- *Corresponding Author:
- Sarita Mallik
School of Biological and Life Sciences, Galgotias University, Greater Noida, Uttar Pradesh 203201, India
E-mail: sarita81@gmail.com
| Date of Received | 01 February 2023 |
| Date of Revision | 20 October 2023 |
| Date of Acceptance | 26 March 2024 |
| Indian J Pharm Sci 2024;86(2):573-579 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Emergence and spread of antibiotics resistance among bacteria poses a serious concern for human health and disease therapy. In addition to this, novel pathogens are also surfacing rapidly. Discovery and development of newer, potent antimicrobial and therapeutic agents is constantly required. Soil is perplexing at different climatic conditions thereby providing variable source for the production of antibiotic producing microorganisms. Bacteriocin is a proteinaceous substance produced by bacteria which inhibits the growth of closely or exactly related bacterial strains. It is natural, safe secondary metabolite and its development and need for better antibiotic spectrum is always in high demand. In the present investigation, the bacteriocin producing microorganisms were isolated from the local rhizospheric soil samples. The isolates were subject to antibacterial susceptibility test against the common pathogenic bacteria i.e., Pseudomonas, Aeromonas, Bacillus and Escherichia coli by agar well diffusion method. The prominent zone of inhibition shown by isolate strains was recorded. Out of the 11 isolates, 5 isolated bacterial strains displayed significant inhibitory activity against the pathogenic bacteria. For further identification, biochemical tests were performed for these strains. These bacterial strains were identified with high similarity to Streptococcus sp., Enterococcus and Gemella berger. This investigation indicates that these bacteria have great potential for the production of antimicrobial property and this can be further characterized and utilized to control the growth of microbes in the future.
Keywords
Antibiotics, rhizosphere, bacteriocin, agar diffusion assay, Streptococcus
Humans are constantly exposed to a variety of microbes in the environment and a small portion of these microbes can interact with the host in ways that cause infections and illnesses. To control disease infection, antimicrobial compounds and antibiotics are generally used therapeutically. Development and spread of antibiotic resistance among bacteria has become a crucial issue in disease therapeutics. The prolonged overuse of antibiotics has allowed pathogens to adapt and to develop antibiotic resistance thereby reducing the effectiveness of antibiotic therapy. Additionally, resistant bacterial strains persist, spread as well as transfer resistance genes to other bacteria in environment. Another major component in the expansion of antibiotic resistance is treatment of livestock with antibiotics similar to those employed to treat human infection[1]. The emergence of novel pathogens further presents a major hazard to public health[2]. To figure out this problem search and development of new therapeutic antimicrobial agents has become increasingly important.
Bacteriocin is a ribosomally encoded proteinaceous substance produced by bacteria which inhibits the growth of closely or exactly related bacterial strains[3-5]. They are usually considered to be narrow spectrum antibiotics produced by both gram negative and gram positive bacteria. Few bacteriocins exhibit a specific antibacterial activity against species that are closely associated to the producers, while others shows broad antibacterial spectrum. Narrow spectrum bacteriocins are of particular interest as targeted therapeutics since they could be expected to have minimal impact on resident microbiota. Also, these peptides show great potential to prohibit the growth of certain antibiotic-resistant bacteria. Since the discovery of first bacteriocin, colicin V more diverse bacteriocins have been identified and describe from variety of bacterial species[6-8].
Currently, the bacteriocin nisin reported in lactic acid bacilli Lactococcus lactis, is approved as food preservative by the European Union (registered with the E number E234 as a food additive), World Health Organization and Food and Drug Administration[9,10]. Also nicin is approved for use in veterinary industry[10]. Besides this, several bacteriocins have been shown to be effective against many pathogenic bacteria[5,11]. Bacteriocins are also reported to inhibit important pathogens of plant and animals, such as Enterotoxigenic Escherichia coli (ETEC), Shiga Toxin-Producing E. coli (STEC), Methicillin-Resistant Staphylococcus aureus (MRSA), Vancomycin-Resistant Enterocci (VRE), Agrobacterium and Brenneria sp.[12-14].
