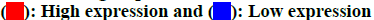- *Corresponding Author:
- Wujie Xia
Department of Cardiology,
The Second Affiliated Hospital and Yuying Children’s Hospital of Wenzhou Medical University, Wenzhou,
Zhejiang 325000, China
E-mail: liwe12421@gmail.com
| Date of Submission | 18 September 2020 |
| Date of Revision | 23 February 2021 |
| Date of Acceptance | 03 April 2022 |
| Indian J Pharm Sci 2022;84(2):415-422 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Glutathione is an important antioxidant that plays an important role in myocardial remodeling. However, the exact mechanisms underlying the effects of glutathione on heart failure are elusive. This study aimed to investigate the mechanisms of glutathione in heart failure treatment based on network pharmacology technology. The gene expression profiles were downloaded from Gene Expression Omnibus database (GSE133054). Differentially expressed genes were identified by Gene Ontology analysis and Kyoto Encyclopedia of Genes and Genomes pathway analysis. A protein protein interaction network was constructed to screen potential regulatory proteins. We identified 970 potential target genes of glutathione and 2381 differentially expressed genes of heart failure, with 80 genes overlapping in both groups. Gene Ontology analysis and Kyoto Encyclopedia of Genes and Genomes pathway analysis showed that the important genes were mainly distributed in phosphatidylinositol 3 kinase-protein kinase B signaling pathway, mitogen-activated protein kinase signaling pathway, lipid and atherosclerosis pathway. In glutathione target genes-heart failure network analysis, 7 targets were identified, including tumor necrosis factor, vascular endothelial growth factor A, fibronectin 1, serine proteinase inhibitor 1, insulin-like growth factor binding protein 3, matrix metalloproteinase-1 and liquid oxygen, with serine proteinase inhibitor 1, insulin-like growth factor binding protein 3, fibronectin 1 and liquid oxygen significantly over expressed in heart failure. We utilized network pharmacology to systematically explore the mechanism of action of glutathione on heart failure and identified genes and pathways as potential therapeutic targets for heart failure.
Keywords
Glutathione, network pharmacology, heart failure, myocardial infarction
Heart Failure (HF) is one of the leading causes of morbidity and mortality worldwide, affecting at least 26 million people globally with annual mortality rate of over 50 % in the end-stage group[1,2]. As a result of the complicated pathologic cascades involved in HF, a variety of pharmacologic remedies for HF have been applied, including diuretics,beta (β)-blockers, angiotensin-converting enzyme inhibitors, angiotensin receptor blockers and spironolactone. While some patients with HF respond to appropriate therapy, many patients do not respond. The reason for the lack of response may be due to differences in clinical characteristics, underlying causes, biomarkers, genetic variants and protein expression. Therefore, effective therapy strategies for HF are urgently needed.
Glutathione (GSH) is an important intracellular antioxidant and redox potential regulator that plays a vital role in drug detoxification and elimination and in cellular protection from damage by free radicals, peroxides and toxins. It has been suggested that reduced GSH plays an important role in the pathogenesis of myocardial remodeling and dysfunction[3-5]. Adamy et al.[6] found that total GSH was depleted by 54 % in the left ventricle of human failing heart. In addition, there was a GSH deficiency in the left ventricle in 2 mo post-myocardial infarction rats with chronic HF. Interestingly, treatment with the GSH precursor N-acetylcysteine for 1 mo normalized GSH in the left ventricle, improved left ventricular contractile function and lessened adverse left ventricular remodeling in 3 mo post-myocardial infarction rats. Modulating GSH levels as well as the activity of GSH related enzymes has held promise in a variety of therapeutic settings. Since GSH cannot be administered clinically, a variety of precursors or chemically modified analogues have been generated to mimic pharmacologic effects of GSH. However, the exact mechanisms underlying the effects of GSH on HF are elusive.
Network pharmacology can decode the mechanisms of drug action with an overall view point, with the focus changing from "single protein target, single drug" to "multiple protein targets, multiple drugs"[7]. Currently, network pharmacology has been extensively utilized to explore the mechanisms of the drug on multiple targets in diverse diseases. Therefore, this study aimed to reveal the mechanisms of GSH in HF therapy based on network pharmacology.
