- *Corresponding Author:
- G. Prakash Yoganandam
Department of Pharmacognosy, College of Pharmacy, Mother Theresa Post Graduate and Research Institute of Health Sciences, (Govt. of Puducherry), Gorimedu, Puducherry 605006, India
E-mail: gprakashyoga@gmail.com
| Date of Received | 29 June 2021 |
| Date of Revision | 24 April 2022 |
| Date of Acceptance | 12 September 2022 |
| Indian J Pharm Sci 2022;84(5):1203-1209 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
In 2019, a new strain of Coronavirus disease was identified in Wuhan, China. Specific therapies are unavailable and investigations regarding Coronavirus disease treatment are lacking. The present study aimed to assess bioactive compounds found in medicinal plant Phyllanthus amarus as potential Coronavirus disease main protease inhibitors, using a molecular docking study. Molecular docking was performed using Autodock 4.2. Coronavirus disease main protease was docked with several compounds, and docking was analysed by Autodock 4.2, Lopinavir was used as standard for comparison. The binding energies obtained from the docking of 6LU7 with native ligand, lopinavir, Phyllanthin, Hypophyllanthin, Hexanedioic acid, bis (2-ethylhexyl) ester, Benzeneethanamine,3,4-dimethoxy-n-((pentafluoro phenyl) methylene), Phenethylamine, 2-methoxy-alpha.-methyl-4,5-(methylenedioxy), Diisooctyl adipate, P-Tert-Octylresorcinol, 4-methyl-2,5-dimethoxy phenethylamine, 3-(2,4-dimethoxy-phenyl)-2-formylaminopropionic acid, ethyl ester, Rutin, Quercetin and Niranthin were -6.08, -4.44, -4.14, -2.32, -4.23, -4.07, -2.5, -4.66, -4.14, -3.38, -7.46, -6.28 and -4.62 kcal/mol, respectively. Therefore, lopinavir may represent potential treatment option and Rutin and Quercetin appeared to have the best potential to act as Coronavirus disease main protease inhibitors. Thus all the compounds have shown significant binding energy and potent inhibitory effect against Coronavirus disease main protease. However, further research is necessary to investigate their potential medicinal use.
Keywords
Coronavirus disease, main protease, 6LU7, Phyllanthus amarus, phytoconstituents, molecular docking
Coronavirus Disease (COVID-19) is an ongoing pandemic of COVID-19 which is caused by Severe Acute Respiratory Syndrome-Corona Virus-2 (SARS-CoV-2). The outbreak of COVID-19 was first identified in Wuhan, china, in December 2019[1]. It is easily transmissible and spread worldwide. Common symptoms are including fever, muscle and body aches, cough, sore throat, loss of smell and shortness of breath. Symptoms may appear 5-6 d after infection but it may range from 2-14 d. Table 1 shows COVID-19 outbreak updates.
| Place | Total cases | Recovered | Total deaths |
|---|---|---|---|
| World wide | 5036 815 | 1990 057 | 326 251 |
| India | 106 750 | 42 298 | 3303 |
| America | 1576 950 | 364 263 | 93 858 |
| England | 248 818 | 344 | 35 341 |
| Italy | 226 699 | 129 401 | 32 169 |
| Spain | 278 803 | 196 958 | 27 778 |
| France | 180 809 | 62 563 | 28 239 |
| China | 82 965 | 78 244 | 4634 |
| Russia | 308 705 | 85 392 | 2972 |
| Pakistan | 45 898 | 13 101 | 985 |
| Tamilnadu | 12 448 | 4895 | 84 |
| Puducherry | 18 | - | 1 |
Table 1: Covid-19 Outbreak Updates on May 20, 2020.
At present, specific therapies such as vaccine, specific antiviral drugs for COVID-19 are not available and antiviral drugs for COVID-19 are not available and acquired immunodeficiency syndrome patients, for the treatment of COVID-19-infected patients[2]. Other reported antiviral treatments form human pathogenic Corona Viruses (CoV) include nucleoside analogues, neuraminidase inhibitors, remdesivir, umifenovir (arbidol), Tenofovirdisoproxil (TDF) and Lamivudine (3TC)[2]. However, the measures that have been implemented remain limited to preventive and supportive therapies, designed to prevent further complications and organ damage. To treat COVID-19 infections, some preliminary studies have investigated potential combinations that include anti-malarial drug chloroquinone and anti-HIV vaccines. The Main protease (Mpro)/3 Chymotrypsin Like protease (3CLpro) from COVID-19 represent a potential target for the CoV replication inhibition[2]. It was observed that genome of CoV encodes two proteins ppla and pplb which are involved in spike, membrane, envelop nucleoprotein, replicase and polymerase activity of viruses. This function is performed by Mpro/3CLpro[3].