Numerous reports of potential bacteriocin producing rhizospheric and plant-associated bacteria have been identified exhibiting wide range antimicrobial activity toward economically important plant pathogens[15]. To date, approximately 500 bacteriocins have been identified and characterized of which majority are produced by rhizosphere bacteria[4]. In this study, bacterial species from local rhizospheric soil samples were isolated and screened for their ability to produce bacteriocins. The cell free culture media of soil isolates was screened for antimicrobial activities against pathogenic bacteria. Following antimicrobial assays, the isolates were subjected to identification using biochemical assays and biochemical test result analysis by ABIS online software.
Materials and Methods
Isolation of rhizospheric bacteria:
A total of 4 soil samples were collected from different location in local area of Noida, Uttar Pradesh, India. The samples were collected in a sterile plastic container and transferred to laboratory Helix biogenesis, Noida, India for further analysis.
For the isolation of bacterial strains from rhizospheric soil samples, serial dilution method was utilized. Briefly, 10 g each of soil samples were serially diluted in 100 ml of sterile distilled water and homogenized. Each of soil solution samples was further diluted to obtain dilution factor of 10-6. 100 ml of appropriate serial dilution were plated with nutrient agar plates and incubated at 30° for 48 h. After incubation, plates were observed for bacterial growth and distinct colonies were randomly selected. Selected colonies that were picked out were repeatedly streaked on nutrient agar plates until pure cultures were obtained. The pure cultures were maintained on nutrient agar plate at 5° and sub-cultured at 2 w intervals. Also glycerol stocks of the cultures were prepared and stored at -20°.
Preparation of a cell free culture supernatant:
All the isolated strains were grown by propagating in Lysogeny broth (LB) for 72 h at 30°. Following this growth, the broth culture was centrifuged at 5000 rpm for 15 min. The supernatant was transferred into a new Eppendorf tube followed by another round of centrifugation until a pure supernatant was obtained. The indicator bacterial strains, Aeromonas, E. coli, Pseudomonas and Bacillus were obtained from the culture collection centre (IMTECH, Chandigarh, India). The strains were maintained on nutrient agar slants at 5° and transferred at 2 w intervals.
Agar well diffusion method:
Antibiotic sensitivity testing using agar well diffusion method is widely used to evaluate the antimicrobial activity of plants or microbial extracts[16]. The different strains isolated from soil sample were sub-cultured and tested for antimicrobial property by agar well diffusion method. Standard bacterial pathogens stains Pseudomonas, E. coli, Aeromonas and Bacillus were cultivated in 10 ml LB and kept overnight in shaking incubator at 37°. A total of 4 petri plates were prepared each containing 30 ml of Luria agar media. Solidified media plates were spread with 200 µl of bacterial suspension of standard strains respectively. The plates were left for 15 min and wells were punched in the inoculated media plates. Freshly prepared culture supernatant (50 µl) of rhizosphric isolates was loaded in the wells. The plates were incubated at 37° for 24-48 h. Inhibition of pathogenic bacteria growth was recorded as the diameter of the inhibition zone (cm).
Identification of isolated microbes by morphological and biochemical characterization:
The colonies obtained after incubation of each isolated rhizospheric strain on nutrient agar were examined for the type of growth, size, shape, elevation, consistency and pigmentation. Selected strains C1-C11 were examined by gram staining and biochemical characterization using the following biochemical test assays viz., aerobic growth, anaerobic growth, growth at 10° and 45°, growth in 6.5 % Sodium chloride (NaCl) medium, Voges-Proskauer test, esculin hydrolysis, glucose test, sorbitol test, lactose test, maltose test, mannitol test, sucrose test and starch test. The results of biochemical assay results were recorded and entered for analysis in ABIS (https://www.tgw1916.net/bacteria_logare_desktop.html) software for confirmation of bacterial species.
Results and Discussion
A total of 12 bacterial colonies were picked from bacterial isolation plates from the 4 soil samples collected in this study. We were able to grow pure culture of 11 isolates and stored the pure cultures as glycerol stocks. Sub-culturing was done from glycerol stock of pure culture for all experimental procedures. In order to identify potential bacteriocin producing strains, all the soil isolates were screened by in vitro antimicrobial activity assay against 4 major pathogenic bacteria-Pseudomonas, E. coli, Aeromonas and Bacillus. All 11 isolates were also subjected to gram staining and biochemical characterization for identification.
