- *Corresponding Author:
- J. Dwivedi
Department of Chemistry, Banasthali Vidyapith, Banasthali, Rajasthan, India
E-mail:155346410@qq.com
| Date of Received | 20 June 2021 |
| Date of Revision | 12 April 2022 |
| Date of Acceptance | 22 February 2023 |
| Indian J Pharm Sci 2023;85(1):217-226 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Obesity is a serious health problem of 21st century. Around 1 billion adults are overweight and more than 300 million people are found to be obese globally. In fact 2.6 million people die per year due to obesity and thus it has now become a major reason of morbidity and mortality. In line to this few novel pyrazole derivatives (Z)-3-(4-subtituted phenyl)-2-(3, 5-dimethyl-1H-pyrazol-1-yl)-1-phenylprop-2-en-1-one (4a-f) were synthesized from 3, 5 dimethyl pyrazole by a simple inexpensive, rapid method. All compounds were authenticated using different physical and analytical techniques such as Infrared, 1H nuclear magnetic resonance, 13C nuclear magnetic resonance and mass spectrometry. Hypolipidemic activity was carried out for all the compounds using Triton WR 1339 induced hyperlipidemia model in rodents. Further, molecular docking was performed to understand the receptor-ligand interactions of synthesized compounds with cannabinoid receptor (protein data bank code: 5TGZ) using Argus lab 4.0, followed by anti-obesity studies of the selected compound (Z)-3-(4-chlorophenyl)-2-(3, 5-dimethyl-1H-pyrazol-1-yl)-1-phenylprop-2-en-1-one (4b). Amongst six compounds four compounds showed significant reduction in cholesterol levels in along with triglyceride levels in hyperlipidemic rats. Compound 4b also caused significant elevation in % high density lipoprotein levels of rats comparable to the standard Gemfibrozil. All these compounds showed good interactions with the target protein. Compound 4b (5 mg/kg and 10 mg/kg orally), caused notable reduction in body weight of high fat diet rats. Compound 4b i.e. (Z)-3-(4-chlorophenyl)-2-(3,5-dimethyl-1H-pyrazol-1- yl)-1-phenylprop-2-en-1-one has ability to reduce body weight and improve lipid profile in rats and requires further studies to establish its safety and efficacy in preclinical and clinical subjects.
Keywords
Synthesis, pyrazole, molecular docking, hypolipidemic, anti-obesity
Rapid prevalence of obesity world over has been recognized as major threat of 21st century[1]. A survey on literature reveals that globally 1 billion adults are overweight and more than 300 million people are found to be obese. In fact 2.6 million people die per year due to obesity[2]. Literature is replete with the examples of pyrazole derivatives exhibiting a wide array of pharmacological properties including anticonvulsant, anti-inflammatory, analgesic, antipyretic, anti-obesity[3-7]. Several derivatives containing this nucleus have been designed synthesized and examined for lipolytic and antiobesity activity[8,9]. Pyrazole derivative such as the 5-methylpyrazole-3-carboxamide, AM251, ibipinabent and rimonabent has immerged as anti-obesity drug acting as cannabinoid receptor antagonist[10-12]. Chalcones derivative are known to have various pharmacological properties[13]. Many chalcones derivatives have been synthesized and screened for their anti-obesity activity. Furanchalcones derivative has been reported to act as protein tyrosine phosphatase inhibitors[14-16]. The generated structure of complex (ligand and protein) where grouped by their scoring functions generated by docking. In the docking studies total energy of the system has calculated and it provides the optimized conformer of the complex, docking studies also help to predict the affinity of the ligand and receptor or protein in respect to amino acid of the proteins[17]. Molecular docking study is used to determine the interaction between ligands and proteins and their three-dimensional structure of the complex. It has been noticed that incorporation of more than one pharmacophore into a single molecular frame work enhances the pharmacological effects of resultant conjugate molecule. In view of these facts and high prevalence of obesity and hyperlipidemic disorders, present study was aimed to synthesize new analogues bearing conjugate features of pyrazole and chalcones and perform molecular docking studies and evaluate the hypolipidemic and anti-obesity potential in albino rats.
Materials and Methods
General:
Melting points of the compound were determined using capillary apparatus and were uncorrected. Preliminary purity of compounds was examined using Thin Layer Chromatography (TLC) silica gel 60 F254 plates (200 mm; Merck, Darmstadt, Germany). Structures of the compounds were regularly analyzed by Infrared (IR), 1H Nuclear Magnetic Resonance (NMR), 13C NMR and mass spectrometer. IR spectra were recorded in Potassium bromide (KBr) on an Agilent tech, Cary 660 Fourier-Transform Infrared Spectroscopy (FTIR) spectrophotometer. Bruker spectroscopy in DPX-300 MHz spectrophotometer was used to determine 1H-NMR and 13C NMR spectra using dimethyl sulfoxide as solvent and tetramethylsilane as an internal standard (chemical shift in δ ppm). Abbreviations s, d, t and m were used to indicate the peak multiplicity as singlet, doublet, triplet and multiplet. Mass spectra were recorded on waters, Q-TOF micro mass spectrometer. Silica gel (Merck) was used in column chromatography. Anhydrous sodium sulfate was utilized as a drying agent for the organic phase. The solvents were used after purification by distillation.
