- *Corresponding Author:
- Pratima Tatke
Department of Pharmaceutical Chemistry, CU Shah College of Pharmacy, Shreemati Nathibai Damodar Thackersey (SNDT) Women’s University, Mumbai, Maharashtra 400049, India
E-mail: drpratimatatke@gmail.com
| Date of Received | 12 March 2020 |
| Date of Revision | 01 December 2021 |
| Date of Acceptance | 18 November 2022 |
| Indian J Pharm Sci 2022;84(6):1398-1406 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Glycyrrhiza glabra (licorice), is used for therapeutic purposes since ages. Neuropathic pain is a disease with complex pathophysiology, involving dysfunction in multiple ion channel receptors such as sodium, transient receptor potential channels, neurotransmitters, inflammatory mediators and reactive oxygen species. Network pharmacology is newly emerging approach dealing with understanding mechanism of actions of druggable molecules with the multiple targets. This approach is suitable to deal with AYUSH based medicines such as extracts containing many bioactives, acting on multiple targets. The aim of the present study is to construct a network of licorice to predict its actions against neuropathic pain. Various knowledge databases were used for collecting information regarding bioactives, their targets and genes associated with these targets or pathophysiology of neuropathic pain. These databases include Dr. Duke's Phytochemical and Ethnobotanical Database, PubMed, Google scholar, PubChem, BindingDB, Kyoto encyclopedia of genes and genomes, DisGeNET and Therapeutic Target Database. The network was constructed using Cytoscape3.7.2. 19 bioactive were shortlisted by extensive search having targets on humans and rats using BindingDB. These targets were searched on Kyoto encyclopedia of genes and genomes, DisGeNET for their association with neuropathic pain. Targets were shown to be part of pathways of neuropathic pain, helping to predict mechanism of action of bioactives. Hence, this approach can prove helpful in modern pharmacological research.
Keywords
Network pharmacology, licorice, neuropathic pain, transient receptor potential, herbal extracts
Network pharmacology is newly emerging approach which deals with understanding mechanism of actions of drug like molecules with its multiple Targets (TARs). This method uses various databases to retrieve information of Bioactives (BAs), their TARs and involvement of the TARs in the pathophysiology of disease. It also includes using software Cytoscape, to construct a network highlighting interrelation between the medicinal plant, its BAs and their TARs. The modern drug discovery process follows one drug-one target approach. This approach well suited for synthetic medicines having a single active ingredient. But there are limitations of this approach when it comes to deal with AYUSH medicines. Ayurvedic drugs as well as extracts of medicinal plants consist of many BAs, which would be acting on multiple TARs at the same time, producing synergistic actions[1]. Many disorders like diabetes, cardiovascular diseases, obesity and even Neuropathic Pain (NP) have complex pathophysiology, which could be treated well with multi targeted approach. Network pharmacology can be an important tool for understanding complex mechanism of action of various BAs present in herbal extracts/polyherbal medicines[2].
Glycyrrhiza glabra (G. glabra) also known as licorice and sweet wood, is native to the Mediterranean and certain areas of Asia. Historically, the dried rhizome and root of this plant were employed medicinally by the Egyptian, Chinese, Greek and Indian civilizations as an expectorant and carminative[3]. Licorice is known to have antibacterial, antiviral, antioxidant, antiinflammatory, anti-tussive, anti-ulcer, anticancer and hepatoprotective activities[4].
NP has been described as “the worst of tortures which a nerve wound may experience”[5]. NP is prominently experienced as a complication of diabetes, herpes-zoster infection, Human Immunodeficiency Virus (HIV) infection and cancer[6]. The pathophysiology of NP is complex. Peripheral NP is divided into two types depending on its pathophysiology, spontaneous pain (stimulus-independent pain) and evoked pain (stimulus dependent pain)[7]. Stimulus dependent pain results due to over activity of nociceptors. These receptors get stimulated by various stimuli such as mechanical stimuli, thermal stimuli and chemical stimuli. Mechanosensitive nociceptors are activated by pressure and mechanical stress. Thermosensitive receptors are sensitive to heat above 43°-45°[8]. Inflammation within the nerve tends to release variety of endogenous and exogenous substances like prostaglandins, bradykinin, histamine, serotonin, Tumor Necrosis Factor-alpha (TNF-α) and nerve growth factor, which also stimulates nociceptors. Nerve injury also induces up regulation of receptors of TRP family, such as TRPV1[8]. Formation of Advanced Glycosylated End products (AGEs) is related to diabetic neuropathy. It is also been found that in diabetic neuropathy, AGEs and Receptors for AGE (RAGE) interactions induce oxidative stress, result in up regulation of Nuclear Factor-Kappa B (NFkB) and various NF-kB-mediated proinflammatory genes, inducing altered pain sensation[9]. Apart from sodium and alpha adrenoceptors, calcium channels are also shown to be involved in pathophysiology of NP. The aim of this paper is to evaluate phytoconstituents of licorice by network pharmacological techniques to predict their mechanism of action against NP.