The Mpro has 3 structural domains; domain I (residues 8-101) and domain II (residues 102-184) both have beta barrel motifs representing chymotrypsin catalytic domain and domain III (residues 185-200) with a helical structure participates in dimerization of protein and active enzyme production. Mpro is considered to be a suitable target for viral inhibitor development as an approach towards SARS. At present, natural compounds have served humans as cheaper and safer drug candidates against several diseases[4,5]. Most of the herbal drugs having antiviral properties which are used as supportive treatment for COVID-19 patients. Phyllanthus amarus (P. amarus) (Keezhkainelli) is one of the herbal drugs has got hepatoprotective properties, antioxidant and anti-inflammatory properties[6] and also antiviral properties against Hepatitis B virus and reverse transcriptase of retroviruses[7,8]. From the above light of information, it was thought worth to investigate the phytoconstituents of methanolic extract of P. amarus as potential inhibitor for COVID-19 Mpro.
Materials and Methods
Ligand preparation:
The bioactive constituents used for docking were obtained from methanolic extract of P. amarus such as as Phyllanthin, Hypophyllanthin, Hexanedioic acid, bis (2-ethylhexyl) ester, Benzeneethanamine,3,4-dimethoxy-n-(pentafluorophenyl) methylene), Phenethylamine, 2-methoxy-alpha.-methyl-4,5-(methylenedioxy), Diisooctyladipate, P-Tert-Octylresorcinol, 4-methyl-2,5-dimethoxy phenethylamine, 3-(2,4-dimethoxy-phenyl)-2-formylamino-propionic acid, ethyl ester, Rutin, Quercetin and Niranthin.
Ligands can be sketched using tools like Chemsketch or Marvinsketch. While selecting the ligand, the Lipinski rule of 5 should be applied (Table 2). The rule is very important for drug development where the pharmacologically active lead structure is optimized stepwise for increased activity and selectivity as well as drug like properties.
| Compounds | Molecular weight | Log P | No. of atoms | H-bond donors | H-bond acceptors | Violations |
|---|---|---|---|---|---|---|
| Std | 628.81 | 5.69 | 46 | 4 | 9 | 2 |
| L1 | 418.53 | 3.92 | 30 | 0 | 6 | 0 |
| L2 | 430.5 | 3.5 | 31 | 0 | 7 | 0 |
| L3 | 370.57 | 7.36 | 26 | 0 | 4 | 1 |
| L4 | 359.29 | 3.75 | 25 | 0 | 3 | 0 |
| L5 | 209.25 | 1.2 | 15 | 2 | 4 | 0 |
| L6 | 370.57 | 6.84 | 26 | 0 | 4 | 1 |
| L7 | 222.33 | 4.5 | 16 | 2 | 2 | 0 |
| L8 | 195.26 | 1.34 | 14 | 2 | 3 | 0 |
| L9 | 281.31 | 1.55 | 20 | 1 | 6 | 0 |
| L10 | 610.52 | -1.06 | 43 | 10 | 16 | 3 |
| L11 | 302.24 | 1.68 | 22 | 5 | 7 | 0 |
| L12 | 432.51 | 3.95 | 31 | 0 | 7 | 0 |
Table 2: Lipinski Rule of 5 for the Docked Phytoconstituents.
Protein preparation:
To investigate the phytochemical analogs of P. amarus against SARS-CoV-2 virus, we have selected COVID-19 3CLpro/Mpro (Protein Data Bank (PDB) ID: 6LU7)[9]. The structure of protein was obtained from PDB (https://www.rcsb.org/), in PDB format (fig. 1). PDB is an archive for the crystal structures of biological macromolecules, worldwide[10].
Active sites for 6LU7:
THR24, THR26, PHE140, ASN142, GLY143, CYS145, HIS163, HIS164, GLU166, HIS172.