Synthesis of target compounds:
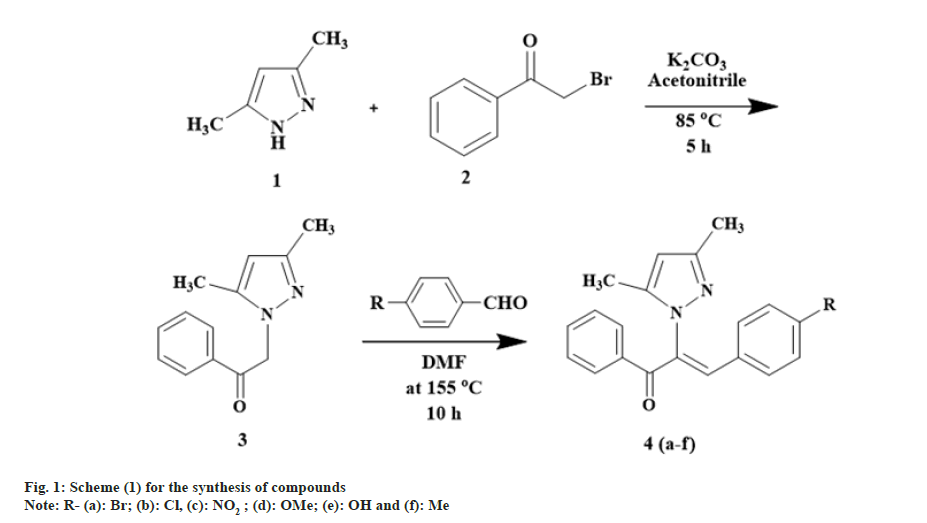
Title compounds were synthesized by following strategy given in fig. 1.
Preparation of 2-(3, 5-dimethyl-1H-pyrazol-1-yl)-1- phenylethanone (3): A mixture of 3, 5-dimethyl-1Hpirazole (0.02 mol), 2-bromacetophenone (0.02 mol) and potassium carbonate (0.020 mol) in acetonitrile was stirred and reflux for 5 h. Completion of the reaction was checked by TLC (hexane:ethyl acetate 7:3). The target compound was crystallized by transferring the reaction mixture into ice-cold water. The mixture was filtered and washed with water to obtain compound 3. The compound was purified by column chromatography using silica with hexane:ethyl acetate in 8:2 ratio as solvent.
Yield 73 %; Melting Point (MP) 76-78°; IR analysis (KBr) cm-1 3058 (C-H strAr), 1691 (C=O), 2934(Ar- Methyl (CH3)), 1578 (C=N); NMR analysis: H1 (δ ppm in Deuterated chloroform (CDCl3)) 7.79-7.99 (m, 5H, Ar), 5.91 (s, 2H, vinylic), 5.45 (s, H, pyrazole -C4), 2.23 (s, 3H, CH3), 2.16 (s, 3H, CH3); C13 (CDCl3, 100 MHz) 192.75 (1C, C=O), 148.36 (1C, C-3), 140.60 (1C, C-5),134.58 (1C, C-10), 134.02 (1C, C-13), 128.95 (2×1C, C-11), 128.12 (2×1C, C-12), 105.88 (1C, C-4),55.27 (1C, C-8), 13.53 (1C, C-6), 11.04 (1C, C-7); Elemental analysis Calcd. for C13H14N2O; C, 72.87; H, 6.59; N, 13.07 Found: C, 72.76; H, 6.60; N, 13.14; Mass spectroscopic analysis m/z 214 [M+].
General method of synthesis of (Z)-3-(4-subtituted phenyl)-2-(3, 5-dimethyl-1H-pyrazol-1-yl)-1-phenylprop-2-en-1-one (4a-f): To a mixture of 2-(3, 5-dimethyl-1H-pyrazol-1-yl)-1-phenylethanone (3) (0.005 mol) and p-substituted benzaldehyde (0.005 mol) was dissolved in N,N Dimethylformamide (DMF), sodium hydride was added and refluxed at 155° for 10 h. Completion of the reaction was checked by TLC (hexane:ethyl acetate 7:3). The target compound was obtained by pouring the reaction mixture into ice-cold water. Crystals were separated by adding hydrochloric acid which were filtered and recrystallized from ethanol. Resultant product was purified by column chromatography using silica with hexane:ethyl acetate 7:3 as solvent.
(Z)-3-(4-bromophenyl)-2-(3, 5-dimethyl-1Hpyrazol- 1-yl)-1-phenylprop-2-en-1-one (4a): Yield 81 %; MP 155-156°; IR analysis (KBr) cm-1 3090 (C-H strAr), 2973 (Ar-CH3), 1679 (C=O), 1586 (C=C), 570 (C-Br); NMR analysis: H1 (δ ppm in CDCl3) 8.42 -7.86 (m, 5H, ArH), 7.61-7.55 ( dd, 4H ), 5.35 (s, 1 H, vinylic), 5.49 (s, 1H, pyrazole), 2.53(s, 3H, CH3), 2.43 (s, 3H, CH3); C13 (CDCl3, 100 MHz) 183.93 (C=O), 148.8 (C-5, C-3), 136.0 (C-10), 135.59 (C-13), 133.14 (C-17, C-20), 131.48 (C-11, C-15), 130.83 (C- 8), 129.02 (C-12,C-14), 128.53 (C-19), 128.28 (C-18), 120.04 (C-16), 106.23 (C-4), 13.50 (C-6), 10.87 (C-7); Elemental analysis Calcd. for C20H17BrN2O; C, 63.00; H, 4.49; N, 7.35; Found: C, 63.08; H, 4.45; N, 7.38; Mass spectroscopic analysis m/z 381 [M+], 383 [M++2] (1:1).
(Z)-3-(4-chlorophenyl)-2-(3, 5-dimethyl-1Hpyrazol- 1-yl)-1-phenylprop-2-en-1-one (4b): Yield 86 %; MP 144-146°; IR analysis (KBr) cm-1 3050 (C-H strAr), 2923 (Ar-CH3), 1683 (C=O), 1573 (C=C), 629 (C-Cl); NMR analysis : H1 (δ ppm in CDCl3) 8.41-7.82 (m, 5H, ArH), 7.61-7.48 (dd, 4H, Ar), 5.34 (s, 1H, pyrazole), 5.21 (s, 1H, vinylic), 2.27 (s, 3H, CH3), 2.19 (s, 3H, CH3); C13 (CDCl3, 100 MHz) 183.93 (C=O),148.8 (C-5, C-3),136.0 (C-10), 135.59 (C-13), 133.14 (C-17,C-20), 131.48 (C-11, C-15), 130.83 (C- 8), 129.02 (C-12, C-14), 128.53 (C-19), 128.28 (C-18), 120.04 (C-16), 106.23 (C-4), 13.50 (C-6), 10.87 (C-7); Elemental analysis Calcd. for C20H17ClN2O; C, 71.32; H, 5.09; N, 8.32; Found: C, 71.35; H, 5.11; N, 8.30; Mass spectroscopic analysis m/z 336 [M+], 338 [M++2] (3:1).