Materials and Methods
The information regarding BAs of licorice, its TARs involved in NP, genes associated with NP were collected from various databases as follows.
BAs:
The list of BAs from licorice was obtained from Dr. Duke's Phytochemical and Ethno botanical databases[10]. (Version is 1.9.12.6-Beta_2019-02-06). This website provides information about different chemicals present in plant. The website also gives list of chemicals in different parts of plants and it also provides a comprehensive list of chemicals with their [11]. For the current study, list of BAs from roots and rhizomes of licorice was downloaded.
The search of BAs was further narrowed down using PubMed and Google Scholar. Articles dealing with Glycyrrhiza uralensis, Glycyrrhiza inflata and Glycyrrhiza echinata were excluded from search as scope of this study were limited to G. glabra. Articles describing chemical analysis of aerial parts of licorice were excluded from search as the aim of the study is restricted to phytoconstituent of licorice root. Further molecular structures of these BAs were searched from databases like PubChem and Chem Spider. The names of BAs, their molecular formula, PubChem IDs were noted down and its Three Dimensional (3D) structures were downloaded in Standard Delay Format (SDF).
In order to search TARs for the BAs, it is necessary to look into the pharmacokinetics of the BAs. Hence these BAs were first searched in a database known as Swiss-Absorption, Distribution, Metabolism and Excretion (ADME). Swiss-ADME is a free web tool developed by Swiss Institute of Bioinformatics. This database helps in predicting about physicochemical properties, lipophilicity and water solubility. It can also predict pharmacokinetic parameters like GI absorption, blood brain barrier permeation, effect of hepatic metabolic enzymes and skin permeation rate. The database can also predict “drug likeness” and “lead likeness” of the compound[12]. The BAs which are predicted to be bioavailable as per Swiss-ADME were considered for TAR search.
TARs:
The TARs for the BAs were searched using Binding Database (DB). BindingDB is a freely available database which focuses mainly on interactions of proteins which can be considered as TARs with small drug like molecules[13]. The compounds can be searched in this database using SD Files or SMILES format. The TARs for the BAs were searched with the help of the tab, “Find my compound’s target”, by submitting the structures in SD files format. BindingDB searches the TARs for drug like molecules based upon structural similarities between the ligand and the target[14]. To get the optimum list of TARs the similarity search was adopted for 85 % of similarity i.e., the ligand and TARs are ≥85 % structurally similar to each other. For each of the obtained target, the information was collected as; target name, number of hits, percentage similarity, UniProt ID, UniProt name, name of the organism and gene associated with target. BindingDB is further connected to UniProt/SwissProt, Kyoto Encyclopedia of Genes and Genomes (KEGG), DrugBank and Google Scholar. UniProtKB was used for collecting UniProt ID, UniProt name, associated gene and name of organism. The BAs were also searched in database ChEMBL. ChEMBL is another database which gives information about TARs of drug like compounds. The BA was searched in this database using name and list of TARs was collected. The common TARs were excluded using filter in Excel file. TARs present in human beings and rats were included in final TARs list.
KEGG:
KEGG pathway database is a database giving detailed information about various pathways involved in pathogenesis of various diseases[15,16]. KEGG has not enlisted any separate pathway for NP. Hence the TARs were manually searched in pathways related to pathophysiology of NP i.e., regulation of TRP channels, arachidonic acid pathway and inflammation etc.
DisGeNET:
DisGeNET is a discovery platform containing one of the largest publicly available collections of genes and variants associated to human diseases. This database integrates data from repositories, GWAS catalogues, animal models and the scientific literature[16]. The version 6.0 was used for this study. This version contains 628 685 Gene-Disease Associations (GDAs), between 17 549 genes and 24 166 diseases, disorders, traits and clinical or abnormal human phenotypes[17]. Gene IDs which were collected from UniProt were submitted to this database, to check if any of these TARs are associated genetically with NP. TARs associated with peripheral neuropathy, sensory neuropathy, diabetic neuropathy, pain, neuralgia, sciatic neuropathy, inflammation were collected and a combined list was prepared.
Therapeutic Target Database (TTD):
TTD is a database which provides information about various therapeutic proteins and nucleic acid TARs of the targeted disease[18]. It also provides pathway information and the corresponding drugs directed at each of these TARs. This database also has links to relevant databases containing information about target function, sequence, 3D structures, ligand binding properties, enzyme nomenclature and drug structure, therapeutic class, clinical development status. All information provided in this database is fully referenced[19]. The list of therapeutic TARs for NP was collected from this database. Out of different collected TARs, two TARs were found to be successful therapeutic TARs for NP.
