- *Corresponding Author:
- V. Murugan
Department of Pharmaceutical Chemistry, J. S. S. College of Pharmacy, Rock lands, Ootacamund-643 001, India
E-mail: murugan62@yahoo.com
| Date of Submission | 22 February 2005 |
| Date of Revision | 06 August 2005 |
| Date of Acceptance | 17 May 2006 |
| Indian J Pharm Sci, 2006, 68 (3): 323-326 |
Abstract
This present work is an investigation of anticancer activities of the nitrogen mustards possessing quinazolinone, benzimidazole, benzoxazole, and benzothiazole nuclei by the three-dimensional Quantitative Structure Activity paradigm, Comparative Molecular Field Analysis. A total of 39 compounds were modelled in SYBYL 6.7 (Tripos, USA). The molecules were aligned by root-mean-square fit of atoms and field fit of the steric and electrostatic molecular fields and the resulting databases analysed by partial least squares analysis with cross-validation, leave-one-out and no validation to extract the optimum number of components. The analysis was then repeated with bootstrapping to give the final Quantitative Structure Activity Relationship models. Eight compounds, which were kept separately as test set, were used to test the predictive ability of the Comparative Molecular Field Analysis models. Out of the two models generated, one was found to be useful. The predicted activities of the test set were in good agreement with experimentally determined values.
The primary goal of any drug design strategy is to predict the biological activity of new compounds prior to their synthesis. Comparative Molecular Field Analysis (CoMFA), introduced by Cramer in 1988, is one of the most robust modern tools for Quantitative Structure Activity Relationship (QSAR) studies [1]. The CoMFA methodology is a 3-D QSAR technique that ultimately allows one to design and predict activities of molecules. This is based on the assumption that the interactions between a receptor and its ligand or an enzyme and its substrate or inhibitor are primarily non-covalent in nature and shape-dependent [2]. Several successful CoMFA studies have been reported, including biphosphonic acid esters [3], somatostatin analogues [4], 2-hetero substituted statin derivatives [5], acyloxy methylketones [6], 36-aryl sulfonamides [7] and N-benzyl piperidines [8].
The biological activity data for the compounds under study, which were previously synthesised and screened for their short-term in vitro antitumor activity in DLA cells, were obtained from the Department of Pharmaceutical Chemistry, J. S. S. College of Pharmacy, Ootacamund.
Materials and Methods
Biological activity data
Short-term antitumor activity data for a series of 39 compounds were subjected for QSAR analysis by CoMFA technique. Anticancer activity of the title compounds was originally given in terms of cytotoxic concentration (CTC50) and concentration required to inhibit the growth of the DLA cells in μg/ml. The CTC50 data were used for QSAR analysis as a dependent parameter, after converting into reciprocal of the logarithm of CTC50 (pCTC50) expressed in μM/ml.
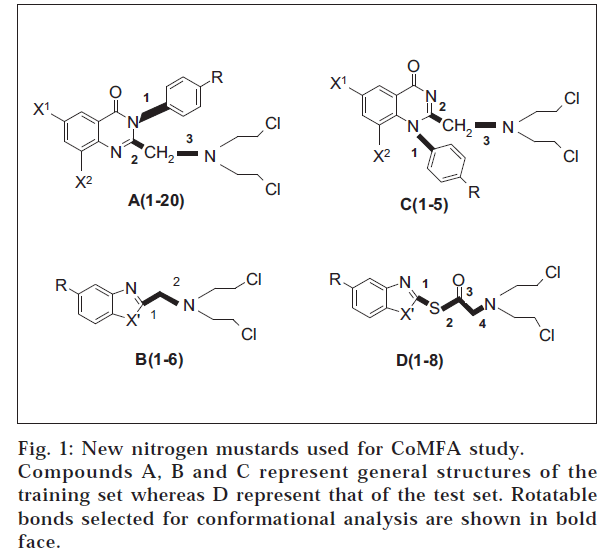
The application of statistical methods depends on a proper experimental design for the training set from which a QSAR model is derived, as well as for the test set, for which biological data shall be predicted [9,10]. Training set was formed by selecting 31 compounds from the series of compounds in hand (fig. 1). Approximately 20% from the series were taken as test set.
Molecular modelling
A database of 31 compounds forming the training set was generated by molecular modelling. Structures of the compounds were built using the sketcher tool provided in the modelling environment of SYBYL 6.7 [11] software package (Tripos, USA). The molecules were subjected for energy minimization (geometry optimization) at a gradient of 1.0 Kcal/M with delta energy change of 0.001 Kcal/M under Tripos standard force field [12]. Finally, the molecules were named and saved in appropriate databases.
