- *Corresponding Author:
- H. Padh
Department of Cell and Molecular Biology, B. V. Patel Pharmaceutical Education and Research Development (PERD) Centre, Thaltej-380 054, India
E-mail: hpadh@yahoo.com
| Date of Submission | 11 June 2016 |
| Date of Revision | 14 December 2016 |
| Date of Acceptance | 27 March 2017 |
| Indian J Pharm Sci 2017;79(2):311-316 |
Abstract
Genome variations in the form of single nucleotide polymorphism, INDELs (insertions or deletions) and, duplications, inversions and copy number variations are more widely prevalent than initially predicted. Mitochondrial tumor suppressor gene maps on chromosome 8p, in which exon 4-specific deletion is associated with susceptibility towards breast and various other forms of cancer. In this study, 41 head and neck cancer, 15 breast cancer patients and 280 healthy control individuals were analyzed for mitochondrial tumor suppressor gene copy number variation. The frequencies of wt/wt (homozygous wild-type), wt/Del (heterozygous variant) and Del/Del (homozygous deletion variant) mitochondrial tumor suppressor gene genotypes were found to be 73.3%, 26.7%, and 0% in breast cancer patients, 41.5%, 58.5%, and 0% in head and neck cancer patients and 43%, 57%, and 0% in healthy individuals, respectively. A significant association of the deletion variant with breast cancer (odds ratio = 0.27, 95% confidence interval = 0.08–0.87, P = 0.0207) was found in our population. In addition, the allele and genotype frequency varied significantly (P<0.0001) among Indian and German populations, reflecting ethnic diversity. This pilot case-control study highlighted the indicative role of mitochondrial tumor suppressor gene deletion in protection from breast cancer in Indian population. However, the findings need to be investigated in larger patient sample size before any conclusive role of mitochondrial tumor suppressor gene copy number variation on cancer risk.
Keywords
Breast cancer, copy number variation, head and neck cancer, health, pharmacogenetic
Copy number variation (CNVs) arise from the genomic rearrangements resulting in the deletion, duplication, insertion and translocations covering about 12% of the human genome [1,2]. Recent studies have highlighted the role of CNVs in complex disorders including susceptibility to cancer [3-5]. The association of CNVs in disease susceptibility involving wide range of common human disorders is well compiled and documented by several researchers [6,7]. In order to detect these changes several approaches have been developed and applied till date [8]. Among them, array-based comparative genomics approach by Hinds et al. in 2006 was used to detect common deletions ranging from 70 bps to 7 Kb [9]. Of the 100 CNVs identified, the mitochondrial tumor suppressor gene 1 (MTUS1) encompassing the deletion of complete coding exon 4 (162 bp) was one of them. The breakpoint of the deleted region was redefined and validated in large case-control samples from German population [10]. In 2005, Lupski and Stankiewicz documented the role of CNVs in genetic disorders but the extent to which CNVs contribute to disease development remains poorly understood [11]. However, the exon-specific deletion of MTUS1 gene is an illustration of a CNV associated with the development of cancer [12].
MTUS1 gene, also designated as MTSG1, GK1 or angiotensin II (AT2) receptor-interacting protein (ATIP1) is located at chromosomal position 8p22 spanning over 112 Kb in size. It is found to be ubiquitously expressed in normal tissues but transiently up-regulated during initiation of cellular quiescence and differentiation process [4]. The tumor suppression function was confirmed by mRNA expression studies in the pancreatic cancer tissues and the pancreatic cancer cell lines MIA PaCa-2, whereas the recombinant expression of MTUS1 in MIA PaCa-2 cells inhibited proliferation [13].
MTUS1 is found to be associated with disease progression in human cancers, including bladder, colorectal, oesophageal, head and neck squamous cell, hepatocellular, lung, ovarian, pancreatic, prostate and especially breast carcinomas[13-17]. Furthermore, it is found that MTUS1/ATIP1 is an early mediator of AT2 receptor activation. Together with AT2, it antagonizes AT1 receptor function, inhibiting epidermal growth factor signalling via autophosphorylation of epidermal growth factor receptor. This leads to altering the activity of growth factor-induced extracellular regulated kinase, phosphorylation of signal transducer and activation of transcription 3 (STAT3) and protein kinase C, involved in apoptosis and proliferation[18-24].
The aim of this study was to determine the frequency distribution of the MTUS1 deletion variant in Indian population. Subsequently in a pilot case-control study we intended to investigate the association with MTUS1 deletion variant with development of cancer.