(Z)-3-(4-nitrophenyl)-2-(3, 5-dimethyl-1H-pyrazol- 1-yl)-1-phenylprop-2-en-1-one (4c): Yield 72 %; MP 138-140°; IR analysis (KBr) cm-1 3061 (C-H strAr), 2917 (Ar-CH3), 1670 (C=O), 1574 (C=C), 1384 (Ar-NO2); NMR analysis : H1 (δ ppm in CDCl3) 8.83-7.95 (m, 5H, ArH), 7.68-7.61 ( dd, 4H, Ar ), 5.57 (s,1H, pyrazole), 5.39 (s, 1H, vinylic), 2.41 (s, 3H, CH3), 2.29(s, 3H, CH3); C13 (CDCl3, 100 MHz) 183.41 (C=O), 148.12 (C-20), 146.55 (C-5, C-3), 141.21 (C-17), 132.05 (C- 10), 131.99 (C-13), 129.88 (C-11, C-15), 129.61 (C-8), 129.14 (C-12, C-14),127.29 (C-18, C-22), 124.03 (C- 19, C-21), 120.12 (C-16), 106.15 (C-4), 13.92 (C-6), 10.80 (C-7); Elemental analysis C20H17N3O3; Calcd. for C20H17N3O3; C, 69.15; H, 4.93; N, 12.10; Found: C, 69.07; H, 4.92; N, 12.13; mass spectroscopic analysis m/z 347 [M+].
(Z)-3-(4-methoxyphenyl)-2-(3, 5-dimethyl-1Hpyrazol- 1-yl)-1-phenylprop-2-en-1-one (4d): Yield 79 %; MP 165-167°; IR analysis (KBr) cm-1 3065 (C-H strAr), 2924 (Ar-CH3), 1695 (C=O), 1597 (C=C), 1029 (C-O-C); NMR analysis : H1 (δ ppm in CDCl3) 8.48-7.96 (m, 5H, ArH), 7.84-7.68 (dd, 4H, Ar), 5.69 (s, 1H, pyrazole), 5.41 (s, 1H, vinylic), 3.71 (s, 3H, -OCH3), 2.12(s, 3H, CH3), 1.89 (s, 3H, CH3); C13 (CDCl3, 100 MHz) 189.77 (C=O), 147.87 (C-20),147.8 (C-5, C-3), 133.70 (C-10), 131.57 (C-13), 129.87 (C-11,C-15), 129.61 (C-8), 128.90 (C-12, C-14), 127.85 (C-17, C-18, C -22), 120.80 (C-16), 114.15 (C- 19, C-21), 106.27 (C-4), 56.93 (O-CH3), 13.55 (C-6), 10.83 (C-7); Elemental analysis Calcd. for C21H20N2O2; C, 75.88; H, 6.06; N, 8.43; Found: C, 75.79; H, 6.04; N, 8.46; Mass spectroscopic analysis m/z 332 [M+].
(Z)-3-(4-hydroxyphenyl)-2-(3, 5-dimethyl-1Hpyrazol- 1-yl)-1phenylprop-2-en-1-one (4e): Yield 70 %; MP 150-151°; IR analysis (KBr) cm-1 3032 (C-H strAr), 2926 (Ar-CH3), 1688 (C=O), 1594 (C=C), 3256 (Ar-OH); NMR analysis : H1 (δ ppm in CDCl3) 8,40 -7.90 (m, 5H, ArH), 7.66-7.52 (dd, 4H, Ar ), 5.41 (s, 1H, vinylic), 5.81 (s,1H, pyrazole), 5.27 (s, 1H, -OH), 2.22 (s, 3H, CH3), 2.14 (s, 3H, CH3); C13 (CDCl3, 100 MHz) 183.41 (C=O), 148.12 (C-20), 146.55 (C- 5, C-3), 141.21 (C-17), 132.05 (C-10), 131.99 (C- 13),129.88 (C-11, C-15), 129.61 (C-8), 129.14 (C-12, C-14), 127.29 (C-18, C-22), 124.03 (C-19, C-21), 120.12 (C-16), 106.15 (C-4), 13.92 (C-6), 10.80 (C-7); Elemental analysis Calcd. for C20H18N2O2; C, 75.45; H, 5.70; N, 8.80 Found; C, 75.43; H, 5.72; N, 8.87; Mass spectroscopic analysis m/z 318 [M+].
(Z)-3-p-tolyl -2-(3, 5-dimethyl-1H-pyrazol-1-yl)-1- phenyl-prop-2-en-1-one (4f): Yield 82 %; MP 170- 173°; IR analysis (KBr) cm-1 3022 (C-H strAr), 2925 (Ar-CH3), 1695 (C=O), 1557 (C=C); NMR analysis : H1 (δ ppm in CDCl3) 8.16 -7.78 (m, 5H, ArH), 7.48- 7.12 (dd, 4H, Ar), 5.78 (s,1H, pyrazole), 5.49 (s, 1H, vinylic), 2.52 (s, 3H, CH3), 2.49 (s, 3H, CH3), 2.12 (s, 3H, CH3); C13 (CDCl3, 100 MHz) 187.41 (1C, C=O), 143.88 (C-5, C-3), 137.81 (C-10), 133.92 (C-13), 132.04 (C-17), 132.04 (C-20), 131.36 (C-11,C-15),129.61 (C- 8),129.02 (C-19,C-21), 128.84 (C-12,C-14), 126.73 (C-18,C-22), 120.13 (C-16), 106.77 (C-4), 20.43 (CH3, C-23), 13.12 (C-6), 10.03 (C-7); Elemental analysis Calcd. For C21H20N2O; C, 79.72; H, 6.37; N, 8.85 Found: C, 79.59; H, 6.34; N, 8.79; Mass spectroscopic analysis m/z 316 [M+].