Molecular docking studies:
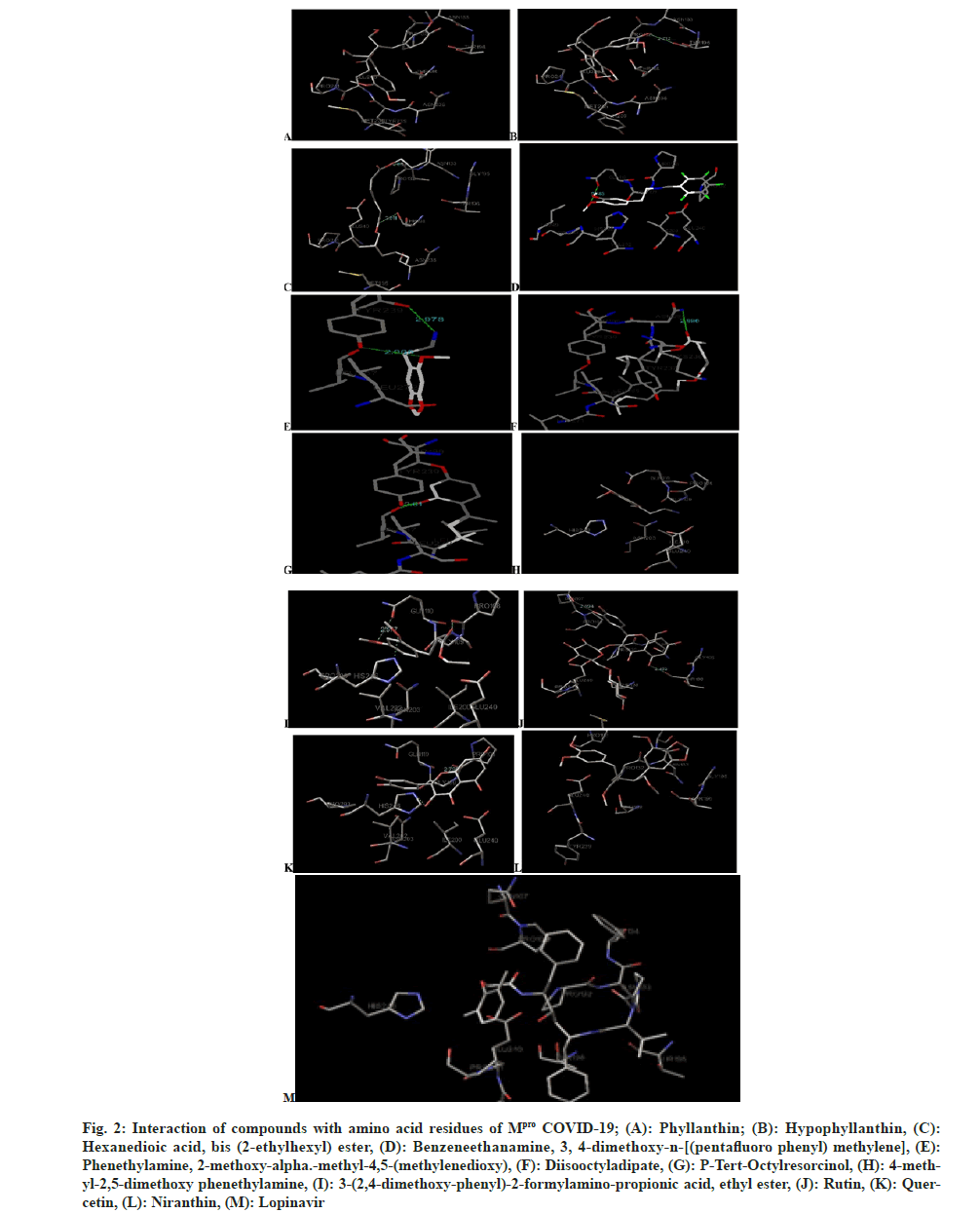
Molecular docking was carried out for standard drug Lopinavir (anti-viral drug) and 12 phytochemical constituents of methanolic extract of P. amarus. The phytochemical analogs were docked with COVID-19 3CLpro/Mpro (PDB ID: 6LU7) by using Autodock software with default settings and the grid box was defined based on trial and error and carried out in normal mode (fig. 2). The crystal structure of protein was obtained from protein data bank. The structures of phytochemical constituents were sketched by using Marvinsketch and the structures were converted into Three Dimensional (3D) images and saved as PDB format. Analysis of docking results was done with Autodock Software and the results are compared with the standard drug and shown in Table 3.