MTUS1 deletion was genotyped in 41 patients with head and neck cancer (HNC), 15 patients with breast cancer (BC) and 280 healthy control individuals. The healthy control population comprised of the individuals 20-50 y of age (245 males and 35 females) mainly from the Western India. They were recruited at B. V. Patel PERD Centre, Ahmedabad. The case (HNC and BC patients) population comprised of the individuals 18 or older in age, mainly from the Western India. The recruited patients were undergoing treatment at local cancer hospital in Ahmedabad. Newly diagnosed BC and HNC solid tumor patients before medication were selected for the study and significant inclusion and exclusion criteria were taken into consideration. The study was approved by the Institutional Ethics Committee and written informed consent form was obtained prior to blood collection from individuals.
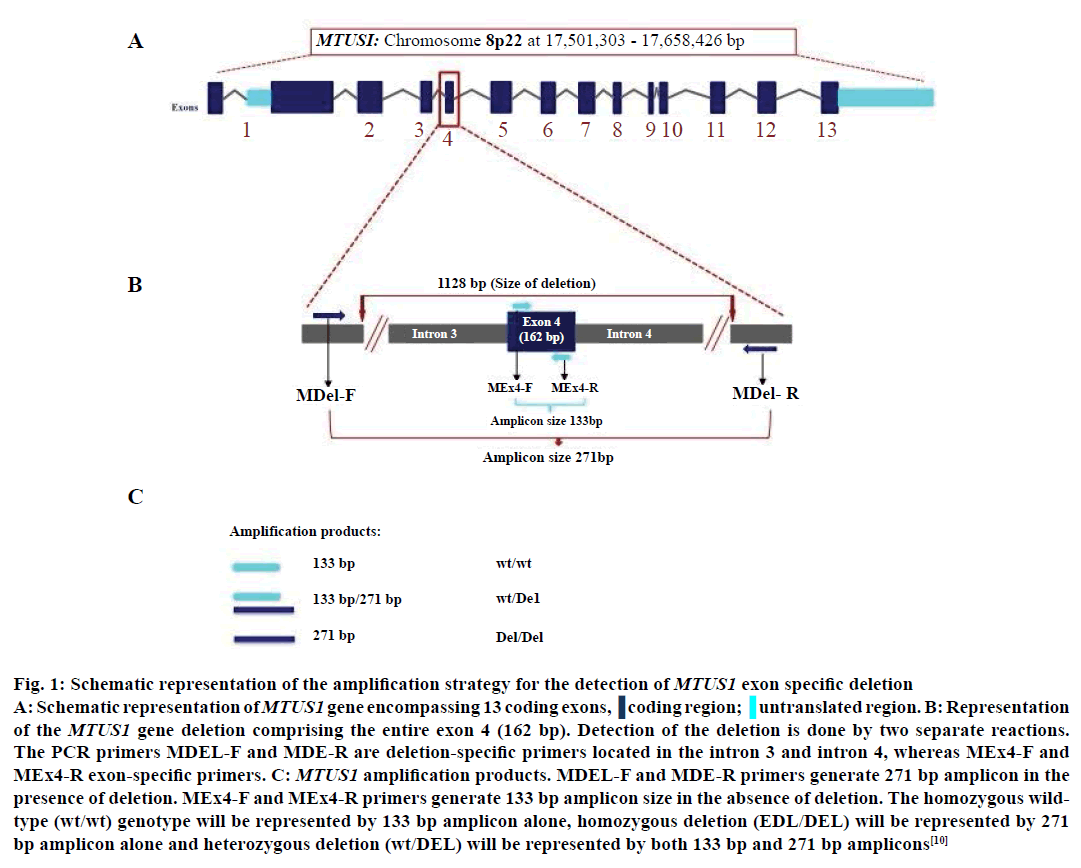
Genomic DNA isolation was carried from 5 ml of blood withdrawn from healthy individuals and cancer patients. Blood (5 ml) was collected in anticoagulant (K2 EDTA or sodium heparin) precoated collection tubes and genomic DNA was extracted using phenol-chloroform method [25]. The RBCs were first ruptured using lysis buffer followed by the lysis of the WBC pellet using digestion buffer. The protein degradation was carried out using proteinase K treatment and the purification of the sample using phenol:chloroform:isoamyl alcohol mixture. The DNA was precipitated using absolute alcohol and subsequently washed with 70% alcohol. The DNA pellet thus obtained was air dried and dissolved in appropriate amount of TE buffer [25]. MTUS1 genotyping was performed by polymerase chain reaction (PCR) amplification. Two separate reactions were designed to detect MTUS1 deletion (Figures 1 and 2) using primer pairs as described earlier [10]. The confirmation of the PCR products were carried out by sequencing deletion-specific and exon-specific amplicons using Applied Biosystems 3730xl sequencer (Macrogen Inc., Korea).