After conformational analysis (CA), the least energy conformation was selected, saved, and used for the charge calculation, assuming that it was the active one. We have used two different types of charges – calculated by Gasteiger-Marsili method and the semi-empirical MOPAC method [13].
Partial least squares analyses (PLS)
The PLS analyses were done by following standard protocols [14]. In order to speed up the analysis and reduce the amount of noise, column filter was used excluding the columns with a variance smaller than 2.0. Equal weights for the steric and electrostatic descriptors were assigned using the CoMFA scaling option.
CoMFA results
Two CoMFA models were generated by using different types of partial atomic charges, which is shown in Table 1. Model A was derived by using charges calculated according to Gasteiger-Marsilli method, while Model B was obtained using MOPAC charges.
| Details | Model A | Model B |
| r2 cv | 0.200 | 0.281 |
| SEP | 0.284 | 0.251 |
| r2 conventional | 0.860 | 0.909 |
| Standard error | 0.096 | 0.079 |
| No. of compounds | 5 | 6 |
| F value | 26.976 | 35.055 |
| P value | 0.000 | 0.000 |
| Steric contribution | 0.481 | 0.525 |
| Electrostatic contribution | 0.519 | 0.475 |
| r2 BS | 0.910 ± 0.051 | 0.953 ± 0.033 |
| SD BS | 0.074 ± 0.049 | 0.048 ± 0.039 |
Table 1: Summary Of Comfa Results
Prediction of activity
The 3-D QSAR analysis obtained as Model B was used for predicting the activity of the 32 compounds in the training set. The results are shown in Table 2. From the table, it can be observed that the predicted activities are very close to the experimental activities with minimum residual activity.
| Compd. No.s | Substituents | pCTC50 | pCTC50 | Residual | |||
|---|---|---|---|---|---|---|---|
| X | X ’ | R | Experimental | Predicted | Activity | ||
| A1 | Br | Br | Cl | 0.742 | 0.692 | 0.048 | |
| A2 | Br | Br | NO2 | 0.824 | 0.860 | -0.040 | |
| A3 | Br | Br | CH3 | 0.461 | 0.461 | -0.001 | |
| A4 | Br | Br | OCH3 | 0.559 | 0.556 | -0.004 | |
| A5 | Br | Br | OC2 H5 | 0.513 | 0.524 | -0.015 | |
| A6 | Br | H | Cl | 0.516 | 0.521 | -0.005 | |
| A7 | Br | H | NO2 | 0.690 | 0.696 | -0.006 | |
| A8 | Br | H | CH3 | 0.400 | 0.459 | -0.059 | |
| A9 | Br | H | OCH3 | 0.454 | 0.409 | 0.041 | |
| A10 | Br | H | OC2 H5 | 0.411 | 0.387 | 0.023 | |
| A11 | H | H | Cl | 0.397 | 0.399 | 0.001 | |
| A12 | H | H | NO2 | 0.564 | 0.569 | -0.009 | |
| A13 | H | H | CH3 | 0.342 | 0.340 | -0.002 | |
| A14 | H | H | OCH3 | 0.338 | 0.293 | 0.047 | |
| A15 | H | H | OC2 H5 | 0.268 | 0.291 | -0.021 | |
| A16 | I | H | Cl | 0.409 | 0.516 | -0.106 | |
| A17 | I | H | NO2 | 0.432 | 0.471 | -0.041 | |
| A18 | I | H | CH3 | 0.678 | 0.535 | 0.0154 | |
| A19 | I | H | OCH3 | 0.406 | 0.510 | -0.010 | |
| A20 | I | H | OC2 H5 | 0.553 | 0.460 | 0.090 | |