Network construction:
Network pharmacology uses various databases to retrieve information of BAs, their TARs and involvement of the TARs in the pathophysiology of disease. It also includes using software Cytoscape, to construct a network highlighting interrelation between the medicinal plant, its BAs and their TARs. Cytoscape, a Java-based open-source software platform, is a useful tool for visualizing molecular interaction networks and integrating them with any type of attribute data[20]. In addition to the basic set of features for data integration, analysis and visualization, additional features are available in the form of apps, including network and molecular profiling analysis and links with other databases. Other software, such as Gephi, an exploration platform for networks and complex systems and cell illustrator, a Java-based tool specialized in biological processes and systems can also be used for building networks[21].
A network consists of nodes and edges. The entities in which interaction is studied are called as nodes and lines connecting nodes are called as edges. The nodes of the networks in the current study include licorice, the BAs of licorice, the TARs of BAs (genes) and various neuropathy related pathways associated with those TARs. The networks were constructed using Cytoscape 3.7.2, Cytoscape is useful open-source software platform for visualizing complex networks and integrating them with any type of attribute data[22]. Excel format was used to input data in Cytoscape and to develop a network. Cytoscape network analyzer tool was used to analyze the networks.
Results and Discussion
The list of BAs from licorice was obtained from Dr. Duke's phytochemical and ethno botanical databases. The plant name G. glabra was chosen. The database gave list of 538 compounds present in licorice, out which 217 have different activities. The search of BAs from rhizomes and roots retrieved 13 and 130 BAs respectively. Further extensive search was carried out using PubMed and Google scholar and articles detailing about component analysis of licorice extracts. The articles enlisting constituents of ethanol, methanol and ethyl acetate extracts of licorice using analytical techniques like high-performance liquid chromatography, liquid chromatography–mass spectrometry, highperformance thin-layer chromatography, electrospray ionization mass spectrometry were searched and list of BAs was prepared. A comprehensive list of 36 BAs was made. These BAs were first searched in Swiss-ADME database in order to predict their bioavailability and drug likeliness as given in Table 1.
| Sl. no | Name of phytoconstituent | Molecular formula | Molecular weight (g/mol) | GI absorption | Drug-likeness (whether following Lipinski Rule) |
|---|---|---|---|---|---|
| 1 | 18βGA | C30H46O4 | 470.68 | High | Yes; 1 violation: MLOGP>4.15 |
| 2 | Isoliquiritegenin | C15H12O4 | 256.25 | High | Yes; 0 violation |
| 3 | Isoliquiritin | C21H22O9 | 418.39 | Low | Yes; 1 violation: NH or OH>5 |
| 4 | Kumatakenin | C17H14O6 | 314.29 | High | Yes; 0 violation |
| 5 | Licoflavone B | C25H26O4 | 390.47 | High | Yes; 0 violation |
| 6 | Licoflavonol | C20H18O6 | 354.35 | High | Yes; 0 violation |