Materials and Methods
Differentially Expressed Gene (DEG) analysis:
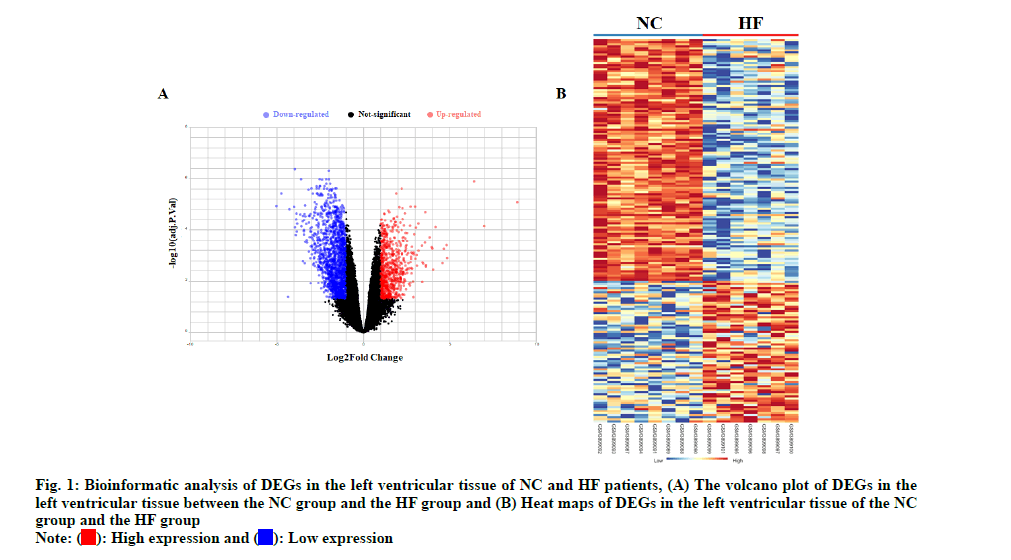
The high throughput sequencing dataset GSE133054 was downloaded from the Gene Expression Omnibus (GEO) database. Difference analysis was performed by R script using Linear Models for Microarray Analysis (LIMMA) R package, p<0.05 and (logFC>1) as cut off values for screening DEGs. The DEGs of the comparison group were shown as the volcano plot. The heat map and the hub gene expression data were generated using NetworkAnalyst online tools (https:// www.networkanalyst.ca/).
GSH target genes identification:
The chemical structure of GSH was obtained from the PubChem database (https://pubchem.ncbi.nlm. nih.gov/). GSH target genes were screened from the PharmMapper database (http://www.lilab-ecust.cn/ pharmmapper/), the Similarity Ensemble Approach (SEA) database (http://sea.bkslab.org/) and the function of the target genes were analyzed by the Swiss Target Prediction database (http://www.swisstargetprediction. ch/).
Gene functional and pathway enrichment analysis:
The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis were based on the Metascape database (https://metascape. org/). The DisGeNET database (https://www.disgenet. org/) was used for gene-disease association analysis. The map of the KEGG signaling pathway was obtained from the KEGG database (https://www.genome.jp/ kegg/pathway.html). Gene Set Enrichment Analysis (GSEA) v4.1.0 was applied for analysis. p<0.05 indicated significant pathways related to HF.
Protein-Protein Interaction (PPI) network establishment:
The interaction of GSH target genes and DEGs of HF patients were obtained by the Venn diagram online tools (https://bioinfogp.cnb.csic.es/tools/venny/index. html). Cytoscape 3.8.0 was used to determine GSH target genes-HF network. Furthermore, the STRING database (https://string-db.org/) was used to analyze PPI network of GSH targets and the link of the genes was calculated by CytoHubba application in Cytoscape software.
Statistical analysis:
All data were shown as mean±Standard Error of Mean (SEM) and analyzed by GraphPad Prism software version 9.0 (CA, USA). The comparison of two groups was analyzed using the two-tailed Student’s t-test. p<0.05 was considered significant.
Results and Discussion
The left ventricles tissues high throughput sequencing dataset GSE133054 was used to analyze DEGs of HF. The gene expression analysis was made on Normal Control (NC) group and HF groups. The volcano plot and heat map of HF DEGs were shown in fig. 1. Total 2381 DEGs were identified, including 749 up-regulated and 1632 down-regulated in HF patients as shown in fig. 2.
The chemical structure of GSH was obtained from the PubChem database as shown in fig. 2A. Based on its structure, PharmMapper database, SEA database and Swiss Target Prediction database were used to predict potential target genes of GSH. A total of 970 potential target genes were obtained from those databases. The function of the target genes was classified according to their biochemical criteria as shown in fig. 2B.