Pharmacology:
Acute toxicity studies: The doses for the test compounds were selected on the basis of acute toxicity studies following OECD 423 guidelines where compounds 4(a-f) were evaluated on fixed doses of 5- 2000 mg/kg administered orally in albino rats.
Hypolipidemic studies: The experimental protocol was approved by the Institutional Animal Ethical Committee (IAEC) of Banasthali University, Banasthali, Rajasthan, India (BU/BT/68/2.5.2012). Triton WR 1339 (Sigma Aldrich, United States of America (USA)) surfactant was employed to induce hyperlipidemia in rats. Wistar Albino rats (180-200 g) were divided into different groups containing six animals in each. Group I served as negative control and administered with vehicle only (2 % acacia solution), Group II served as positive control and administered Triton WR 1339 at 200 mg/ kg; i.p, Group III administered Triton WR 1339 (200 mg/kg i.p.) along with standard gemfibrozilat 400 mg/ kg p.o. in 2 % w/v acacia and group IV-IX were treated with Triton WR 1339 (200 mg/kg i.p.) along with test compound 4 (a-f) at a dose of 10 mg/kg p.o. in 2 % w/v acacia (aq.) respectively. Lipid profile of treated rats was determined using commercially available kits (Auto span, Mumbai, India)[18].
Anti-obesity studies: The Normal Diet (ND) consisted of simple laboratory chow only (Ashirwad Industries, Chandigarh, India). The High Fat Diet (HFD) was prepared and presented to different groups of rats for induction of obesity. The composition of HFD included the condensed milk 20 %, bread 20 %, chocolate 10 %, dried coconut 10 %, cheese 20 %, boiled potatoes 20 % with one multi-vitamin tablet Supradyn (Piramal Enterprises Limited, Maharashtra, India).
Wistar Albino rats (180-200 g) were divided into five groups containing six rats in each. Group I acted as negative control and administered with ND, group II served as positive control and received HFD, group III received HFD as well as treated with standard rimonabent at a dose of 10 mg/kg p.o., group IV were administered with HFD along with test compound (5 mg/kg p.o.) and group V were administered with HFD along with test compound (10 mg/kg p.o.). All the treatments were given for 40 d. The body weight (gm) was recorded before and after 40 d of the using rat digital weighing balance. Rimonabant (Cipla Pharmaceuticals Limited, India.) was used as standard[18].
Docking study:
In order to validate the findings of pharmacological studies, molecular docking has been employed to understand the receptor-ligand interactions of synthesized compounds with CB receptor (Protein Data Bank (PDB) Code: 5TGZ) using Argus lab 4.0. The ligand structures were generated using the Chem draw software. Three-dimensional optimizations of the ligand structures were done and saved as PDB file format. Geometry optimizations of the ligands were performed according to the Hartree-Fock (HF) calculation method using Argus Lab 4.0.1 (Mark A. Thompson, Planaria Software LLC, Seattle, WA, USA) software[19]. Argus Lab has work on quantum mechanics based program. It predicts the potential energies, molecular structures; geometry optimization of structure, vibration frequencies of coordinates of atoms, bond length and bond angle. CB was docked against six compounds the interaction was carried out to find the favorable binding geometries of the ligand with the protein. Docking simulations were performed by selecting “Argus Dock” as the docking engine. The selected residues of the receptor were defined to be a part of the binding site. A spacing of 0.4 Å between the grid points was used and an exhaustive search was performed by enabling “High precision” option in Docking precision menu, “Dock” was chosen as the calculation type, “flexible” for the ligand. A maximum of 150 poses were allowed to be analyzed. All the compounds were docked into the active site of protein, using the same protocol. The docking poses saved for each compound were ranked according to their dock score function. The pose having the highest dock score was selected for further analysis. The two-dimensional Pose where generated by using the software discovery studio[20].
Results and Discussion
The goal of this study was to obtain various pyrazole substituted chalcones 4(a-f), by Claisen-Schmidt condensation of 2-(3, 5-dimethyl-1H-pyrazol-1-yl)-1- phenylethanone (3) and 4 substituted benzaldehyde. DMF was used as solvent as it impart a better medium for reaction and Sodium hydride (NaH) used as strong base. The synthesized compound gave satisfactory results of their microanalysis. IR, 1H NMR, 13C NMR and mass spectrometry spectral data which were found to be consistent to the assigned structure. The IR spectral analysis of the products was validated using reactant spectra.
In the first step 3,5 dimethyl pyrazole was allowed to react with phenacyl bromide to yield 2-(3,5-dimethyl- 1H-pyrazol-1-yl)-1-phenylethanone (3) and its formation ascertained by IR absorption band at 1691 cm-1 assignable to >C=O Stretching (str.) and a band at 1578 cm-1 due to C=N str. and by absence of -NH peak at 3500-3400 cm-1 1H NMR of the compound (3) was exhibited a sharp up field singlet at δ 2.23 and δ 2.16 to the six proton of two -CH3 group attached to pyrazole ring and up field singlet which appeared at δ 5.45 due to two proton of CH2 (methylene group) attached to pyrazole ring. 13C NMR spectrum of compound (3) displayed the three signals at 48.29, 105.91 and 140.73 due to C-3, C-4 and C-5 respectively, which are unique to pyrazole system[21]. Along with this the 1H NMR spectrum of compound (3) showed multiplet for Ar-H at 8.05-7.45 ppm (m, 5H, Ar). Presence of a singlet at 5.9 ppm assignable to CH2-C=O moiety. Further, the formation of compound (4a-f) was ascertained by the appearance of peak at 1679 cm-1 (>C=O str.), and also by the other peaks which appeared at 1586 cm-1 (C=C str.) due to C=C of Alpha (α), Beta (β) conjugated ketone structure, 3090 cm-1 (C-H str., Ar), 2973 cm-1 (C-H str., Ar-CH3), 570 cm-1 (C-Br).