| 7 | Licoflavone A | C20H18O4 | 322.35 | High | Yes; 0 violation |
| 8 | Neoisoliquiritine | C21H22O9 | 418.4 | Low | Yes; 1 violation: NH or OH>5 |
| 9 | Neoliquiritin | C21H22O9 | 418.4 | Low | Yes; 0 violation |
| 10 | Liquiritin | C21H22O9 | 418.4 | Low | Yes; 0 violation |
| 11 | Glycyrrhizin | C42H62O16 | 822.93 | Low | No; 3 violations: MW>500, N or O>10, NH or OH>5 |
| 12 | Glabrene | C20H18O4 | 322.4 | High | Yes; 0 violation |
| 13 | Licochalcone | C21H22O4 | 338.4 | High | Yes; 0 violation |
| 14 | Glabridin | C20H20O4 | 324.37 | High | Yes; 0 violation |
| 15 | Glabrol | C25H28O4 | 392.49 | High | Yes; 0 violation |
| 16 | Glycycoumarin | C21H20O6 | 368.4 | High | Yes; 0 violation |
| 17 | Glisoflavone | C21H20O6 | 368.4 | High | Yes; 0 violation |
| 18 | Licoisoflavanone | C20H18O6 | 354.4 | High | Yes; 0 violation |
| 19 | Isowighteone | C20H18O5 | 338.4 | High | Yes; 0 violation |
| 20 | Isoangustone A | C25H26O6 | 422.5 | Low | Yes; 0 violation |
| 21 | Glyasperin D | C22H26O5 | 370.4 | High | Yes; 0 violation |
| 22 | Glicophenone | C20H22O6 | 358.4 | High | Yes; 0 violation |
| 23 | Liquiritin | C21H22O9 | 418.4 | Low | Yes; 0 violation |
| 24 | Liquiritigenin | C15H12O4 | 256.25 | High | Yes; 0 violation |
| 25 | Licoisoflavone A | C20H18O6 | 354.4 | High | Yes; 0 violation |
| 26 | Glabridin | C20H20O4 | 324.4 | High | Yes; 0 violation |
| 27 | Licoricone | C22H22O6 | 382.4 | High | Yes; 0 violation |
| 28 | Neoliquiritin 2''-apioside | C26H30O13 | 550.5 | Low | No; 3 violations: MW>500, N or O>10, NH or OH>5 |
| 29 | Licorice glycoside A | C36H38O16 | 726.7 | Low | No; 3 violations: MW>500, N or O>10, NH or OH>5 |
| 30 | Isopropyl apiosylglucoside | C14H26O10 | 354.35 | Low | Yes; 1 violation: NH or OH>5 |
| 31 | Glycyrrhizaisoflavone C | C21H20O6 | 368.4 | High | Yes; 0 violation |
| 32 | Glycyrrhizaisoflavone C | C21H20O6 | 368.4 | High | Yes; 0 violation |
| 33 | Liquiritic acid | C30H46O4 | 470.7 | High | Yes; 1 violation: MLOGP>4.15 |
| 34 | Licoagroside B | C18H24O12 | 432.4 | Low | Yes; 1 violation: N or O>10 |
| 35 | Licorice glycoside B | C35H36O15 | 696.6 | Low | No; 3 violations: MW>500, N or O>10, NH or OH>5 |
| 36 | Licorice saponin A3 | C48H72O21 | 985.1 | Low | No; 3 violations: MW>500, N or O>10, NH or OH>5 |
| 37 | Licorice saponin G2 | C42H62O17 | 838.9 | Low | No; 3 violations: MW>500, N or O>10, NH or OH>5 |
Table 1: Predicted Bioavailability of Licorice Phytoconstituents as per Swiss-Adme Database
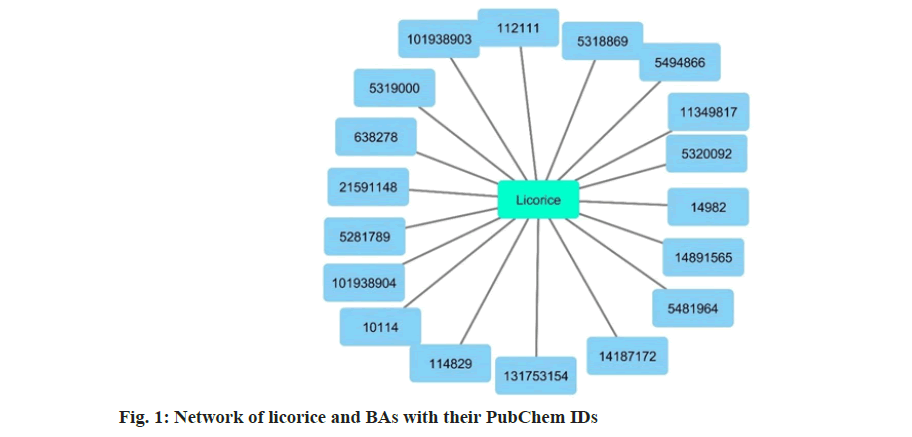
The BAs listed in Table 1 were searched in BindingDB for the TARs, using the parameters mentioned above. Out of 36 BAs, 19 were found to be having TARs in pathways related to pathophysiology of NP and diabetic neuropathy. The network of licorice and its BAs were prepared using Cytoscape, as shown in fig. 1. The list of BAs with their TARs is given in Table 2.
| Sl. no | Name of bioactive | TARs involved in NP |
|---|---|---|
| 1 | 18βGA | HSD11B1, HSD11B2, Ar, CYP19A1, CACNA1C, PTGES, PRKCH |
| 2 | Isoliquiritigenin | AKR1B1, Alox5, ABCG2, BACE1, KCNA3 |
| 3 | Kumatakenin | Adora1,ADORA3,Adora2a,Akr1b1,ABCG2,CA2, HSD17B2, NOX4 |
| 4 | Licoflavone B | CYP19A1, PPARG, Htr2a |
| 5 | Licoflavonol | Adora1, ADORA3, Adora2a, ABCB1 |