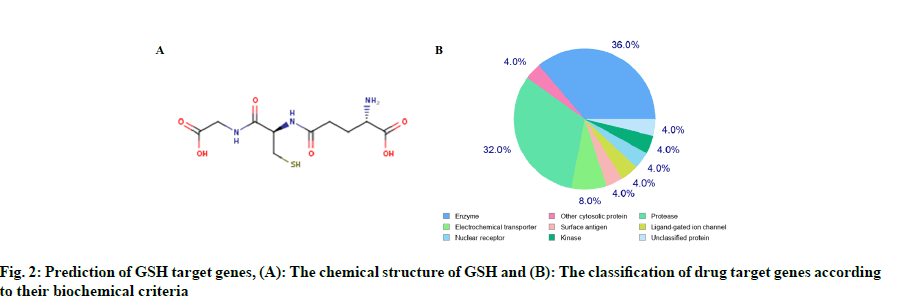
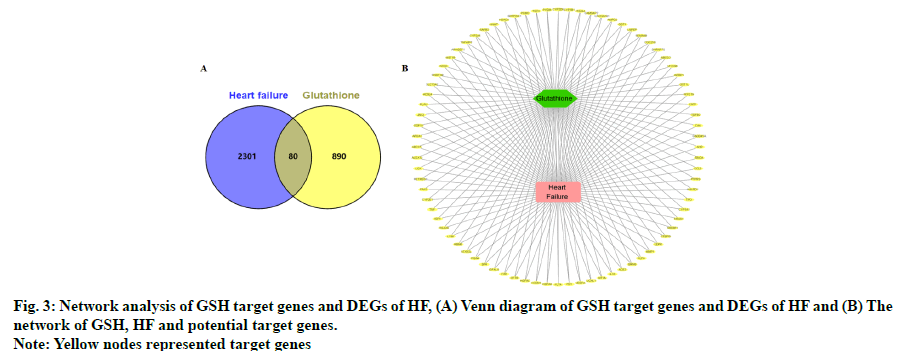
We found 970 potential target genes of GSH and 2381 DEGs of HF. As shown in fig. 3A, there were 80 genes in both groups and these target genes were not only HF related but also the targets of GSH. We constructed interactive GSH target genes-HF network as shown in fig. 3B, which indicated that GSH might have effect on HF by stimulating or inhibiting these target genes.
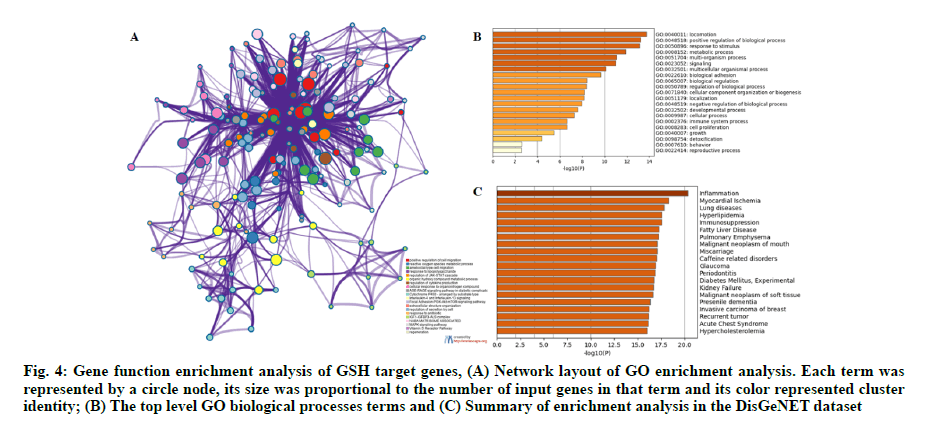
A serial functional enrichment analysis was conducted to gain a comprehensive understanding of the overlapped genes. A variety of GO enrichment terms were enriched and a subset of representative terms from this cluster were selected and converted into a network layout. As shown in fig. 4A, the different colors indicated different GO terms, positive regulation of cell migration, reactive oxygen species metabolic process, focal adhesion, Phosphatidylinositol 3-Kinase- Protein Kinase B-mammalian Target of Rapamycin (PI3KAkt- mTOR) signaling pathway and Mitogen-Activated Protein Kinase (MAPK) signaling pathway were enriched. The top level GO Biological Processes (BP) terms were shown in fig. 4B. We found that the biological processes such as positive regulation of biological process, metabolic process, cellular component organization or biogenesis and cell proliferation were enriched. Summary of enrichment analysis in the DisGeNET dataset was shown in fig. 4C. We found that the diseases, such as inflammation, myocardial ischemia, hyperlipidemia and hypercholesterolemia, may be closely related to the cause of HF.