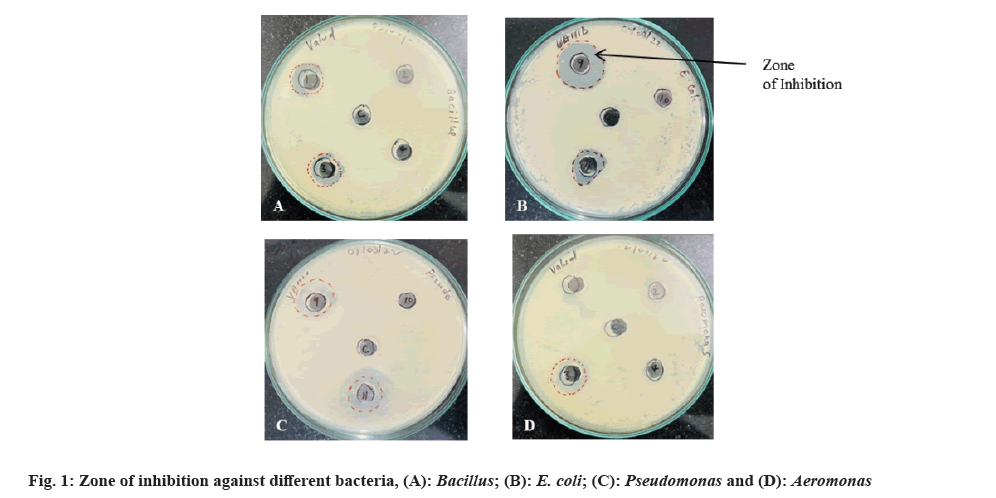
Antimicrobial Susceptibility Testing (AST) is a measurement of the susceptibility of bacteria to antimicrobial agents. Bacteria are becoming increasingly resistant to conventional antibiotics; hence, has become an important requirement to find out newer and better antimicrobial agents. Agar well diffusion method is a generally used technique to evaluate the antimicrobial activity of plants or microbial extracts[16]. In this study, the different bacterial strains isolated from soil sample were sub-cultured and culture supernatant was tested for antimicrobial property by agar well diffusion method. The strains showing the distinct inhibition of growth of standard pathogenic bacteria were recorded and diameter of zone of inhibition was measured. The results are shown in fig. 1 and Table 1.
| Isolate name | Diameter of inhibition zone (cm) | |||
|---|---|---|---|---|
| E. coli | Aeromonas | Pseudomonas | Bacillus | |
| C1 | 1.5 cm | - | - | 1.7 cm |
| C2 | - | - | - | - |
| C3 | 1.5 cm | 1.65 cm | - | 1.4 cm |
| C4 | - | - | - | - |
| C5 | - | - | - | - |
| C6 | 1.6 cm | - | - | - |
| C7 | - | - | - | - |
| C8 | - | - | - | - |
| C9 | 2.3 cm | 1.9 cm | 1.7 cm | - |
| C10 | - | - | - | - |
| C11 | 1.5 cm | - | 2.5 cm | - |
Note: (-): No zone of inhibition
Table 1: Details of zone of inhibition observed in assay for antimicrobial property by agar well diffusion method.
Of all 11 isolated bacterial strains, 5 strains (C1, C3, C6, C9 and C11) showed distinct zone of inhibition against at least one pathogen with inhibition zone diameter ranging from 1.4-2.5 cm (Table 1). All the 5 isolates inhibited the bacterium E. coli and isolate C9 exhibited the highest zone of inhibition (2.3 cm). Inhibition against Aeromonas was shown by isolates C3 (zone of inhibition=1.65 cm) and C9 (zone of inhibition=1.9 cm) only. Pseudomonas was inhibited by isolates C9 (zone of inhibition=1.7 cm) and C11 (zone of inhibition=2.5 cm) while, Bacillus growth was inhibited by isolates C1 and C3.