| 6 | Licoflavone A | Htr2a, Adora1, Adora2a |
| 7 | Neoisoliquiritine | AKR1B1 |
| 8 | Glycyrrhizin | HSD11B1, HSD11B2 |
| 9 | Isowighteone | PTPN1 |
| 10 | Isoangustone A | PTPN2 |
| 11 | Liquiritigenin | Ar, CA2, HSP90AB1, RIPK1, PSMB5 |
| 12 | Licoisoflavone A | PTPN1 |
| 13 | Licoricone | PTPN1 |
| 14 | Licorice glycoside A | AKR1B1 |
| 15 | Isopropyl apiosylglucoside | FGF1, ABL1, CCNA2, FGF2, HDAC10 HDC3, HDC5, HDC8, HDC9 |
| 16 | Liquiritic acid | HSD11B1, HSD11B2, HSD11B3, HSD11B4, CACNA1C, PTGES, PRKCH |
| 17 | Licorice glycoside B | AKR1B1 |
| 18 | Licorice saponin A3 | HSD11B1, HSD11B2 |
| 19 | Licorice saponin G2 | HSD11B1, HSD11B2 |
Table 2: List of Bas and their Tars Involved in Np (Gene Names)
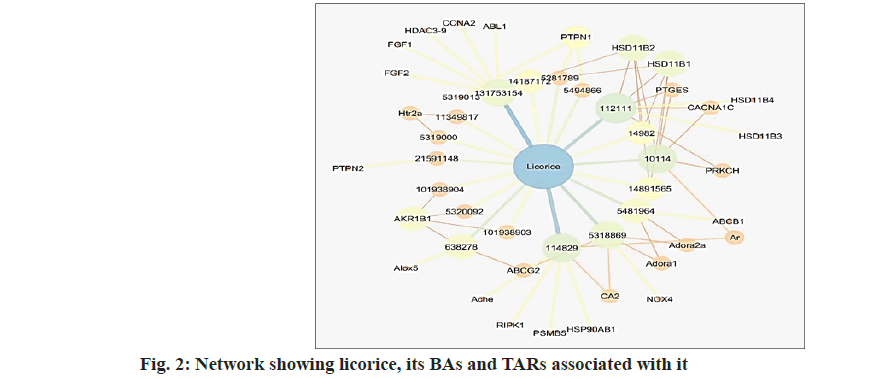
Each of the TARs was further searched in the KEGG database for their involvement in neuropathy related pathophysiology such as inflammation (TNF-α, bradykinin, serotonin, prostaglandin, histamine) arachidonic acid pathway, Na channel blocking, TRP channel regulation, mitogen-activated protein kinase pathway etc. The TARs were searched on DisGeNET for their association with NP. The list of TARs for NP was also retrieved from TTD. As per KEGG database, TARs Protein Kinase C Eta (PRKCH) and 5-Hydroxytryptamine Receptor 2A (HTR2A) were found to be associated with regulation of TRP channels. TARs Prostaglandin E Synthase (PTGES) and Arachidonate 5-Lipoxygenase (ALOX5) were found to be associated with arachidonic pathway. TARs Protein Tyrosine Phosphatase Non-Receptor Type (PTPN)-1, PTPN2 and Nicotinamide Adenine Dinucleotide Phosphate Oxidase 4 (NOX4) were found to be associated with insulin resistance and AGE-RAGE pathway involved in diabetic complications such as diabetic neuropathy. Two TARs with UniProt name, adenosine receptor A1 and adenosine receptor A2a are included in list of TARs on TTD as successful TARs. Genes Androgen Receptor (AR), PTGES, Aldo-Keto Reductase Family 1 Member B (AKR1B1), Adenosine Triphosphate (ATP) binding cassette subfamily G member 2, potassium voltage-gated channel subfamily A member 3, adenosine A1 receptor, carbonic anhydrase 2, NOX4, peroxisome proliferator activated receptor gamma, HTR2A, ATP binding cassette subfamily B member 1 were found to be associated with NP as per DisGeNet database. TARs Hydroxysteroid 11-Beta Dehydrogenase 1 (HSD11B1), HSD11B1, HSD11B2, acetylcholinesterase, heat shock protein 90 alpha family class B member 1, receptor interacting serine/threonine kinase 1, proteasome 20S subunit beta 5, Fibroblast Growth Factor (FGF) 1, ABL proto-oncogene 1, non-receptor tyrosine kinase, cyclin A2, FGF2, histone deacetylase 10, Histidine Decarboxylase (HDC) 3, HDC5, HDC8, HDC9, PTGES, Calcium Voltage-Gated Channel Subunit Alpha1 C (CACNA1C) were found be novel TARs for treatment of NP as per literature search[23-26]. The network showing the plant licorice, its BAs and TARs associated with are shown in fig. 2.
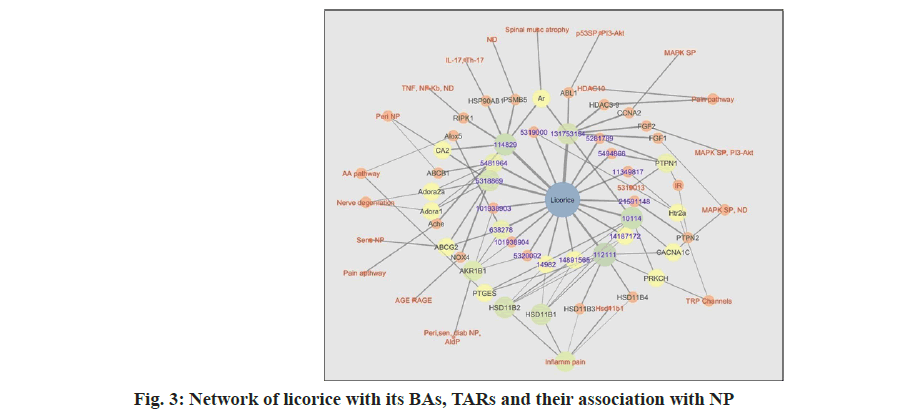
Finally, a network of licorice BAs and their TARs involved in pathophysiology of NP was constructed fig. 3. These networks were constructed using Excel data sheets. TARs having involvement with pathophysiology of NP only were included in the network. Other TARs were excluded.