Fig. 4: Gene function enrichment analysis of GSH target genes, (A) Network layout of GO enrichment analysis. Each term was represented by a circle node, its size was proportional to the number of input genes in that term and its color represented cluster identity; (B) The top level GO biological processes terms and (C) Summary of enrichment analysis in the DisGeNET dataset
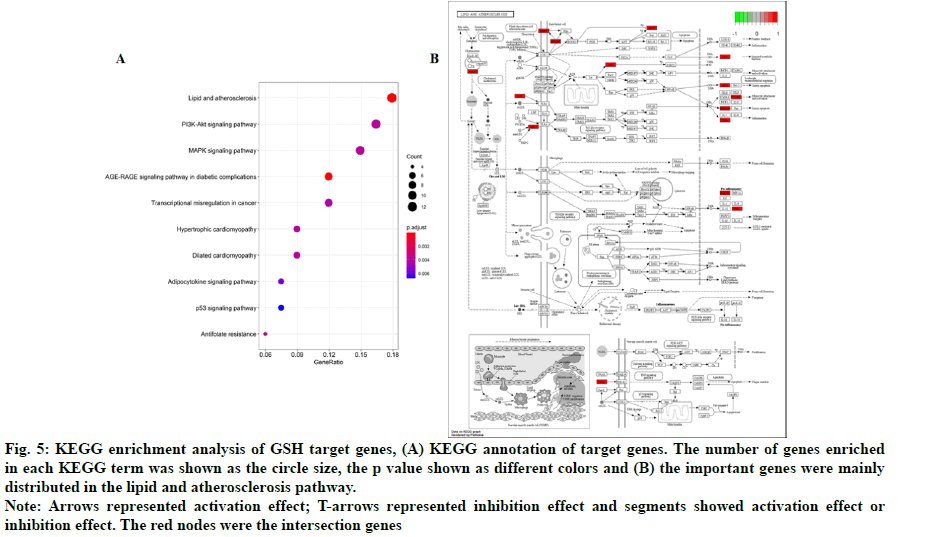
In addition, 18 KEGG pathways were enriched and top 10 KEGG pathways were shown in fig. 5A. We found that the important genes were mainly distributed in the lipid and atherosclerosis pathway (fig. 5B), indicating that GSH might affect HF through the lipid and atherosclerosis pathway. We also found that PI3KAkt and MAPK signaling pathways were enriched, consistent with the results of GO analysis.
Fig. 5: KEGG enrichment analysis of GSH target genes, (A) KEGG annotation of target genes. The number of genes enriched in each KEGG term was shown as the circle size, the p value shown as different colors and (B) the important genes were mainly distributed in the lipid and atherosclerosis pathway.
Note: Arrows represented activation effect; T-arrows represented inhibition effect and segments showed activation effect or inhibition effect. The red nodes were the intersection genes
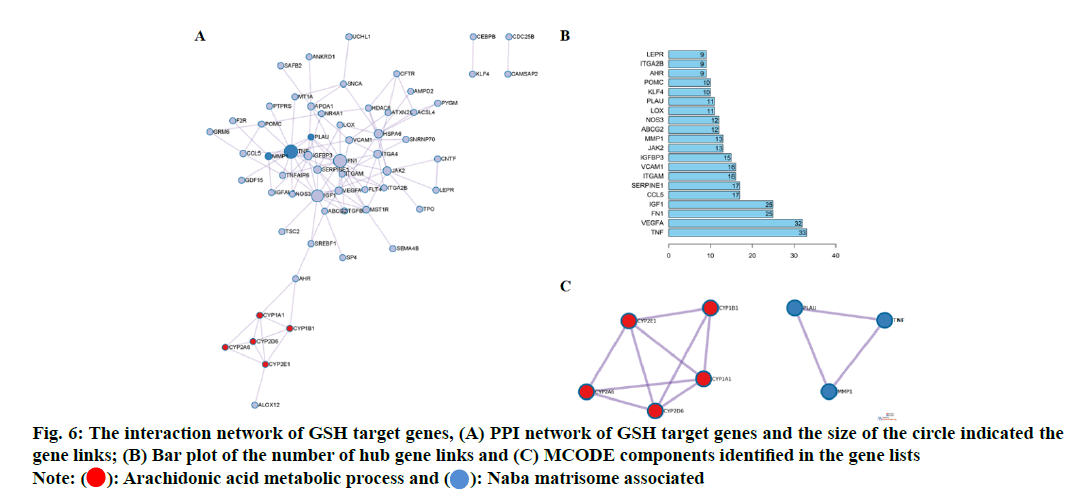
In order to explore the relationship among 80 potential target genes, PPI network analysis was performed to construct network. The size of the circle indicated the gene links (fig. 6A) and a bar plot of the number of hub gene links was shown in fig. 6B. The hub genes such as Tumour Necrosis Factor (TNF), Vascular Endothelial Growth Factor A (VEGFA), Fibronectin 1 (FN1), Serine Proteinase Inhibitor 1 (SERPIN1), Insulin- Like Growth Factor Binding Protein 3 (IGFBP-3), Matrix Metalloproteinase-1 (MMP-1) and Liquid Oxygen (LOX) may play important role in biological activity of GSH. Combined with GO network analysis, the Molecular Complex Detection (MCODE) algorithm was then applied to this network to identify neighborhoods where proteins were densely connected. We found two MCODE enriched in the PPI network as shown in fig. 6A. MCODE 1 (red) was the arachidonic acid metabolic process and MCODE2 (blue) was Naba matrisome associated as shown in fig. 6C.