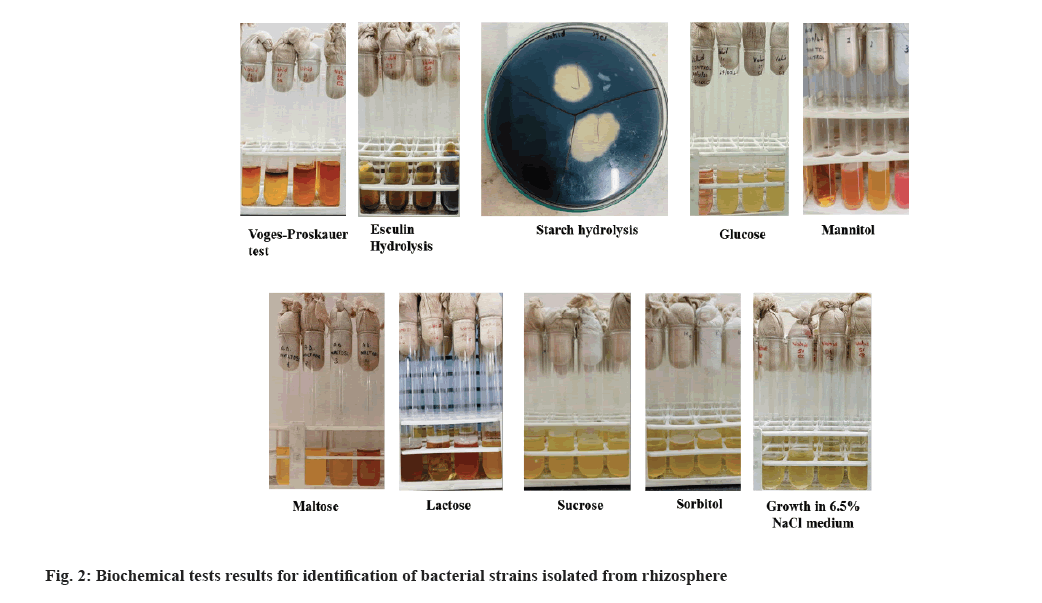
All 11 isolates were subjected to biochemical tests and morphological test which included gram staining. Gram staining results showed that all 11 pure cultures isolated from total 4 soil samples, were positive. Fig. 2 shows the results of biochemical tests performed on the rhizosphric soil isolates. Table 2 summarizes the observation for all tests. Most bacteria isolated were aerobic in nature and two of them are anaerobic too. Many sugars like lactose, sorbitol, sucrose and glucose were fermented by these bacteria and resulted in the production of acid. Some bacteria were positive for the starch hydrolysis test and show the capacity to hydrolyze the starch present in the medium and most bacteria were halotolerant as they can grow in high salt concentrations (6.5 % NaCl).
| S no. | Tests | Bacterial cultures | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C1 | C2 | C3 | C4 | C5 | C6 | C7 | C8 | C9 | C10 | C11 | ||
| 1 | Glucose | + | + | + | + | + | + | + | + | + | + | + |
| 2 | Starch | + | - | - | + | - | + | - | - | - | + | + |
| 3 | Lactose | - | + | - | + | - | + | + | + | + | + | + |
| 4 | Mannitol | - | + | - | + | - | + | + | + | - | + | + |
| 5 | Sucrose | + | + | + | + | + | + | + | + | + | + | + |
| 6 | Sorbitol | + | + | + | + | + | + | + | + | + | + | + |
| 7 | Maltose | + | + | + | - | + | - | + | + | + | + | + |
| 8 | Aerobic growth | + | + | + | + | + | + | + | + | + | + | + |
| 9 | Anaerobic growth | + | + | - | - | - | - | - | - | - | + | - |
| 10 | Growth at 10° | - | - | - | - | - | - | - | - | - | - | - |
| 11 | Growth at 45° | + | + | + | + | + | + | + | + | + | + | + |
| 12 | Growth in 6.5 % NaCl medium | + | + | + | + | + | + | + | + | + | + | + |
| 13 | Esculin hydrolysis | - | + | - | - | - | - | + | - | + | + | - |
| 14 | VP Test | - | + | + | + | + | + | + | + | + | + | - |
Note: (+): Production and (-): No production
Table 2: Details of biochemical tests results for identification of bacterial strains isolated from rhizosphere.