It is noteworthy that all the compounds (4a-4f) caused rats showed a state of lethargy at the doses 300 mg p.o. and considered unsafe, thus less than 1/30th and less of the doses were selected to omit any kind of toxicity precipitation in the experimental rats for the present study. The synthesized compounds (4a-4f) were evaluated for possible lipid lowering activity using triton WR 1339 induced hyperlipidemic rats. Results of hypolipidemic activity study are presented in fig. 2. Amongst six compounds (4a-4f), compounds 4a, 4b, 4d, and 4e showed significant reduction in cholesterol levels in rats i.e. 49.07 %±3.09 %, 52.20 %±1.92 %, 47.16 %±1.29 % and 43.42 %±2.57 %, respectively, which was comparable gemfibrozil (58.79 %± 4.3 %).
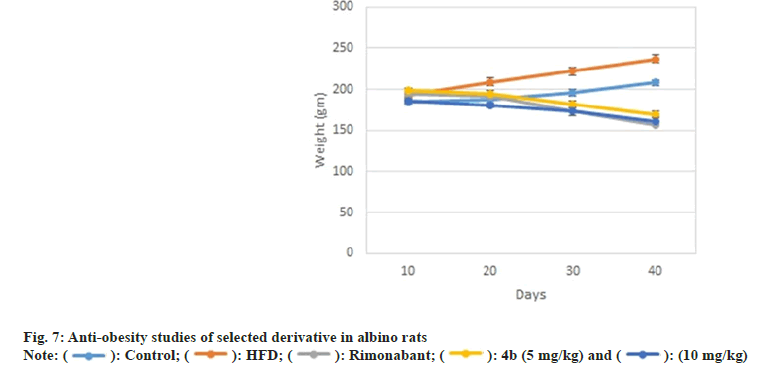
Similarly, compounds 4a, 4b, 4d and 4e also showed impressive reduction in triglyceride levels i.e. 43.40 %±1.48 %, 45.40 %±1.27 %, 39.35 %±1.41 % and 36.11 %±1.32 %, respectively, which was comparable to standard gemfibrozil (49.39 %±3.6 %). Notably, compound 4b also caused significant elevation (33.26 %±1.32 %) in % High Density Lipoprotein (HDL) levels of rats comparable to the standard gemfibrozil (36.66±4.7 %) (fig. 3). Compound 4b was recorded as most effective in ameliorating the lipid profiles in albino rats, thus it was further evaluated for weight reduction activity in HFD induced obesity model in rats as shown in Table 1.
| Body weights (gm) | Normal diet | HFD | Compound 4 (b) (5 mg)+HFD | Compound 4 (b) (10 mg)+HFD | Stand.+HFD |
|---|---|---|---|---|---|
| Weight before treatment | 154±7.8 | 157±5.4 | 155±3.3 | 154±5.4 | 155±4.8 |
| Weight after treatment | 171±4.0 | 218±5.4 | 194±5.0 | 185±6.2 | 173±3.5 |
Table 1: Effect of Rimonabant and Compound 4b on Body Weight of Hfd Rats
Note: *Values represent mean±SEM of six rats
Amongst six, three compounds showed promising receptor-ligand binding interactions with high docking score (Table 2). All the amino acids involved to form different bond with ligand are given in Table 3. Standard compound Rimonabent has used to compare the docking results as this compound has also the pryazole nucleolus and results clearly reveals that docking score of most of the synthesized compounds were more than the standard its means binding affinity of synthesized compounds are more as compare to standard for selected protein, in standard compound LYS-232, ALA-233, GLY-205,PHE-208, ASP-213, ILE-216, MET-240, THR-242 total 8 amino acids are form Vander Waal and interactions and LEU-209 amino acid residue form pi sigma bond t is also observed that the ILE-212, VAL-228, ALA-236, LEU-239, LEU-286 amino residues were responsible for pi and pi-alkyl bond formations. Compound 4b (Z)-3-(4-chlorophenyl)- 2-(3,5-dimethyl-1H-pyrazol-1-yl)-1-phenylprop-2- en-1-one (fig. 4) illustrated seven Vander Waal and interactions with docking score of -15.31A. Phenyl propenone depicted four Vander Waal interactions with SER-265, VAL-196, PHE-200 and TRP-356 amino acid residues. Dimethyl pyrazol showed two Vander Waal interactions with ASP-366, THR- 197 amino acid residues whereas one pi-sulfur bonds was also showed with MET-363 and total six alkyl bonds were observed with different amino acid residue.
| S. no | Compound | Docking score |
|---|---|---|
| 1 (4a) | 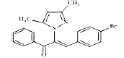 |
-15.31 |
| 2 (4b) | 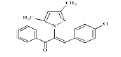 |
-15.27 |
| 3 (4c) | 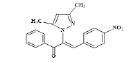 |
-13.95 |
| 4 (4d) | 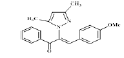 |
-12.75 |
| 5 (4e) | 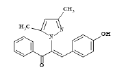 |
-12.81 |
| 6 (4f) | 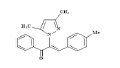 |
-15.25 |
| Standard (Rimonabant) | 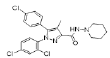 |
-11.20 |
Table 2: Best Docking Score Derivatives With Cannabinoid Receptor
| S. No | Vander waal | Pi-Sigma | Pi-Sulfur | Pi-pi T-shaped | Alkyl and pi-alkyl |
|---|---|---|---|---|---|
| 4a | VAL-196, PHE-200, TRP-356, ASP-272, THR-197, ASP-366, SER-265 | MET-363 | TRP-279, PHE-268 | LEU-360, PRO-251, ILE-271, LEU-193,ILE-280, ILE-267 | |
| 4b | VAL-196, PHE-200, TRP-356, ASP-272, THR-197, ASP-366, SER-265 | LEU-276 | TRP-279, PHE-268 | LEU-359, LEU-360, ILE-271, LEU-193, PRO-251, ILE-280, ILE-267 | |
| 4c | VAL-367, THR-197, ASP-366, SER-265, TRP-356, ASP-272, PRO-252, LEU-196, TYR-272, ALA-248 | MET-363 | PHE-268, TRP-279 | ILE-267, ILE-271, LEU-193, ILE-360, ILE-280 | |
| 4e | MET-363, THR-197, ILE-169, GLY-166, ILE-267, PRO-269, PHE-189, ASP-104, ASN-101, SER-383, | Phe-170 | LEU-193, PHE-379, ILE-105, LEU-359, LEU-193 | ||
| 4f | ASP-366, SER-165, ASP-272, LEU-190, VAL-196, PHE-200, THR-197, TRP-356 | LEU-276 | TRP-279, PHE-268 | ILE-267, PRO-251, LEU-193, LEU-360, LEO-259, ILE-280 | |
| Standard Rimonabant | LYS-232, ALA-233, GLY-205,PHE-208, ASP-213, ILE-216, MET-240, THR-242 | LEU-209 | - | ILE-212, VAL-228, ALA-236, LEU-239, LEU-286 |
Table 3: Different Interactions Of Amino Acid Residues
In addition (Z)-3-(4-chlorophenyl)-2-(3, 5-dimethyl- 1H-pyrazol-1-yl)-1-phenylprop-2-en-1-one (4b) (fig. 5) illustrated seven Vander Waal interactions with that of the receptor acquiring docking score of -15.27. Phenyl propenone moiety showed Vander Waal interaction with PHE-200, TRP-356, and VAL-196 amino acid residues. Methoxy phenyl on other hand showed a Vander Waal interaction with ASP-272 and THR-197. In addition, dimethyl pyrazol illustrated third Vander Waal interaction with SER-265 and ASP-366.