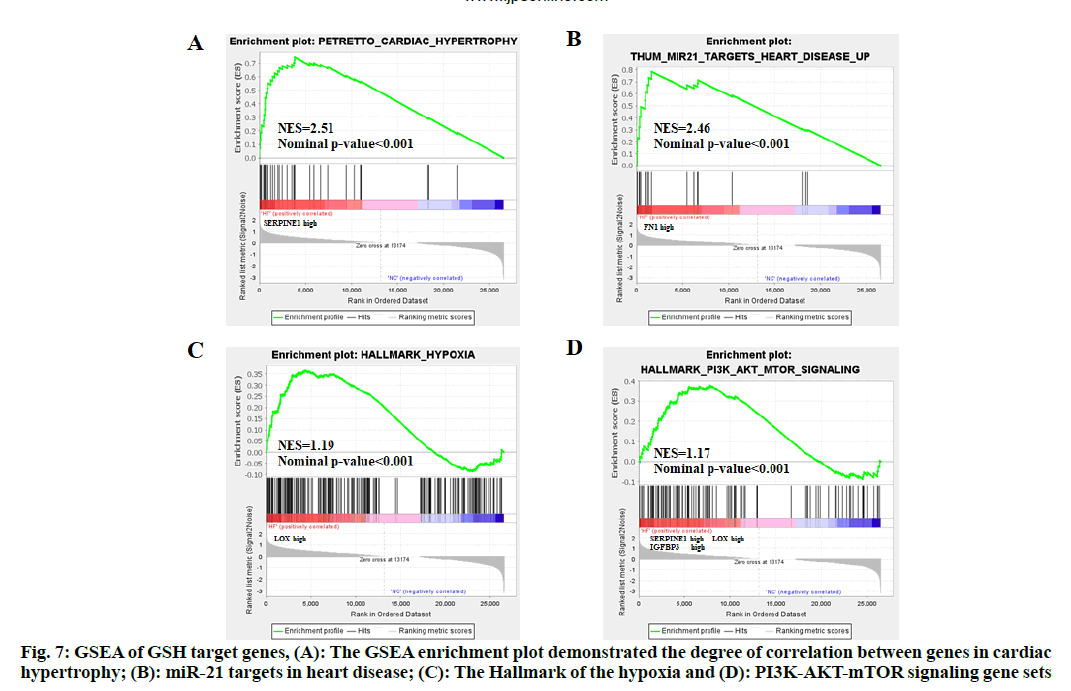
GSEA analysis of the overlapped genes was conducted to show changes in gene expression. GSEA enrichment plot demonstrated the degree of correlation between gene changes in cardiac hypertrophy (fig. 7A, SERPINE1 high expression) and MicroRNA-21 (miR-21) targets in heart disease (fig. 7B, FN1 high expression), the Hallmark of the hypoxia (fig. 7C, LOX high expression) and PI3K/AKT/mTOR signaling (fig. 7D, SERPINE1, LOX and IGFBP3 high expression) gene sets.