Figure 1: Schematic representation of the amplification strategy for the detection of MTUS1 exon specific deletion A: Schematic representation of MTUS1 gene encompassing 13 coding exons. B: Representation of the MTUS1 gene deletion comprising the entire exon 4 (162 bp). Detection of the deletion is done by two separate reactions. The PCR primers MDEL-F and MDE-R are deletion-specific primers located in the intron 3 and intron 4, whereas MEx4-F and MEx4-R exon-specific primers. C: MTUS1 amplification products. MDEL-F and MDE-R primers generate 271 bp amplicon in the presence of deletion. MEx4-F and MEx4-R primers generate 133 bp amplicon size in the absence of deletion. The homozygous wild-type (wt/wt) genotype will be represented by 133 bp amplicon alone, homozygous deletion (EDL/DEL) will be represented by 271 bp amplicon alone and heterozygous deletion (wt/DEL) will be represented by both 133 bp and 271 bp amplicons (adapted from Frank et al. 2007)
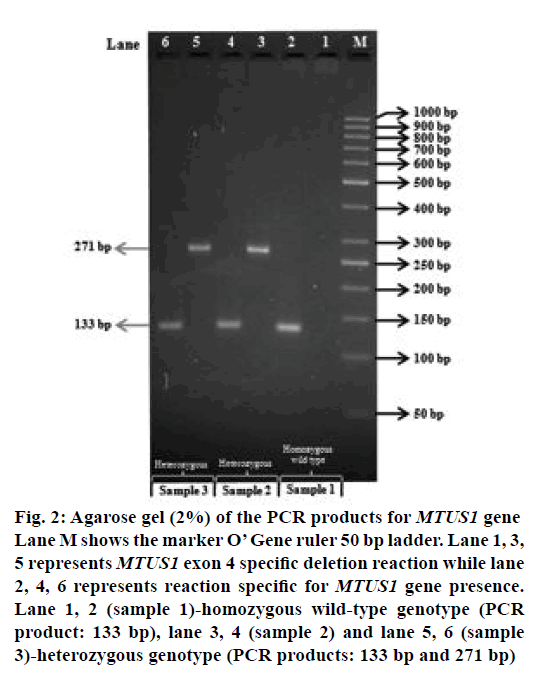
Figure 2: Agarose gel (2%) of the PCR products for MTUS1 gene Lane M shows the marker O’ Gene ruler 50 bp ladder. Lane 1, 3, 5 represents MTUS1 exon 4 specific deletion reaction while lane 2, 4, 6 represents reaction specific for MTUS1 gene presence. Lane 1, 2 (sample 1)-homozygous wild-type genotype (PCR product: 133 bp), lane 3, 4 (sample 2) and lane 5, 6 (sample 3)-heterozygous genotype (PCR products: 133 bp and 271 bp)
The difference between the genotype frequencies of HNC and BC cases with control group was calculated by χ2 test for statistical significance. Hardy-Weinberg equilibrium (HWE) was examined using χ2 test with one degree of freedom. Genotype-specific odds ratios (ORs), 95% confidence intervals (CI) and P-values were calculated by unconditional logistic regression using IBM SPSS 16.0 software.
The distribution of MTUS1 allele and genotypes was assessed in 280 healthy Indian control individuals. The frequency for wt allele was 0.71 and that of Del allele was 0.29. The genotype frequency was found to be 49.2% for wt/wt (homozygous wild-type) genotype, 57.1% for wt/Del (heterozygous variant) genotype while no healthy individual carried Del/Del (homozygous deletion variant) genotype. However, it was observed that the genotype frequencies studied in the control individual were not in the agreement with the HWE. Deviation from HWE could have been because of a number of reasons, but sequencing and confirming the results ruled out the technical error resulting in such conclusion. However, the reason for lack of HWE is still not clear.