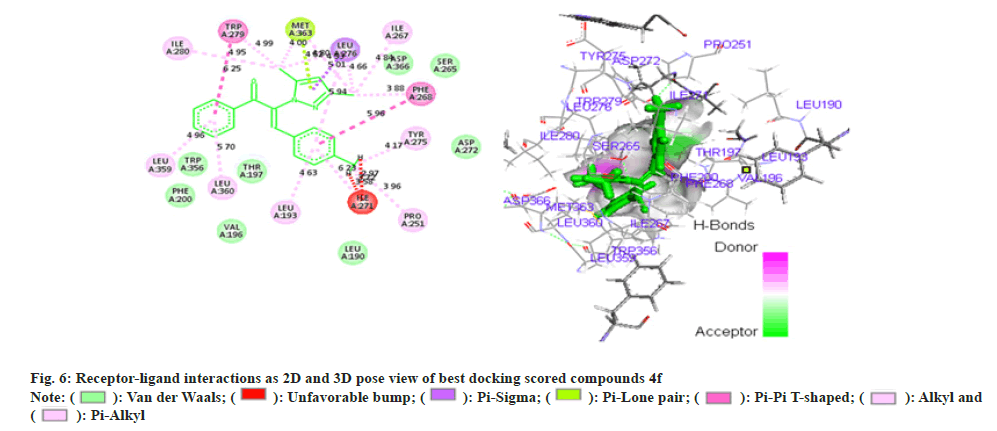
Phenyl propenone of 4f (Z)-2-(3,5-dimethyl-1Hpyrazol- 1-yl)-1-phenyl)-3-p-tolylprop-2-en-1-one (fig. 6) demonstrated eight Vander Waal interaction with different amino acid residues with docking score of -15.25. Toluene moiety on other hand illustrated two Vander Waal interactions with ASP-272 and LEU-190 amino acid residues. Dimethyl pyrazol showed the Vander Waal interaction with ASP-366 and SER-265.
Amongst all compounds 4a, 4b and 4f have illustrated better binding patterns including hydrogen bond interaction which obtained high docking score (Table 2). Some amino acid residues are play a crucial role for binding with ligands such as VAL-196, PHE-200, TRP-356, ASP-272, THR-197, ASP-366 and SER- 265 amino acids are essential for better binding. We can conclude that 4b (Z)-3-(4-chlorophenyl)-2-(3,5- dimethyl-1H-pyrazol-1-yl)-1-phenylprop-2-en-1-one have been found effective in binding with CB receptor. Many of pyrazole and chalcones derivatives are important pharmacophore in exhibiting anti-obesity activity. Structure activity relationship of various antiobesity drug revealed that the presence of electron withdrawing group, α, β unsaturated ketone and chlorophenyl group are responsible for the anti-obesity properties[22]. Moreover, results of docking studies were found to be consistent that the compound 4b (Z)- 3-(4-chlorophenyl)-2-(3,5-dimethyl-1H-pyrazol-1-yl)- 1-phenylprop-2-en-1-one has better ability to interact with CB receptor , thus has substantial potential to act as anti-obesity agent. On the basis of results of hypolipidemic and docking studies compound 4b (Z)-3-(4-chlorophenyl)-2-(3,5-dimethyl-1H-pyrazol- 1-yl)-1-phenylprop-2-en-1-onewas selected forantiobesity. Anti-obesity study was carried out using HFD induced obesity model. A 40 d HFD exposure showed impressive weight gain in group II rats whereas there was no significant increase in bodyweight of rats in group I. Administration of compound 4b (Z)-3-(4- chlorophenyl)-2-(3, 5-dimethyl-1H-pyrazol-1-yl)-1- phenylprop-2-en-1-one at dose of 5mg/kg and 10 mg/ kg orally, caused marked reduction in body weight of rats when compared to group II (fig. 7). It is noteworthy that during treatment, no behavioral alteration and mortality of test animal was observed that indicated its high therapeutic index and approved its suitability for prolonged use of the therapy.
Pyrazoles have been documented to possess wide array of pharmacological activities including hypolipidemic activities. In view of this in the present study, various novel pyrazole derivatives (4a-f) have been successfully synthesized using simple and robust protocols and characterized using physical and spectral methods. Amongst all the synthesized compounds,(Z)- 3-(4-chlorophenyl)-2-(3,5-dimethyl-1H-pyrazol-1- yl)-1-phenylprop-2-en-1-one(compound 4b) showed significant reduction in body weight together with hypolipidemic activity in experimental rats. There was no behavioral alteration and mortality was observed which indicates its safety and its suitability for prolonged course of therapy in obesity and hyperlipidemic disorders. However the present preliminary studies calls for comprehensive toxicological and pharmacological studies to establish its therapeutic safety and efficacy in preclinical and clinical subjects.