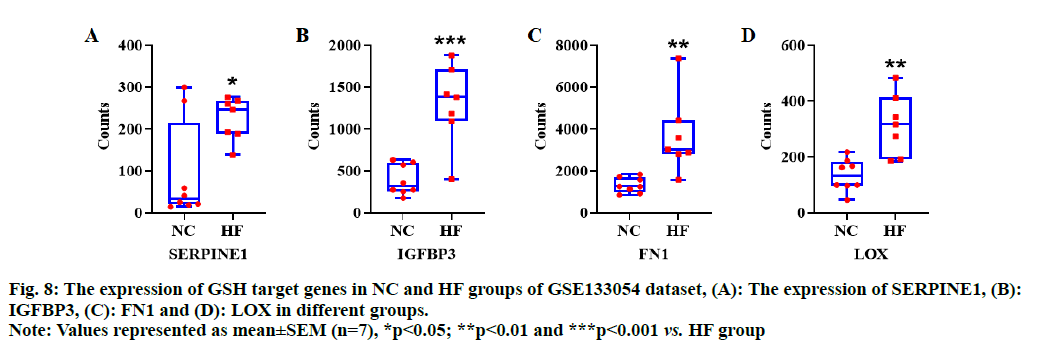
Based on bioinformatic analysis of GSH target genes, and the results of the network and functional enrichment analysis, four hub genes including SERPINE1, IGFBP3, FN1 and LOX were selected to observe the expression pattern of HF dataset. We found that all the hub genes were significantly over expressed in disease group of HF dataset (p<0.05, fig. 8A-fig. 8D), indicating that these genes could be potential therapeutic targets for HF.
GSH is an antioxidant that plays an important role in vascular and cardiac function[8-10]. GSH deficiency is involved with HF progression and cardiac remodeling. Damy et al. found that GSH deficiency was involved in functional and structural cardiac abnormalities of patients with cardiac diseases. However, the pharmacological mechanism of GSH against HF has not been clearly clarified. To our knowledge, this is the first study to systematically explore the mechanism of the action of GSH on HF using network pharmacology method.
With the strategy of database mining, we identified 80 genes related to HF pathology and GSH treatment. To further understand the function of these genes, we performed PPI analysis and functional enrichment analysis. We found that the hub genes such as TNF, VEGFA, FN1, SERPINE1, IGFBP3, MMP1 and LOX may play a crucial role in biological activity of GSH on HF. In particular, four genes SERPINE1, IGFBP3, FN1 and LOX were selected to validate the efficacy of GSH on HF.
SERPINE1 encodes plasminogen activator inhibitor type 1 (PAI-1), which serves as the primary inhibitor of uridylyl Phosphate Adenosine (uPA) and tissue plasminogen activator (tPA)[11,12]. SERPINE1 was involved in dilated ischemic cardiomyopathy[13]. In addition, SERPINE1 loss-of-function mutations were associated with cardiac fibrosis[14,15]. IGFBPs play a pivotal role in regulating physiological function of Insulin-Like Growth Factors (IGFs). As an important member of IGFBPs, IGFBP3 could induce hypertrophic response in neonatal rat cardiac myocytes[16]. IGFBP3 has been linked to hypertension, obesity and cardiovascular disease[17]. After acute myocardial infarction, IGFBP3 level significantly increased, which was associated with improved outcomes and echocardiographic parameters[18]. FN1 gene encodes for FN protein, a glycoprotein present as the soluble dimeric form in plasma and as a dimeric or multimeric form at the cell surface and in the Extracellular Matrix (ECM)[19]. FN functions in cell adhesion and migration, metastasis and other cellular processes such as embryogenesis, wound healing and blood coagulation[20]. It has been shown that FN1 participates in ECM remodeling and its inhibition attenuates fibrosis and improves HF[21]. Increased expression of FN1 in the diseased hearts suggested that FN1 could contribute to the pathology of HF[22]. LOX is an extracellular matrix-embedded protein. Animal studies showed that myocardial LOX could regulate the abundance of collagen type I[23,24]. A recent study suggested that LOX was associated with both formation and assembly of collagen network and subsequent LV adverse remodeling[25].We found that all four genes were differentially expressed in the GSE133054 dataset. Meanwhile, the expression of all four genes markedly changed after GSH treatment, suggesting that they are potential therapeutic targets for HF.
GO analysis showed that potential targets of GSH were mainly associated with various biological processes, such as the positive regulation of cell migration, reactive oxygen species metabolic process, focal adhesion, PI3K-Akt-mTOR signaling pathway and MAPK signaling pathways, which have been shown to be involved in the pathogenesis of HF[26-30]. Similarly, KEGG pathway enrichment analysis showed that some enriched pathways are known to exert anti-HF potential, such as PI3K-Akt signaling pathway[31], MAPK signaling pathway[32] and lipid and atherosclerosis pathway[33]. These signaling pathways could be the targets of GSH against HF.
In this study, we utilized network pharmacology to systematically explore the mechanism of action of GSH on HF. We identified genes and pathways as candidate therapeutic targets for HF and provided the basis for follow-up experiments to evaluate the potential of these genes and pathways in the treatment of HF.
Authors' contributions:
Wujie Xia designed the study, Jing Song and Weiqian Lin collected and analyzed the data. All authors read and approved the manuscript.
Conflict of interests:
The authors declared no conflicts of interest.