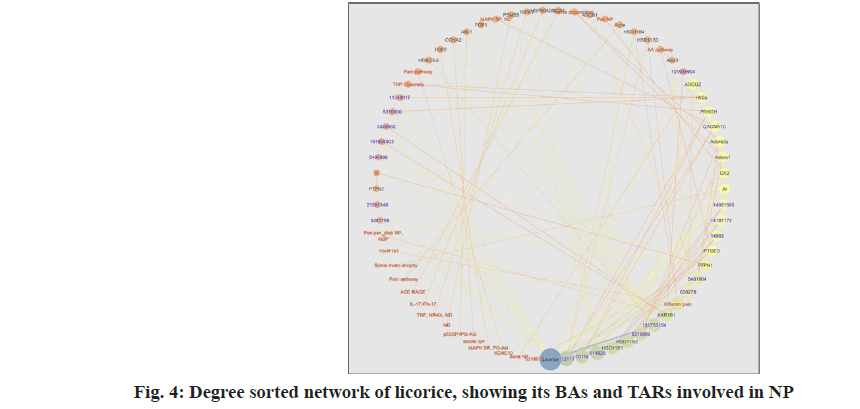
Further the network was analyzed with the help of tools available with network analyzer and taking help of various layout styles. The network was viewed in degree sorted circle layout fig. 4. Node degree of a node is defined as number of edges linked to a node. Degree sorted circle layout arranges a network depending on node degree distribution. The node with highest degree appears at 6’o clock position, with biggest circle. The nodes with decreasing degrees appear in anticlockwise direction, with decreasing size of circle. The network analysis is shown in Table 3.
| Licorice Network | |
|---|---|
| Number of nodes | 69 |
| Network density | 0.042 |
| Network heterogeneity | 0.862 |
| Self-loops | 0 |
| Network diameter | 8 |
| Network radius | 4 |
| Isolated nodes | 0 |
| Average number of neighbors | 2.841 |
Table 3: Analysis of Licorice Network Using Cytoscape Network Analyzer
The current study generated network provides a molecular overview of antinociceptive effect of licorice by bringing the BAs, their TARs, interacting proteins and pain pathways together. Licorice is known to have various active constituents with proven preclinical anti-inflammatory and antioxidant activities. Chen et al., has reported mechanism of action licorice by network pharmacological techniques. As per the network reported by them, licorice constituents can work on TARs involved in anti-inflammatory process, neuroactive and vasoactive amine metabolisms, hormone transcription regulation and glyceraldehyde oxidoreductase activity[27].
From the degree sorted circle layout, it can be seen that 18β Glycyrrhetinic Acid (GA), glycyrrhizin, liquiritin and liquiritigenin are few of the important constituents of the licorice extract. 18β GA has shown sodium channel blocking activity and antiinflammatory activity[28,29]. Liquiritin and liquiritigenin have shown to reduce lipopolysaccharide induced release pro-inflammatory mediators such as TNF-α, Interleukin (IL)-1β and IL-6, in various pre-clinical models[30]. The network pharmacology technique, helped to predict various TARs through which BAs are working to attenuate NP. Phytoconstituent 18βGA was found to work through HSD11B1, HSD11B2, CACNA1C, PTGES and PRKCH receptors. HSD11B1, HSD11B2 are associated with enzyme 11β-Hydroxysteroid Dehydrogenase (11-βOHSD). It is reported that 18βGA is a potent inhibitor of 11-βOHSD. It is postulated that its antiinflammatory activity is the expression of reduced conversion of cortisol to its biologically inactive product cortisone[31]. A research report by Kiso et al., also claims involvement of HSD11B1, HSD11B2 in pathophysiology of NP, further straightening the claim[23]. CACNA1C is associated with voltage dependent L-Type Calcium Channels (LTCs). LTCs are mostly post-synaptic channels regulating neuronal firing and gene expression. Their involvement in pain processes was also recognized recently. When applied locally in the dorsal horn of the spinal cord, the three families of LTC pharmacological blockers–dihydropyridines, phenylalkylamines and benzothiazepines proved effective in altering short-term sensitization to pain, inflammationinduced hyper excitability and neuropathy-induced allodynia[32]. Target PRKCH is associated with TRP channels. TRP channels are activated in damaging pressure, extreme temperature and toxic chemicals. TRP channels including TRPV1, TRPA1 and TRPM8 play a central role in the sensitization of nociceptive transduction as mentioned earlier. TRPM8 is also listed as a successful target in TTD. Target PTGES is associated with Prostaglandin E synthase. Prostaglandin E2 (PGE2) is considered to be the principal proinflammatory prostanoid and to play an important role in nociceptive processing and sensitization in the spinal cord as well as in the periphery by acting through four PGE receptor. Blockade of PGE2 have shown to attenuate symptoms of NP[24].