Author’s contributions:
Authors are grateful to Department of Science and Technology (DST), New Delhi for the project, “Banasthali Center for Education and Research in Basic Sciences” under their CURIE (Consolidation of University Research for Innovation and Excellence in Women Universities) program for their financial support. Authors are also thankful to SAIF, Punjab University Chandigarh for providing the 1H-NMR, 13C-NMR and mass spectrometry data of the compounds and to the Department of Pharmacy Banasthali Vidyapith for providing facility to conduct anti-obesity studies.
Conflict of interests:
The authors declared no conflict of interests.
References
- Derosa G, Maffioli P. Anti-obesity drugs: A review about their effects and their safety. Exp Opin Drug Saf 2012;11(3):459-71.
[Crossref] [Google Scholar] [PubMed]
- Trigueros L, Peña S, Ugidos AV, Sayas-Barberá E, Pérez-Álvarez JA, Sendra E. Food ingredients as anti-obesity agents: A review. Crit Rev Food Sci Nutr 2013;53(9):929-42.
[Crossref] [Google Scholar] [PubMed]
- Yonetoku Y, Kubota H, Okamoto Y, Ishikawa J, Takeuchi M, Ohta M, et al. Novel potent and selective calcium-release-activated calcium (CRAC) channel inhibitors. Part 2: Synthesis and inhibitory activity of aryl-3-trifluoromethylpyrazoles. Bioorg Med Chem 2006;14(15):5370-83.
[Crossref] [Google Scholar] [PubMed]
- Griebenow N, Schirok H, Mittendorf J, Straub A, Follmann M, Stasch JP, et al. Identification of acidic heterocycle-substituted 1H-pyrazolo [3, 4-b] pyridines as soluble guanylate cyclase stimulators. Bioorg Med Chem Lett 2013;23(5):1197-200.
[Crossref] [Google Scholar] [PubMed]
- Gökhan-Kelekçi N, Yabanoğlu S, Küpeli E, Salgın U, Özgen Ö, Uçar G, et al. A new therapeutic approach in Alzheimer disease: Some novel pyrazole derivatives as dual MAO-B inhibitors and antiinflammatory analgesics. Bioorg Med Chem 2007;15(17):5775-86.
[Crossref] [Google Scholar] [PubMed]
- Chauhan M, Kumar R. Medicinal attributes of pyrazolo [3, 4-d] pyrimidines: A review. Bioorg Med Chem 2013;21(18):5657-68.
[Crossref] [Google Scholar] [PubMed]
- Kumar GG, Vikas K, Vinod K. Pyrazoles as potential anti-obesity agents. Res J Chem Environ 2011;15(3):90-103.
- Alvarado M, Goya P, Macías-González M, Pavón FJ, Serrano A, Jagerovic N, et al. Antiobesity designed multiple ligands: Synthesis of pyrazole fatty acid amides and evaluation as hypophagic agents. Bioorg Med Chem 2008;16(23):10098-105.
[Crossref] [Google Scholar] [PubMed]
- Bifulco M, Pisanti S. End of the line for cannabinoid receptor 1 as an anti-obesity target? An opinion. Nat Rev Drug Discov 2009;8(7):594.
- Fong TM, Heymsfield SB. Cannabinoid-1 receptor inverse agonists: Current understanding of mechanism of action and unanswered questions. Int J Obes 2009;33(9):947-55.
[Crossref] [Google Scholar] [PubMed]
- Doggrell SA. Will the new CB1 cannabinoid receptor antagonist SR-147778 have advantages over rimonabant? Exp Opin Investig Drugs 2005;14(3):339-42.
[Crossref] [Google Scholar] [PubMed]
- Christopoulou FD, Kiortsis DN. An overview of the metabolic effects of rimonabant in randomized controlled trials: Potential for other cannabinoid 1 receptor blockers in obesity. J Clin Pharm Ther 2011;36(1):10-8.
[Crossref] [Google Scholar] [PubMed]
- K Sahu N, S Balbhadra S, Choudhary J, V Kohli D. Exploring pharmacological significance of chalcone scaffold: A review. Curr Med Chem 2012;19(2):209-25.
[Crossref] [Google Scholar] [PubMed]
- Birari RB, Gupta S, Mohan CG, Bhutani KK. Antiobesity and lipid lowering effects of Glycyrrhiza chalcones: Experimental and computational studies. Phytomedicine. 2011;18(8-9):795-801.
[Crossref] [Google Scholar] [PubMed]
- Sun LP, Jiang Z, Gao LX, Sheng L, Quan YC, Li J, Piao HR. Synthesis and biological evaluation of furan-chalcone derivatives as protein tyrosine phosphatase inhibitors. Bull Korean Chem Soc 2013;34(4):1023-4.
- Aly MR, Fodah HH, Saleh SY. Antiobesity, antioxidant and cytotoxicity activities of newly synthesized chalcone derivatives and their metal complexes. Eur J Med Chem 2014;76:517-30.
[Crossref] [Google Scholar] [PubMed]
- Arya N, Munde MK, Dwivedi J, Jagdale AY, Jain KS. Design, Synthesis, and Antihyperlipidemic Evaluation of Novel 2‐[1‐(S ubstitutedphenyl)‐4‐oxo‐azetidin‐2‐yl]‐5, 6‐disubstitutedthieno [2, 3‐d] pyrimidin‐4 (3 H)‐ones. Arch Pharm 2013;346(8):588-95.
[Crossref] [Google Scholar] [PubMed]
- Lim SM, Goh YM, Mohtarrudin N, Loh SP. Germinated brown rice ameliorates obesity in high-fat diet induced obese rats. BMC Complement Alter Med 2016;16(1):140.
[Crossref] [Google Scholar] [PubMed]
- ArgusLab 4.0.1 (Mark A. Thompson, Planaria Software LLC, Seattle, WA, USA.) software.