References
- Balling L, Gustafsson F. Copeptin in heart failure. Adv Clin Chem 2016;73:29-64.
- Savarese G, Lund LH. Global public health burden of heart failure. Card Fail Rev 2017;3(1):7-11.
[Crossref] [Google Scholar] [PubMed]
- Damy T, Kirsch M, Khouzami L, Caramelle P, Le Corvoisier P, Roudot-Thoraval F, et al. Glutathione deficiency in cardiac patients is related to the functional status and structural cardiac abnormalities. PloS one 2009;4(3):e4871.
[Crossref] [Google Scholar] [PubMed]
- Ardanaz N, Yang XP, Cifuentes ME, Haurani MJ, Jackson KW, Liao TD, et al. Lack of glutathione peroxidase 1 accelerates cardiac-specific hypertrophy and dysfunction in angiotensin II hypertension. Hypertension 2010;55(1):116-23.
[Crossref] [Google Scholar] [PubMed]
- Rajasekaran NS, Connell P, Christians ES, Yan LJ, Taylor RP, Orosz A, et al. Human αB-crystallin mutation causes oxido-reductive stress and protein aggregation cardiomyopathy in mice. Cell 2007;130(3):427-39.
[Crossref] [Google Scholar] [PubMed]
- Adamy C, Mulder P, Khouzami L, Andrieu-Abadie N, Defer N, Candiani G, et al. Neutral sphingomyelinase inhibition participates to the benefits of N-acetylcysteine treatment in post-myocardial infarction failing heart rats. J Mol Cell Cardiol 2007;43(3):344-53.
[Crossref] [Google Scholar] [PubMed]
- Isgut M, Rao M, Yang C, Subrahmanyam V, Rida PC, Aneja R. Application of combination high‐throughput phenotypic screening and target identification methods for the discovery of natural product‐based combination drugs. Med Res Rev 2018;38(2):504-24.
[Crossref] [Google Scholar] [PubMed]
- Yucel D, Aydogdu S, Cehreli S, Saydam G, Canatan H, Senes M, et al. Increased oxidative stress in dilated cardiomyopathic heart failure. Clin Chem 1998;44(1):148-54.
[Crossref] [Google Scholar] [PubMed]
- Haddad JJ, Harb HL. l-γ-Glutamyl-l-cysteinyl-glycine (glutathione; GSH) and GSH-related enzymes in the regulation of pro-and anti-inflammatory cytokines: A signaling transcriptional scenario for redox (y) immunologic sensor (s)? Mol Immunol 2005;42(9):987-1014.
[Crossref] [Google Scholar] [PubMed]
- Franco R, Schoneveld OJ, Pappa A, Panayiotidis MI. The central role of glutathione in the pathophysiology of human diseases. Arch Physiol Biochem 2007;113(4-5):234-58.
[Crossref] [Google Scholar] [PubMed]
- Huang J, Sabater-Lleal M, Asselbergs FW, Tregouet D, Shin SY, Ding J, et al. Genome-wide association study for circulating levels of PAI-1 provides novel insights into its regulation. Blood 2012;120(24):4873-81.
[Crossref] [Google Scholar] [PubMed]
- Li S, Wei X, He J, Tian X, Yuan S, Sun L. Plasminogen activator inhibitor-1 in cancer research. Biomed Pharmacother 2018;105:83-94.
[Crossref] [Google Scholar] [PubMed]
- Polyakova V, Loeffler I, Hein S, Miyagawa S, Piotrowska I, Dammer S, et al. Fibrosis in endstage human heart failure: Severe changes in collagen metabolism and MMP/TIMP profiles. Int J Cardiol 2011;151(1):18-33.
[Crossref] [Google Scholar] [PubMed]
- Iwaki T, Urano T, Umemura K. PAI‐1, progress in understanding the clinical problem and its aetiology. Br J Haematol 2012;157(3):291-8.
[Crossref] [Google Scholar] [PubMed]
- Khan SS, Shah SJ, Strande JL, Baldridge AS, Flevaris P, Puckelwartz MJ, et al. Identification of cardiac fibrosis in young adults with a homozygous frameshift variant in SERPINE1. JAMA Cardiol 2021;6(7):841-6.
[Crossref] [Google Scholar] [PubMed]
- Henson M, Damm D, Lam A, Garrard LJ, White T, Abraham JA, et al. Insulin-like growth factor-binding protein-3 induces fetalization in neonatal rat cardiomyocytes. DNA Cell Biol 2000;19(12):757-63.
- Bayes-Genis A, Conover CA, Schwartz RS. The insulin-like growth factor axis: A review of atherosclerosis and restenosis. Circ Res 2000;86(2):125-30.