Fig. 2: Interaction of compounds with amino acid residues of Mpro COVID-19; (A): Phyllanthin; (B): Hypophyllanthin, (C): Hexanedioic acid, bis (2-ethylhexyl) ester, (D): Benzeneethanamine, 3, 4-dimethoxy-n-[(pentafluoro phenyl) methylene], (E): Phenethylamine, 2-methoxy-alpha.-methyl-4,5-(methylenedioxy), (F): Diisooctyladipate, (G): P-Tert-Octylresorcinol, (H): 4-methyl-2,5-dimethoxy phenethylamine, (I): 3-(2,4-dimethoxy-phenyl)-2-formylamino-propionic acid, ethyl ester, (J): Rutin, (K): Quercetin, (L): Niranthin, (M): Lopinavir.
| Compounds | Binding energy (kcal/mol) | Ligand efficiency | Inhibitory constant (µM) | Vanderwaals desolvation energy | Intermolecular energy |
|---|---|---|---|---|---|
| Std | -6.08 | -0.13 | 34.74 | -10.38 | -10.56 |
| L1 | -4.44 | -0.15 | 559.12 | -8.2 | -8.32 |
| L2 | -4.14 | -0.13 | 919.66 | -6.62 | -6.53 |
| L3 | -2.32 | -0.09 | 19.87 | -8.01 | -7.99 |
| L4 | -4.23 | -0.17 | 794.62 | -5.84 | -6.02 |
| L5 | -4.07 | -0.27 | 1.03 | -4.62 | -4.97 |
| L6 | -2.5 | -0.1 | 14.82 | -8.04 | -8.16 |
| L7 | -4.66 | -0.27 | 384.34 | -5.91 | -5.85 |
| L8 | -4.14 | -0.3 | 920.17 | -3.69 | -5.34 |
| L9 | -3.38 | -0.17 | 3.34 | -6.03 | -6.06 |
| L10 | -7.46 | -0.17 | 3.4 | -9.15 | -9.25 |
| L11 | -6.28 | -0.29 | 24.87 | -6.48 | -6.58 |
| L12 | -4.62 | -0.15 | 407.56 | -8.1 | -8.2 |
Table 3: Molecular Docking Analysis of Selected Phytoconstituents against 6LU7.
Results and Discussion
Computational biology means in silico investigation which is an essential arm of biotechnology focused at enhancing a deeper insight of bimolecular interactions in order to address cellular disease pathogenesis while having immense contribution towards design and development of possible therapeutic candidates[11]. Indeed, this technique has assisted in identifying lead compounds for various diseases[12].
The exponential increase in cases and mortality of COVID-19 has called for a need to develop drugs to treat this infection. Using in silico and molecular docking approaches, this study investigated the inhibitory effects of Pradimicin A, Lamivudine, Plerixafor and Lopinavir against SARS-CoV-2 Mpro[13]. Recent study on Spondias mombim L molecular docking of the studied enzymes with chlorogenic acid, lutein and zeaxanthin were carried out using PatchDock. SMEAF had remarkable enzyme inhibitory effects against Phosphodiesterase-5 (PDE-5), arginase, Angiotensin I-Converting Enzyme (ACE), cholinesterase, Monoamine Oxidase A (MAO), Ecto-5’ Nucleotidase (E-NTDase), tyrosinase and stimulated Sodium (N+)-Potassium (K+) Adenosine-Triphosphatase (ATPase) activities[14].
On the other hand, the emergence and sporadic spread of COVID-19 has called for the urgent identification of novel drug and repurposing of existing available chemotherapeutic agents against this pandemic. Corona viruses have a long history of infecting animals and humans and causing respiratory, digestive, liver and central nervous system diseases in them[15]. Currently, COVID-19 has emerged in the human population and is a potential threat to global health, worldwide[15]. The virus polyprotein encodes two proteases, the 3CLpro (Mpro) and a papain-like protease; both are vital targets for drug discovery platforms against coronaviruses. Accordingly, the Mpro is the most probable antiviral candidate due to its crucial role in self-maturation and ensuing development of polyproteins[16].