All the microscopic and biochemical test result were analyzed by ABIS online software. Table 3 shows the results of analysis. Based on biochemical test results, the strain of bacteria isolated from the soil sample which was identified with high similarity (81.4 %-91.9 %) to Streptococcus sp. (8 cultures), Enterococcus sp. (2 cultures) and Gemella berger (G. berger) (1 culture). The 5 isolates that were found to exhibit antimicrobial activity in agar well diffusion assay were identified as follows, isolate C1 was identified as G. berger (81.4 % similarity) and isolate C3 as Streptococcus agalactiae (S. agalactiae) with 90 % similarity. Isolate C6 showed high similarity (86 %) to Streptococcus thoraltensis (S. thoraltensis), isolate C9 showed high similarity (90.6 %) to Streptococcus equinus (S. equinus) and isolate C11 was highly similar (90.6 % similarity) to Streptococcus parasuis (S. parasuis).
| S no. | Bacterium | Similarity % | Organism |
|---|---|---|---|
| 1 | C1* | 81.4 | G. bergerie |
| 2 | C2 | 91.9 | Streptococcus salivarius subsp. salivarius |
| 3 | C3* | 90 | S. agalactiae (S. difficilis) |
| 4 | C4 | 82.9 | Streptococcus ferus |
| 5 | C5 | 81.5 | Streptococcus salivarius subsp. thermophilus |
| 6 | C6* | 85 | S. thoraltensis |
| 7 | C7 | 85 | Enterococcus thailandicus |
| 8 | C8 | 85 | Enterococcus thailandicus |
| 9 | C9* | 90.6 | S. equinus |
| 10 | C10 | 84.4 | Streptococcus alactolyticus |
| 11 | C11* | 87.9 | S. parasuis |
Note: Isolates C1, C3, C6, C9 and C11 exhibiting positive antimicrobial activity
Table 3: Identification of bacteria by analysis of biochemical test results with ABIS software.
Several bacteria produce bacteriocins which are biologically active against other gram-positive and gram-negative bacteria and inhibit growth selectively[17]. Lactic acid bacteria represent a group of gram-positive bacteria, that produce lactic acids, organic acids, secondary metabolites, inhibitory enzymes and bacteriocins as well[18,19]. Notably, bacteriocins production has been reported in several species of the lactic acid bacteria genus Streptococcus such as Bovicin HJ50 from Streptococcus bovis[20]; Mutacins B-Ny266, II, 1140/III from Streptococcus mutans[20,21]; Streptococcins A-FF22, A-M49 and A-M57, Streptin, Salivaricin A, SpbMN, Blp1, and Blp2 from Streptococcus pyogenes[22]; Salivaricin-A sa, Salivaricin A3, Salivaricin 9 from Streptococcus salivarius[21,23]; Thermophilin 347 from Streptococcus thermophilus[21] and Ubericin A, and Uberolysin from Streptococcus uberis[21,25]. In present study 4 out of 5 isolates that exibited antimicrobial activity belonged to Streptococcus genus.
S. equinus is an amylolytic lactic acid bacterium commonly associated with ruminal acidosis in cattle and gastrointestinal infections in humans[26]. In silico genome analysis of S. equinus has revealed presence of large spectrum of bactriocin encoding gene clusters such as bovine HC5, bovicin HJ50 bovicin 255, wo-component system nsuRK and the immunity genes nsuFEG, SB15, which have further validated with in vitro protein characterization and assay studies[20,27,28]. S. parasuis, a potential opportunistic zoonotic pathogen is also close relative of Streptococcus suis (S. suis), initially classified as S. suis serotypes 20, 22 and 26[29,30]. S. suis is reported to produce Suicin 3908 and suicin 90-1330[31,32].
S. agalactiae is a Group B Beta-hemolytic Streptococcus (GBS). Genome analysis identified production of the bacteriocin Agalacticin by the S. agalactiae type strain ATCC 13813[22]. Agalacticin has shown high similarity to Nisin, and possessed antimicrobial activity against several gram-positive species. Another bacteriocin Nisin P, identified in S. agalactiae strain DPC7040, which restricted several gram-positive species, especially Lactobacilli and Staphylococcus[6]. In another study, genome assessments for secondary metabolites and antimicrobial peptides detected the presence of bacteriocin zoocin A in S. agalactiae strains[33]. Furthermore, the production of bacteriocin-like substances in S. agalactiae strain isolated from vaginitis in Iraq was also reported.