[Crossref] [Google Scholar] [PubMed]
- Lee WL, Chen JW, Ting CT, Lin SJ, Wang PH. Changes of the insulin-like growth factor I system during acute myocardial infarction: Implications on left ventricular remodeling. J Clin Endocrinol Metab 1999;84(5):1575-81.
[Crossref] [Google Scholar] [PubMed]
- Chaffer CL, San Juan BP, Lim E, Weinberg RA. EMT, cell plasticity and metastasis. Cancer Metastasis Rev 2016;35(4):645-54.
[Crossref] [Google Scholar] [PubMed]
- Feng X, Xu ES. Alectinib and lorlatinib function by modulating EMT-related proteins and MMPs in NSCLC metastasis. Bosn J Basic Med Sci 2021;21(3):331-8.
[Crossref] [Google Scholar] [PubMed]
- Valiente-Alandi I, Potter SJ, Salvador AM, Schafer AE, Schips T, Carrillo-Salinas F, et al. Inhibiting fibronectin attenuates fibrosis and improves cardiac function in a model of heart failure. Circulation 2018;138(12):1236-52.
[Crossref] [Google Scholar] [PubMed]
- Patel M, Rodriguez D, Yousefi K, John-Williams K, Mendez AJ, Goldberg RB, et al. Osteopontin and LDLR are upregulated in hearts of sudden cardiac death victims with heart failure with preserved ejection fraction and diabetes mellitus. Front Cardiovasc Med 2020;7:610282.
[Crossref] [Google Scholar] [PubMed]
- Galán M, Varona S, Guadall A, Orriols M, Navas M, Aguiló S, et al. Lysyl oxidase overexpression accelerates cardiac remodeling and aggravates angiotensin II–induced hypertrophy. FASEB J 2017;31(9):3787-99.
[Crossref] [Google Scholar] [PubMed]
- Lu M, Qin Q, Yao J, Sun L, Qin X. Induction of LOX by TGF‐β1/Smad/AP‐1 signaling aggravates rat myocardial fibrosis and heart failure. IUBMB life 2019;71(11):1729-39.
[Crossref] [Google Scholar] [PubMed]
- Bi X, Song Y, Song Y, Yuan J, Cui J, Zhao S, et al. Collagen cross‐linking is associated with cardiac remodeling in hypertrophic obstructive cardiomyopathy. J Am Heart Assoc 2021;10(1):e017752.
[Crossref] [Google Scholar] [PubMed]
- Zuo L, Chuang CC, Hemmelgarn BT, Best TM. Heart failure with preserved ejection fraction: Defining the function of ROS and NO. J Appl Physiol 2015;119(8):944-51.
[Crossref] [Google Scholar] [PubMed]
- Wadey K, Lopes J, Bendeck M, George S. Role of smooth muscle cells in coronary artery bypass grafting failure. Cardiovasc Res 2018;114(4):601-10.
[Crossref] [Google Scholar] [PubMed]
- Samarel AM. Focal adhesion signaling in heart failure. Pflugers Arch 2014;466(6):1101-11.
[Crossref] [Google Scholar] [PubMed]
- Yang W, Wu Z, Yang K, Han Y, Chen Y, Zhao W, et al. BMI1 promotes cardiac fibrosis in ischemia-induced heart failure via the PTEN-PI3K/Akt-mTOR signaling pathway. Am J Physiol Heart Circ Physiol 2019;316(1):H61-9.
[Crossref] [Google Scholar] [PubMed]
- Zhang Q, Lu L, Liang T, Liu M, Wang ZL, Zhang PY. MAPK pathway regulated the cardiomyocyte apoptosis in mice with post-infarction heart failure. Bratisl Lek Listy 2017;118(6):339-46.
[Crossref] [Google Scholar] [PubMed]
- Aoyagi T, Matsui T. Phosphoinositide-3 kinase signaling in cardiac hypertrophy and heart failure. Curr Pharm Des 2011;17(18):1818-24.
[Crossref] [Google Scholar] [PubMed]
- Romero-Becerra R, Santamans AM, Folgueira C, Sabio G. p38 MAPK pathway in the heart: New insights in health and disease. Int J Mol Sci 2020;21(19):7412.
[Crossref] [Google Scholar] [PubMed]
- Kader T, Porteous CM, Williams MJ, Gieseg SP, McCormick SP. Ribose-cysteine increases glutathione-based antioxidant status and reduces LDL in human lipoprotein (a) mice. Atherosclerosis 2014;237(2):725-33.
[Crossref] [Google Scholar] [PubMed]