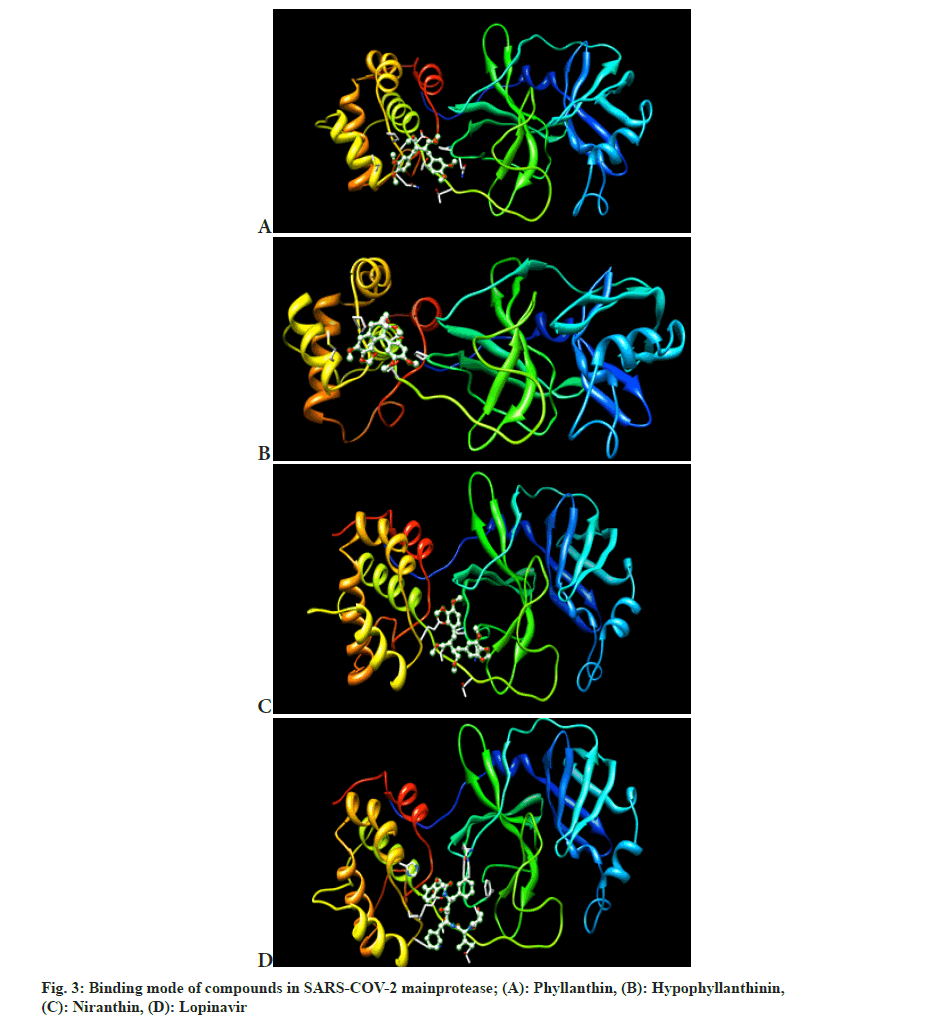
However, no approved drug currently exists to treat the disease. The currently available drugs for COVID-19 treatment primarily act on the Mpro. The aim of this study was to examine the phytoconstituents of medicinal plant P. amarus that may be used to inhibit the COVID-19 infection pathway. Table 2 shows that Lipinski’s rule of five for tested compounds. As per docking score obtained in energy kcal/mol as shown in Table 3. The highest docking score was obtained for the derivative L10 with score of -7.46 kcal/mol followed by compound L11 with score of -6.28 kcal/mol. Our study revealed that all the phytoconstituents present in P. amarus had satisfactory binding affinity and potent inhibitory effect for the SARS-CoV-2 Mpro (fig. 3). The docking analysis in the present study showed the inhibition potential of several compounds, ranked by affinity (ΔG); Rutin (L10)>Quercetin (L11)>Lopinavir (Std)>P-Tert-Octylresorcinol (L7)>Niranthin (L12)>Phyllanthin (L1)>Benzeneethanamine, 3,4-dimethoxy-n-((pentafluoro phenyl) methylene) (L4)>Hypophyllanthin (L2)>4-methyl-2,5-dimethoxy phenethylamine (L8)>Phenethylamine, 2-methoxy-alpha.-methyl-4,5-(methylenedioxy) (L5)>3-(2,4-dimethoxy-phenyl)-2-formylamino-propionic acid, ethyl ester (L9)>Diisooctyladipate (L6)>Hexanedioic acid, bis (2-ethylhexyl) ester (L3). Table 4 shows that hydrogen bond formation and its distance and amino acid involved in H-bond interactions. However, further research is necessary to investigate the potential uses of the phytoconstituents of P. amarus plant.