S. thoraltensis is known to cause zoonotic disease and recently identified as human pathogen that generally isolated in oral flora of humans[34,35]. Genomic analysis has revealed presence of nicin like genes[36]. Gemella is a genus of gram-positive facultatively anaerobic hemolytic bacteria, i.e., indigenous in the oral cavity. Bacterial culture supernatants of Gemella haemolysans have been reported to inhibit the growth of Porphyromonas gingivitis a periodontal pathogens[37]. G. berger is catalase-negative, facultatively anaerobic cocci reported from clinical sources[38,39].
Considering the growing potential of many new bacteriocins being discovered in Streptococcus, it is pivotal to follow up such findings with molecular and functional analysis studies. Additional confirmation of the stain identities is required to be done using molecular identification tools such as 16S Ribosomal Deoxyribonucleic Acid (16S rDNA) amplification, sequencing and phylogenetic analysis for complete identification of the isolates. Also future work is required for characterization of the antimicrobial substance/bacteriocin produced by these organisms using gene amplification, sequencing and isolation and characterization of antimicrobial protein from culture supernatant.
In this study, strains of bacteria were isolated from the soil sample and analyzed for antimicrobial activity and biochemical test analysis for strain identification. Bacteria isolated are aerobic in nature and two of them are anaerobic too. The isolated bacteria give a clear zone of inhibition against pathogenic test organisms. From the above-performed experiments, and biochemical test result analysis by ABIS online software, the isolated strain of bacteria from the soil sample which shows antimicrobial activity were identified with high similarity to Streptococcus sp. (4 cultures), Enterococcus and 1 culture of G. berger. Confirmation of the stain identities is required to be done using molecular identification tools such as 16S rDNA amplification, sequencing and phylogenetic analysis for complete identification of the isolates. Also future work is required for characterization of the antimicrobial substance/bacteriocin produced by these organisms using gene amplification, sequencing and isolation and characterization of antimicrobial protein from culture supernatant.
Acknowledgement:
The authors would like to thank Mr. Ajeet Singh, Helix Biogenesis, Noida for providing valuable guidance in this work.
Conflict of interests:
The authors declared no conflict of interests.
References
- Manyi-Loh C, Mamphweli S, Meyer E, Okoh A. Antibiotic use in agriculture and its consequential resistance in environmental sources: Potential public health implications. Molecules 2018;23(4):795.
[Crossref] [Google Scholar] [PubMed]
- Piret J, Boivin G. Pandemics throughout history. Front Microbiol 2021;11:631736.
[Crossref] [Google Scholar] [PubMed]
- Blaszczyk U, Moczarny J. Bacteriocins of gram-negative bacteria-structure, mode of action and potential applications. Postepy Mikrobiol 2016;55(2):157-71.
- Mojgani N. Bacteriocin-producing rhizosphere bacteria and their potential as a biocontrol agent. Rhizotrophs Plant Growth Promotion Bioremed 2017:165-81.
- Soltani S, Hammami R, Cotter PD, Rebuffat S, Said LB, Gaudreau H, et al. Bacteriocins as a new generation of antimicrobials: Toxicity aspects and regulations. FEMS Microbiol Rev 2021;45(1):fuaa039.
[Crossref] [Google Scholar] [PubMed]
- Karaya K, Shimizu T, Taketo A. New gene cluster for lantibiotic streptin possibly involved in streptolysin S formation. J Biochem 2001;129(5):769-75.
[Crossref] [Google Scholar] [PubMed]
- Martinez FA, Balciunas EM, Converti A, Cotter PD, de Souza Oliveira RP. Bacteriocin production by Bifidobacterium spp. A review. Biotechnol Adv 2013;31(4):482-8.
[Crossref] [Google Scholar] [PubMed]
- Simons A, Alhanout K, Duval RE. Bacteriocins, antimicrobial peptides from bacterial origin: Overview of their biology and their impact against multidrug-resistant bacteria. Microorganisms 2020;8(5):639.
[Crossref] [Google Scholar] [PubMed]
- Molloy EM, Ross RP, Hill C. ‘Bac’to the future: Bioengineering lantibiotics for designer purposes. Biochem Soc Trans 2012;40(6):1492-7.