Apart from these TARs, other phytoconstituent were shown to interact with two more TARs, namely PTPN1 and AKRB1, associated with insulin resistance and AR pathway respectively. During hyperglycemia, these TARs are associated with insulin resistance and aldose reductase pathway. This pathway is responsible for the reduction of glucose to sorbitol via the enzyme AR and subsequent accumulation of sorbitol in cells. Excessive accumulation of sorbitol in peripheral leads to decrease in nerve conduction, which is a prime symptom of neuropathy. AR inhibitors have shown to produce effect in case diabetic neuropathy. Hence association BAs with target AKR1B1 could be beneficial for treating diabetic neuropathy [33].
Network pharmacology approach helps to predict mechanism of action of drug like molecules. This technique is helpful for studying medicinal plants or AYUSH medicines having multiple ingredients and acting on various TARs. This study reports potential of licorice in treatment of NP.
Network pharmacology is a modern approach to understand the multi-target working of Indian traditional medicinal system. One of the major hindrances in acceptance of herbal therapies in global market is proof of efficacy with respect of mechanism of action. This proof can be obtained by in vitro and in vivo studies of medicinal plants, but these methods are time consuming and costly. This in silico mechanistic study offers a rapid and reliable way of predicting BA-TAR interaction and a lead molecule for further potential pharmacological activity or drug discovery.
Evaluation of plants using this approach will help in understanding various therapeutic uses, with providing insight into its mechanism of action. Network pharmacology approach can also be looked as a preliminary screening technique to predict the TARs/pathways. The extract would act on and can help in reducing number of animals in pre-clinical studies. Further, inclusion of databases like Search Tool for the Retrieval of Interacting Genes (STRING), into this approach can also help to predict the protein-protein interactions of TARs, giving deeper insights into molecular mechanisms. This approach is cost-effective, time saving and as well as alternative to animal experimentation, hence can be a boon to modern pharmacological research.
This article attempted to evaluate effectiveness of licorice constituents for NP by applying network pharmacology. Licorice contains various phytoconstituent working on multiple TARs involved in NP. In our laboratory, we have tested ethanol, methanol and ethyl acetate extracts of licorice in preclinical models of NP and they have shown beneficial effects[34]. Hence this plant should be explored further for treatment of NP.
Acknowledgements
The authors are grateful to Dr. Uma Chandran and Dr. Bhushan Patwardhan, Interdisciplinary School of Sciences, SPPU, Pune, Maharashtra, for their support and guidance.
Conflict of Interest
The authors declared no conflict of interests.
References
- Chandran U, Mehendale N, Tillu G, Patwardhan B. Network pharmacology of ayurveda formulation triphala with special reference to anti-cancer property. Comb Chem High Throughput Screen 2015;18(9):846-54.
[Crossref] [Google Scholar] [PubMed]
- Patwardhan B, Chandran U. Network ethnopharmacology approaches for formulation discovery. Indian J Tradit Knowledge 2015;14(4):574-80.
- Lakshmi T, Geetha RV. Glycyrrhiza glabra Linn. commonly known as licorice: A therapeutic review. Int J Pharm Pharm Sci 2011;3(4):20-5.
- Sharma VA, Agrawal RC. Glycyrrhiza glabra-A plant for the future. Mintage J Pharm Med Sci 2013;2(3):15-20.
- Kaur G, Jaggi AS, Singh N. Exploring the potential effect of Ocimum sanctum in vincristine-induced neuropathic pain in rats. J Brachial Plex Peripher Nerve Inj 2010;5(1):e3-11.
[Crossref] [Google Scholar] [PubMed]
- Smith HS, Torrance N. Epidemiology of NP and Its impact on life. Curr Pain Headache Rep 2012:2-8.
- Woolf CJ, Mannion RJ. NP: Etiology, symptoms, mechanisms and management. Lancet Pain 1999;353:1959-64.
- Nickel FT, Seifert F. Mechanism of NP. Eur Psychopharmacol 2012;22:81-91.
- Sugimoto K, Yasujima M, Yagihashi S. Role of advanced glycation end products in diabetic neuropathy. Curr Pharm Des 2008;14(10):953-61.
[Crossref] [Google Scholar] [PubMed]
- Dr. Duke's Phytochemical and Ethnobotanical Databases. U.S. Department of Agriculture; 2021.
- Gu J, Gui Y, Chen L, Yuan G, Lu HZ, Xu X. Use of natural products as chemical library for drug discovery and network pharmacology. PloS One 2013;8(4):e62839.
[Crossref] [Google Scholar] [PubMed]
- Daina A, Michielin O, Zoete V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 2017;7(1):42717.
[Crossref] [Google Scholar] [PubMed]
- Gilson MK, Liu T, Baitaluk M, Nicola G, Hwang L, Chong J. BindingDB in 2015: A public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucl Acids Res 2016;44(D1):D1045-53.