Furthermore, a comparison of the allele and genotype frequency among Indian and German population highlighted a significant (P<0.0001) difference, reflecting the ethnic diversity between individuals of different population (Table 1).
| Genotype | Indian population n (%) (data from this study) |
German population n (%) (data[10]) |
|---|---|---|
| wt/wt | 120 (42.9) | 668 (91.3) |
| wt/Del | 160 (57.1) | 63 (8.6) |
| Del/Del | 0 (0) | 1 (0.1) |
| DEL/DEL + wt/DEL | 160 (57.1) | 64 (8.7) |
Table 1: Frequency distribution of mtus1 deletion variants in Indian and German population[10] in healthy individuals
The MTUS1 genotype frequency in BC patients and HNC patients was found to be 73.3 and 41.5% for wt/ wt (homozygous wild-type) genotype, 26.7 and 58.5% for wt/Del (heterozygous variant) genotype while no BC or HNC patient carried Del/Del (homozygous deletion variant) genotype. A significant association of the deletion variant with a decreased risk to breast cancer was observed (OR=0.27, 95% CI=0.08–0.87, P=0.0207). However, a non-significant association was observed between MTUS1 deletion variant and risk to HNC (OR=1.06, 95% CI=0.85–1.33, P=0.8671) (Table 2) in Indian population. Since, this study is limited by smaller sample size, the results need to be validated using larger population.
| Population | Genotype | Cases n (%) |
Control n (%) |
OR (95% CI), P value | |
|---|---|---|---|---|---|
| Indian population |
Breast cancer | wt/wt | 11 (73.3) | 120 (42.9) | 0.27 (0.08–0.87), 0.0207 |
| wt/Del | 4 (26.7) | 160 (57.1) | |||
| Del/Del | 0 (0) | 0 (0) | |||
| Head and neck cancer |
wt/wt | 17 (41.5) | 120 (42.9) | 1.06 (0.85–1.33), 0.8671 | |
| wt/Del | 24 (58.5) | 160 (57.1) | |||
| Del/Del | 0 (0) | 0 (0) | |||
| German population* | Familial BC | wt/wt | 562 (94.8) | 668 (91.3) | 0.58 (0.37-0.90), 0.01 |
| wt/Del | 30 (5.1) | 63 (8.6) | |||
| Del/Del | 1 (0.1) | 1 (0.1) | |||
| High-risk familial BC |
wt/wt | 355 (96.2) | 668 (91.3) | 0.41 (0.23-0.74), 0.003 | |
| wt/Del | 14 (3.8) | 63 (8.6) | |||
| Del/Del | 0 (0) | 1 (0.1) |
Table 2:Genotype frequencies of mtus1 deletion variants in patients and control subjects in Indian and German population[10]
CNV has become an integral part of genetic variation contributing to the susceptibility towards various diseases [26]. CNV in terms of exon-specific deletion in the tumor suppressor gene (MTUS1) is well studied in German population [10]. As reported, this phenomenon of exon-specific deletion provokes an increased protein activity leading to enhanced tumor suppressor activity. Further analysis revealed that the deleted exon was rich in polyproline-motifs, which are usually involved in interactions with the SH3 or WW functional domains implying that the wild type and deleted variants of MTUS1 may interact with distinct intracellular partners and exhibit different cellular functions, such as tumor suppression [27-29]. In conjunction with the previous reports of MTUS1 gene deletion variant with a decreased risk for both familial and high-risk familial BC in German population we examined the frequency of the deletion variant in Indian population. The frequency of the deletion variant was quite different when compared to the German population (Table 1). Further the 1128 bp deletion was genotyped in healthy Indian population and patients with either HNC or BC.
The association of the MTUS1 deletion variant was found to be significant (P=0.0207) in case of BC while for HNC non-significant association was highlighted in Indian population. However, no conclusive inference can be drawn with the small sample size and hence, validation in a large sample size of cancer patients is warranted.
Acknowledgements
The authors thank Dr. Dipali Dhawan for collection of the patient’s blood samples and acknowledge all the volunteers for participation in the study. Authors also thank the Council of Scientific and Industrial Research (CSIR) New Delhi, India for providing Senior Research Fellowship to Ms. Suhani Almal.
Conflict of interests
Authors report no conflict of interests.
Financial support and sponsorship
Nil.
References
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, et al. Global variation in copy number in the human genome. Nature 2006;444:444-54.
- Perry GH, Ben-Dor A, Tsalenko A, Sampas N, Rodriguez-Revenga L, Tran CW, et al. The fine-scale and complex architecture of human copy-number variation. Am J Hum Genet 2008;82:685-95.
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, et al. Large-scale copy number polymorphism in the human genome. Science 2004;305:525-8.
- Sharp AJ, Locke DP, McGrath SD, Cheng Z, Bailey JA, Vallente RU, et al. Segmental duplications and copy-number variation in the human genome. Am J Hum Genet 2005;77:78-88.
- Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet 2006;7:85-97.
- Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med 2010;61:437-55.
- Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet 2009;10:451-81.
- Dhawan D, Padh H. Pharmacogenetics: technologies to detect copy number variations. CurrOpinMolTher 2009;11:670-80.
- Hinds DA, Kloek AP, Jen M, Chen X, Frazer KA. Common deletions and SNPs are in linkage disequilibrium in the human genome. Nat Genet 2006;38:82-5.
- Frank B, Bermejo JL, Hemminki K, Sutter C, Wappenschmidt B, Meindl A, et al. Copy number variant in the candidate tumor suppressor gene MTUS1 and familial breast cancer risk. Carcinogenesis 2007;28:1442-5.
- Lupski JR, Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet 2005;1:e49.
- Shlien A, Malkin D. Copy number variations and cancer. Genome Med 2009;1:62.
- Seibold S, Rudroff C, Weber M, Galle J, Wanner C, Marx M, et al. Identification of a new tumor suppressor gene located at chromosome 8p21.3-22. FASEB J 2003;17:1180-2.
- Pineau P, Nagai H, Prigent S, Wei Y, Gyapay G, Weissenbach J, et al. Identification of three distinct regions of allelic deletions on the short arm of chromosome 8 in hepatocellular carcinoma. Oncogene 1999;18:3127-34.
- Pils D, Horak P, Gleiss A, Sax C, Fabjani G, Moebus VJ, et al. Five genes from chromosomal band 8p22 are significantly down-regulated in ovarian carcinoma: N33 and EFA6R have a potential impact on overall survival. Cancer 2005;104:2417-29.
- Chaib H, MacDonald JW, Vessella RL, Washburn JG, Quinn JE, Odman A, et al. Haploinsufficiency and reduced expression of genes localized to the 8p chromosomal region in human prostate tumors. Genes Chromosomes Cancer 2003;37:306-13.
- Yokota T, Yoshimoto M, Akiyama F, Sakamoto G, Kasumi F, Nakamura Y, et al. Localization of a tumor suppressor gene associated with the progression of human breast carcinoma within a 1-cM interval of 8p22-p23.1. Cancer 1999;85:447-52.
- Kerangueven F, Noguchi T, Coulier F, Allione F, Wargniez V, Simony-Lafontaine J, et al. Genome-wide search for loss of heterozygosity shows extensive genetic diversity of human breast carcinomas. Cancer Res 1997;57:5469-74.
- Nouet S, Amzallag N, Li JM, Louis S, Seitz I, Cui TX, et al. Trans-inactivation of receptor tyrosine kinases by novel angiotensin II AT2 receptor-interacting protein, ATIP. J BiolChem 2004;279:28989-97.
- Nouet S, Nahmias C. Signal transduction from the angiotensin II AT2 receptor. Trends EndocrinolMetab 2000;11:1-6.
- Di Benedetto M, Bièche I, Deshayes F, Vacher S, Nouet S, Collura V, et al. Structural organization and expression of human MTUS1, a candidate 8p22 tumor suppressor gene encoding a family of angiotensin II AT2 receptor-interacting proteins, ATIP. Gene 2006;380:127-36.
- Deshayes F, Nahmias, C. Angiotensin receptors: a new role in cancer? Trends EndocrinolMetab 2005;16:293-9.
- Wruck CJ, Funke-Kaiser H, Pufe T, Kusserow H, Menk M, Schefe JH, et al. Regulation of transport of the angiotensin AT2 receptor by a novel membrane-associated Golgi protein. ArteriosclerThrombVascBiol 2005;25:57-64.
- Greco S, Muscella A, Elia MG, Salvatore P, Storelli C, Marsigliante S. Activation of angiotensin II type I receptor promotes protein kinase C translocation and cell proliferation in human cultured breast epithelial cells. J Endocrinol 2002;174:205-14.
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1989.
- Almal SH, Padh H. Implications of gene copy-number variation in health and diseases. J Hum Genet 2012;57:6-13.
- Di Benedetto M, Pineau P, Nouet S, Berhouet S, Seitz I, Louis S. et al. Mutation analysis of the 8p22 candidate tumor suppressor gene ATIP/MTUS1 in hepatocellular carcinoma. Mol Cell Endocrinol 2006;252:207-15.
- Macias MJ, Wiesner S, Sudol M. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett 2002;513:30-7.
- Tchatchou S, Burwinkel B. Chromosome copy number variation and breast cancer risk. Cytogenet Genome Res 2008;123:183-7.