| Compounds | No. of hydrogen bonds formed | Amino acid involved in hydrogen bond interactions | Distance between donor and acceptor (A°) |
|---|---|---|---|
| Std | 0 | - | - |
| L1 | 0 | - | - |
| L2 | 0 | - | - |
| L3 | 2 | THR198 | 2.811 |
| L4 | 1 | GLN110 | 2.783 |
| L5 | 2 | THR199;TYR239 | 2.985; 2.978 |
| L6 | 1 | ASN238 | 2.996 |
| L7 | 1 | TYR239 | 2.61 |
| L8 | 0 | - | - |
| L9 | 1 | GLN110 | 2.577 |
| L10 | 1 | GLN107 | 2.694 |
| L11 | 0 | - | - |
| L12 | 0 | - | - |
Table 4: Hydrogen Bond Formation and its Distance and Amino Acid involved in H-Bond Interactions.
References
- Lee PI, Hsueh PR. Emerging threats from zoonotic coronaviruses-from SARS and MERS to 2019-nCoV. J Microbiol Immunol Infect 2020;53(3):365.
[Crossref] [Google Scholar] [PubMed]
- Lu H. Drug treatment options for the 2019-new coronavirus (2019-nCoV). Biosci Trends 2020;14(1):69-71.
[Crossref] [Google Scholar] [PubMed]
- Liu X, Wang XJ. Potential inhibitors against 2019-nCoV coronavirus M protease from clinically approved medicines. J Genet Genomics 2020;47(2):119-121.
[Crossref] [Google Scholar] [PubMed]
- Shen B. A new golden age of natural products drug discovery. Cell 2015;163(6):1297-300.
[Crossref] [Google Scholar] [PubMed]
- Thomford NE, Senthebane DA, Rowe A, Munro D, Seele P, Maroyi A, et al. Natural products for drug discovery in the 21st century: Innovations for novel drug discovery. Int J Mol Sci 2018;19(6):1578.
[Crossref] [Google Scholar] [PubMed]
- Patel A, Singh P, Khan S. Standardization of protocol for in vitro micropropagation of phyllanthus niruri: An important medicinal plant. Pharma Biosci J 2018;6(1):42-7.
- Adusei-Fosu K, Elegba W, Annor C, Klu GY, Danso KE. In vitro regeneration and morphogenesis in Phyllanthus niruri L., an anti-plasmodial herb. Afr J Biotechnol 2012;11(80):14542-52.
- Bagchi GD, Srivastava GN, Singh SC. Distinguishing features of medicinal herbaceous species of Phyllanthus occurring in Lucknow District (UP) India. Int J Pharmacog 1992;30(3):161-8.
- Liu X, Zhang B, Jin Z, Yang H, Rao Z. The crytal structure of 2019-nCoV main protease in complex with an inhibitor N3. RCSB Protein Data Bank 2020;10.
- Rosidi A, Khomsan A, Setiawan B, Riyadi H, Briawan D. Antioxidant potential of temulawak (Curcuma xanthorrhiza roxb). Pakistan J Nutr 2016;15(6):556-60.
- To KK, Hung IF, Chan JF, Yuen KY. From SARS coronavirus to novel animal and human coronaviruses. J Thorac Dis 2013;5(2):S103-8.
[Crossref] [Google Scholar] [PubMed]
- Abdullahi M, Olotu FA, Soliman ME. Dynamics of allosteric modulation of lymphocyte function associated antigen-1 closure-open switch: Unveiling the structural mechanisms associated with outside-in signaling activation. Biotechnol Lett 2017;39(12):1843-51.
[Crossref] [Google Scholar] [PubMed]
- Aruleba RT, Adekiya TA, Oyinloye BE, Kappo AP. Structural studies of predicted ligand binding sites and molecular docking analysis of Slc2a4 as a therapeutic target for the treatment of cancer. Int J Mol Sci 2018;19(2):386.
[Crossref] [Google Scholar] [PubMed]
- Fadaka AO, Pretorius A, Klein A. Functional prediction of candidate MicroRNAs for CRC management using in silico approach. Int J Mol Sci 2019;20(20):5190.
[Crossref] [Google Scholar] [PubMed]
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020;382(8):727-33.
[Crossref] [Google Scholar] [PubMed]
- Muralidharan N, Sakthivel R, Velmurugan D, Gromiha MM. Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. J Biomol Struct Dyn 2021;39(7):2673-8.
[Crossref] [Google Scholar] [PubMed]