[Crossref] [Google Scholar] [PubMed]
- Singh VP. Recent approaches in food bio-preservation-a review. Open Vet J 2018;8(1):104-11.
[Crossref] [Google Scholar] [PubMed]
- Hanchi H, Hammami R, Gingras H, Kourda R, Bergeron MG, Ben Hamida J, et al. Inhibition of MRSA and of Clostridium difficile by durancin 61A: Synergy with bacteriocins and antibiotics. Future Microbiol 2017;12(3):205-12.
[Crossref] [Google Scholar] [PubMed]
- Ahmad V, Khan MS, Jamal QM, Alzohairy MA, Al Karaawi MA, Siddiqui MU. Antimicrobial potential of bacteriocins: In therapy, agriculture and food preservation. Int J Antimicrobial Agents 2017;49(1):1-1.
- Cotter PD, Ross RP, Hill C. Bacteriocins-a viable alternative to antibiotics? Nat Rev Microbiol 2013;11(2):95-105.
[Crossref] [Google Scholar] [PubMed]
- Grinter R, Milner J, Walker D. Bacteriocins active against plant pathogenic bacteria. Biochem Soc Trans 2012:40: 1498–502.
[Crossref] [Google Scholar] [PubMed]
- Timothy B, Sharma S. Isolation and characterization of microbes producing bacteriocin from curd, raw milk and soil and its preservative effects. Int J Pharm Sci Res 2014;5(5):1942.
- Magaldi S, Mata-Essayag S, De Capriles CH, Perez C, Colella MT, Olaizola C, et al. Well diffusion for antifungal susceptibility testing. Int J Infect Dis 2004;8(1):39-45.
[Crossref] [Google Scholar] [PubMed]
- Chernov VM, Chernova OA, Mouzykantov AA, Lopukhov LL, Aminov RI. Omics of antimicrobials and antimicrobial resistance. Expert OpinDrug Discov 2019;14(5):455-68.
[Crossref] [Google Scholar] [PubMed]
- Sahl HG, Bierbaum G. Lantibiotics: Biosynthesis and biological activities of uniquely modified peptides from gram-positive bacteria. Ann Rev Microbiol 1998;52(1):41-79.
[Crossref] [Google Scholar] [PubMed]
- Venegas-Ortega MG, Flores-Gallegos AC, Martinez-Hernández JL, Aguilar CN, Nevarez-Moorillon GV. Production of bioactive peptides from lactic acid bacteria: A sustainable approach for healthier foods. Compr Rev Food Sci Food Saf 2019;18(4):1039-51.
[Crossref] [Google Scholar] [PubMed]
- Mantovani HC, Hu H, Worobo RW, Russell JB. Bovicin HC5, a bacteriocin from Streptococcus bovis HC5. Microbiology 2002;148(11):3347-52.
[Crossref] [Google Scholar] [PubMed]
- Balakrishnan M, Simmonds RS, Carne A, Tagg JR. Streptococcus mutans strain N produces a novel low molecular mass non-lantibiotic bacteriocin. FEMS Microbiol Lett 2000;183(1):165-9.
[Crossref] [Google Scholar] [PubMed]
- Nes IF, Diep DB, Holo H. Bacteriocin diversity in Streptococcus and Enterococcus. J Bacteriol 2007;189(4):1189-98.
[Crossref] [Google Scholar] [PubMed]
- Vogel V, Spellerberg B. Bacteriocin production by beta-hemolytic streptococci. Pathogens 2021;10(7):867.
[Crossref] [Google Scholar] [PubMed]
- Tagg JR. Prevention of streptococcal pharyngitis by anti-Streptococcus pyogenes bacteriocin-like inhibitory substances (BLIS) produced by Streptococcus salivarius. Indian J Med Res 2004;119:13-6.
[Google Scholar] [PubMed]
- Marciset O, Jeronimus-Stratingh MC, Mollet B, Poolman B. Thermophilin 13, a nontypical antilisterial poration complex bacteriocin, that functions without a receptor. J Biol Chem 1997;272(22):14277-84.