[Crossref] [Google Scholar] [PubMed]
- BindingDB. NIH's National Institute of General Medical Sciences; 2021.
- Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucl Acids Res 2016;44(D1):D457-62.
- Piñero J, Bravo À, Queralt-Rosinach N, Gutiérrez-Sacristán A, Deu-Pons J, Centeno E, et al. DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants. Nucl Acids Res 2016;45(D1):D833-9.
[Crossref] [Google Scholar] [PubMed]
- DisGeNET. Gene-Disease Associations; 2021.
- Wang Y, Zhang S, Li F, Zhou Y, Zhang Y, Wang Z, et al. Therapeutic target database 2020: Enriched resource for facilitating research and early development of targeted therapeutics. Nucl Acids Res 2020;48(D1):D1031-41.
- Yang H, Qin C, Li YH, Tao L, Zhou J, Yu CY, et al. Therapeutic target database update 2016: Enriched resource for bench to clinical drug target and targeted pathway information. Nucl Acids Res 2016;44(D1):D1069-74.
[Crossref] [Google Scholar] [PubMed]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13(11):2498-504.
[Crossref] [Google Scholar] [PubMed]
- Bastian M, Heymann S, Jacomy M. Gephi: An open source software for exploring and manipulating networks. Proc Int AAAI Conference Web Social Media 2009;3(1):361-2.
- Hopkins AL. Network pharmacology: The next paradigm in drug discovery. Nat Chem Biol 2008;4(11):682-90.
[Crossref] [Google Scholar] [PubMed]
- Kiso T, Sekizawa T, Uchino H, Tsukamoto M, Kakimoto S. Analgesic effects of ASP 3662, a novel 11β-hydroxysteroid dehydrogenase 1 inhibitor, in rat models of neuropathic and dysfunctional pain. Br J Pharmacol 2018;175(19):3784-96.
[Crossref] [Google Scholar] [PubMed]
- Kunori S, Matsumura S, Okuda-Ashitaka E, Katano T, Audoly LP, Urade Y, et al. A novel role of prostaglandin E2 in neuropathic pain: Blockade of microglial migration in the spinal cord. Glia 2011;59(2):208-18.
[Crossref] [Google Scholar] [PubMed]
- Khangura RK, Sharma J, Bali A, Singh N, Jaggi AS. An integrated review on new targets in the treatment of neuropathic pain. Korean J Physiol Pharmacol 2019;23(1):1-20.
[Crossref] [Google Scholar] [PubMed]
- Ciotu CI, Fischer MJ. Novel analgesics with peripheral targets. Neurotherapeutics 2020;17(3):784-825.
[Crossref] [Google Scholar] [PubMed]
- Chen M, Zhu J, Kang J, Lai X, Gao Y, Gan H, et al. Exploration in the mechanism of action of licorice by network pharmacology. Molecules 2019;24(16):2959.
- Du YM, Xia CK, Zhao N, Dong Q, Lei M, Xia JH. 18β-Glycyrrhetinic acid preferentially blocks late Na current generated by ΔKPQ Nav 1.5 channels. Acta Pharmacol Sin 2012;33(6):752-60.
[Crossref] [Google Scholar] [PubMed]
- Jeong HG, You HJ, Park SJ, Moon AR, Chung YC, Kang SK, et al. Hepatoprotective effects of 18β-glycyrrhetinic acid on carbon tetrachloride-induced liver injury: Inhibition of cytochrome P450 2E1 expression. Pharmacol Res 2002;46(3):221-7.
[Crossref] [Google Scholar] [PubMed]
- Yu JY, Ha JY, Kim KM, Jung YS, Jung JC, Oh S. Anti-inflammatory activities of licorice extract and its active compounds, glycyrrhizic acid, liquiritin and liquiritigenin, in BV2 cells and mice liver. Molecules 2015;20(7):13041-54.
[Crossref] [Google Scholar] [PubMed]
- Teelucksingh S, Mackie AD, Burt D, Edwards CR, Mclntyre MA, Brett L. Potentiation of hydrocortisone activity in skin by glycyrrhetinic acid. Lancet 1990;335(8697):1060-3.
[Crossref] [Google Scholar] [PubMed].
- Roca-Lapirot O, Radwani H, Aby F, Nagy F, Landry M, Fossat P. Calcium signalling through L-type calcium channels: Role in pathophysiology of spinal nociceptive transmission. Br J Pharmacol 2018;175(12):2362-74.
[Crossref] [Google Scholar] [PubMed]
- Schemmel KE, Padiyara RS, D'Souza JJ. Aldose reductase inhibitors in the treatment of diabetic peripheral neuropathy: A review. J Diabetes Complications 2010;24(5):354-60.
[Crossref] [Google Scholar] [PubMed]
- Joshi TA, Tatke P. Neuroprotective effects of licorice extracts in tibial and sural transection induced neuropathic pain in rats. Indian J Pharm Edu Res 2020;54(2):S285-94.