- Discovery studio.com
- Kumar V, Kaur K, Karelia DN, Beniwal V, Gupta GK, Sharma AK, Gupta AK. Synthesis and biological evaluation of some 2-(3, 5-dimethyl-1H-pyrazol-1-yl)-1-arylethanones: Antibacterial, DNA photocleavage, and anticancer activities. Eur J Med Chem 2014;81:267-76.
[Crossref] [Google Scholar] [PubMed]
- Repas T. Obesity and dyslipidemia. South Dakota Medicine. 2011;88: 241-9.
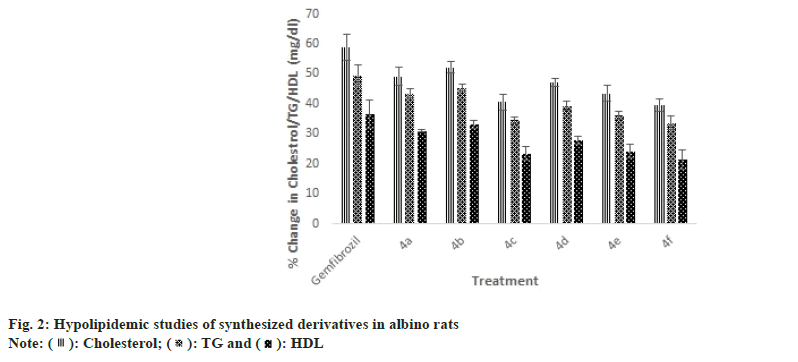
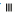 ): Cholesterol; (
): Cholesterol; (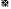 ): TG and (
): TG and (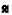 ): HDL
): HDL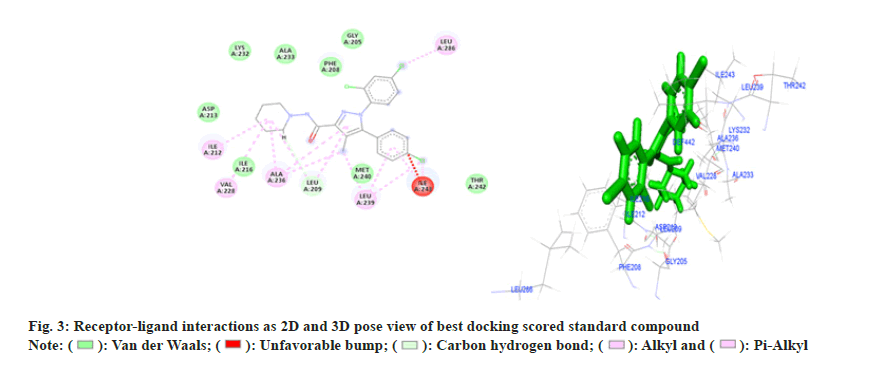
 ): Van der Waals; (
): Van der Waals; ( ): Unfavorable bump; (
): Unfavorable bump; ( ): Carbon hydrogen bond; (
): Carbon hydrogen bond; ( ): Alkyl and (
): Alkyl and ( ): Pi-Alkyl
): Pi-Alkyl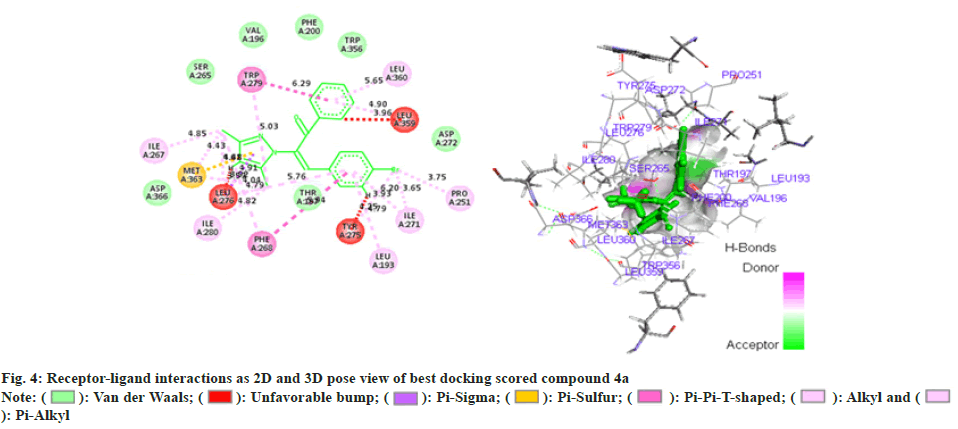
 ): Pi-Sigma; (
): Pi-Sigma; ( ): Pi-Sulfur; (
): Pi-Sulfur; ( ): Pi-Pi-T-shaped; (
): Pi-Pi-T-shaped; (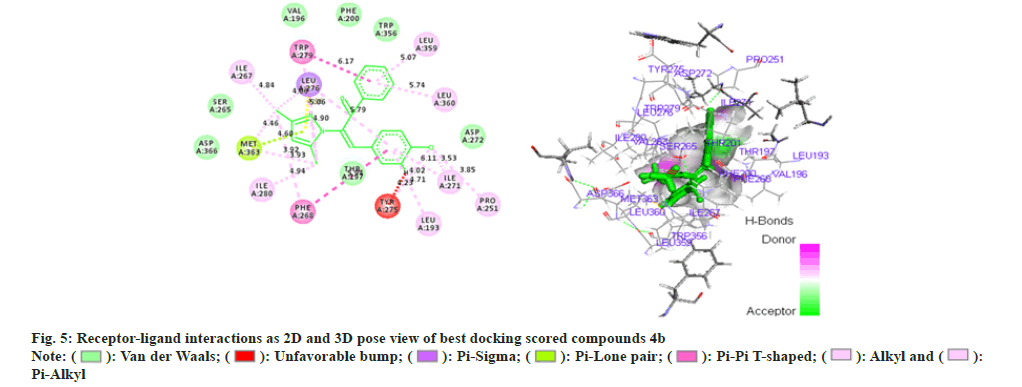
 ): Control; (
): Control; ( ): HFD; (
): HFD; ( ): Rimonabant; (
): Rimonabant; ( ): 4b (5 mg/kg) and (
): 4b (5 mg/kg) and ( ): (10 mg/kg)
): (10 mg/kg)