[Crossref] [Google Scholar] [PubMed]
- Wirawan RE, Klesse NA, Jack RW, Tagg JR. Molecular and genetic characterization of a novel nisin variant produced by Streptococcus uberis. Appl Environ Microbiol 2006;72(2):1148-56.
[Crossref] [Google Scholar] [PubMed]
- Azevedo AC, Bento CB, Ruiz JC, Queiroz MV, Mantovani HC. Draft Genome Sequence of Streptococcus equinus (Streptococcus bovis) HC5, a lantibiotic producer from the bovine rumen. Genome Announcements 2015;3(2):e00085-15.
[Crossref] [Google Scholar] [PubMed]
- Joachimsthal EL, Reeves RK, Hung J, Nielsen LK, Ouwerkerk D, Klieve AV, et al. Production of bacteriocins by Streptococcus bovis strains from Australian ruminants. J Appl Microbiol 2010;108(2):428-36.
[Crossref] [Google Scholar] [PubMed]
- Wang J, Ma H, Ge X, Zhang J, Teng K, Sun Z, et al. Bovicin HJ50-like lantibiotics, a novel subgroup of lantibiotics featured by an indispensable disulfide bridge. PLoS One 2014;9(5):e97121.
[Crossref] [Google Scholar] [PubMed]
- Guo G, Wang Z, Li Q, Yu Y, Li Y, Tan Z, et al. Genomic characterization of Streptococcus parasuis, a close relative of Streptococcus suis and also a potential opportunistic zoonotic pathogen. BMC genomics. 2022 Jun 25;23(1):469.
- Wang J, Yi X, Liang P, Tao Y, Wang Y, Jin D, et al. Investigation of the genomic and pathogenic features of the potentially zoonotic Streptococcus parasuis. Pathogens 2021;10(7):834.
[Crossref] [Google Scholar] [PubMed]
- Athey TB, Vaillancourt K, Frenette M, Fittipaldi N, Gottschalk M, Grenier D. Distribution of suicin gene clusters in Streptococcus suis serotype 2 belonging to sequence types 25 and 28. Biomed Res Int 2016;2016:6815894.
[Crossref] [Google Scholar] [PubMed]
- LeBel G, Vaillancourt K, Frenette M, Gottschalk M, Grenier D. Suicin 90-1330 from a nonvirulent strain of Streptococcus suis: A nisin-related lantibiotic active on gram-positive swine pathogens. Appl Environ Microbiol 2014;80(17):5484-92.
[Crossref] [Google Scholar] [PubMed]
- Vidal Amaral JR, Juca Ramos RT, Almeida Araujo F, Bentes Kato R, Figueira Aburjaile F, de Castro Soares S, et al. Bacteriocin producing Streptococcus agalactiae strains isolated from bovine mastitis in Brazil. Microorganisms 2022;10(3):588.
[Crossref] [Google Scholar] [PubMed]
- Devriese LA, Pot B, Vandamme P, Kersters K, Collins MD, Alvarez N, Haesebrouck F, Hommez J. Streptococcus hyovaginalis sp. nov. and Streptococcus thoraltensis sp. nov., from the genital tract of sows. Int J Syst Microbiol 1997;47(4):1073-7.
[Crossref] [Google Scholar] [PubMed]
- Wazir M, Grewal M, Jain AG, Everett G. Streptococcus thoraltensis bacteremia: A case of pneumonia in a postpartum patient. Cureus 2019;11(9):e5659.
[Crossref] [Google Scholar] [PubMed]
- Heng NC, Wescombe PA, Burton JP, Jack RW, Tagg JR. The diversity of bacteriocins in Gram-positive bacteria. In Bacteriocins: Ecology and evolution. Berlin, Heidelberg: Springer Berlin Heidelberg; 2007. p. 45-92.
- Miyoshi T, Oge S, Nakata S, Ueno Y, Ukita H, Kousaka R, et al. Gemella haemolysans inhibits the growth of the periodontal pathogen Porphyromonas gingivalis. Sci Rep 2021;11(1):11742.
[Crossref] [Google Scholar] [PubMed]
- Collins MD, Hutson RA, Falsen E, Sjoden B, Facklam RR. Gemella bergeriae sp. nov., isolated from human clinical specimens. J Clin Microbiol 1998;36(5):1290-3.
[Crossref] [Google Scholar] [PubMed]