.
- Bauer F, Schweimer K, Klüver E, Conejo‐Garcia JR, Forssmann WG, Rösch P, et al. Structure determination of human and murine β‐defensins reveals structural conservation in the absence of significant sequence similarity. Protein Sci 2001;10:2470-79.
- Mattar EH, Almehdar HA, Yacoub HA, Uversky VN, Redwan EM. Antimicrobial potentials and structural disorder of human and animal defensins. Cytokine Growth Factor Rev 2016;28:95-11.
- Yamaguchi Y, Nagase T, Makita R, Fukuhara S, Tomita T, Tominaga T, et al. Identification of multiple novel epididymis-specific β-defensin isoforms in humans and mice. J Immunol 2002;169:2516-23.
- Li Q, Wang C, Tang C, Zhao X, He Q, Li J. Identification and characterization of blood and neutrophil-associated microbiomes in patients with severe acute pancreatitis using next-generation sequencing. Front Cell Infect Microbiol 2018;8:5.
- Priyadharshini VS, Ramírez-Jiménez F, Molina-Macip M, Renteria-Rosales C, Santiago-Cruz J, Zarate-Segura P, et al. Human Neutrophil Defensin-1,-3, and-4 Are Elevated in Nasal Aspirates from Children with Naturally Occurring Adenovirus Infection. Canadian Resp J 2018;2018:1038593.
- Zhang L, Yu W, He T, Yu J, Caffrey RE, Dalmasso EA, et al. Contribution of human α-defensin 1, 2, and 3 to the anti-HIV-1 activity of CD8 antiviral factor. Science 2002;298:995-00.
- Németh BC, Várkonyi T, Somogyvári F, Lengyel C, Fehértemplomi K, Nyiraty S, et al. Relevance of α-defensins (HNP1-3) and defensin β-1 in diabetes. World J Gastroenterol 2014;20:9128.
- Wallace AM, He JQ, Burkett KM, Ruan J, Connett JE, Anthonisen NR, et al. Contribution of alpha-and beta-defensins to lung function decline and infection in smokers: an association study. Respir Res 2006;7:76.
- Chertov O, Michiel DF, Xu L, Wang JM, Tani K, Murphy WJ, et al. Identification of defensin-1, defensin-2, and CAP37/azurocidin as T-cell chemoattractant proteins released from interleukin-8-stimulated neutrophils. J Biol Chem 1996;271:2935-40.
- Premratanachai P, Joly S, Johnson GK, McCray Jr PB, Jia HP, Guthmiller JM. Expression and regulation of novel human β‐defensins in gingival keratinocytes. Oral Microbiol Immunol 2004;19:111-17.
- Pazgier M, Hoover DM, Yang D, Lu W, Lubkowski J. Human β-defensins. Cell Mol Life Sci 2006;63:1294-13.
- Radhakrishnan Y, Hamil KG, Yenugu S, Young SL, French FS, Hall SH. Identification, characterization, and evolution of a primate β-defensin gene cluster. Gene Immunol2005;6:203.