| B1 | NH | - | H | 0.062 | 0.126 | -0.66 | |
| B2 | NH | - | NO2 | 0.478 | 0.467 | 0.013 | |
| B3 | NH | - | CH3 | 0.361 | 0.325 | 0.035 | |
| B4 | O | - | H | 0.009 | 0.785 | -0.776 | |
| B5 | O | - | NO2 | 0.505 | 0.809 | -0.304 | |
| B6 | O | - | CH3 | 0.154 | 0.673 | -0.519 | |
| C1 | H | H | Cl | 0.652 | 0.528 | 0.122 | |
| C2 | H | H | NO2 | 0.848 | 0.817 | 0.033 | |
| C3 | H | H | OCH3 | 1.131 | 1.320 | -0.011 | |
| C4 | H | H | Br | 0.295 | 0.497 | -0.197 | |
| C5 | H | H | H | 0.441 | 0.379 | 0.061 | |
Table 2: Experimental and Predicted Anticancer Activity Of Compounds In Training Set
CoMFA contour maps
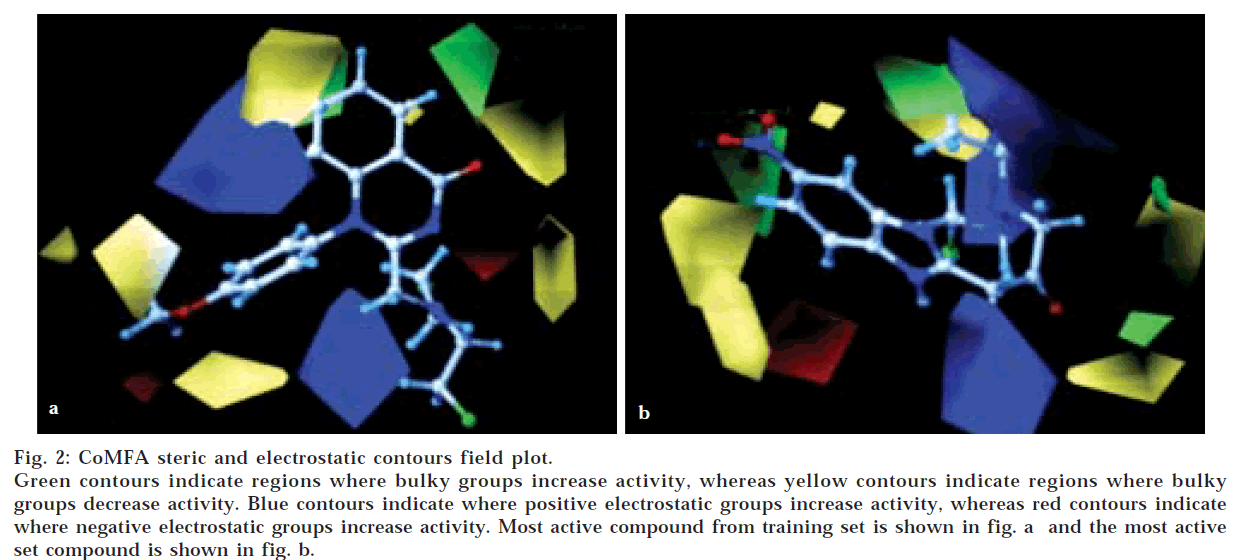
The QSAR produced by CoMFA were represented as a 3-D coefficient contour map. To visualize the CoMFA steric and electrostatic fields from PLS analysis, contour maps of the product of the standard deviation associated with CoMFA column and coefficient (SD Coeff) at each lattice point were generated. The contour maps were plotted as percentage contribution to the QSAR equation and were associated with difference in biological activity. The CoMFA contour maps generated for model B were used to explain the structure activity relationship of anticancer agents.
In CoMFA contour maps, the regions of high and low steric tolerance are shown in green and yellow polyhedra, respectively. CoMFA electrostatic fields are shown as blue and red polyhedra in fig. 2. A low electron density within the inhibitors near blue and red polyhedra, respectively increased or decreased the activity.
Results and Discussion
The validity of Model B was further enhanced by bootstrapping process. Bootstrapping of 10 runs gave r2 of 0.953±0.033 with a very low standard error of 0.048± 0.039. This added to the high confidence limit to this analysis. In this analysis, both steric and electrostatic fields contributed to the QSAR equation by 52.5 and 47.5%, respectively. This suggested that variation in the anticancer activity is predominantly determined by the steric properties. Thus, the results suggested that there is a good internal consistency in the data set generated in model B.
Model B performed exceptionally well in predicting the activity of most compounds used in the test set. However, it must be emphasized that the molecular alignment and conformations used in this study were selected in the absence of X-ray crystallographic coordinates of these molecules; still, the CoMFA model generated in the study showed very good prediction capability.
From the Table 3, it can be observed that the predictions made using CoMFA model were satisfactory in most cases. In general, the percentage difference in the predicted activities of the synthesised compounds ranges from 3.3 to 26.2%. The relative difference in the predictions is not unexpected and is within the From these results, it is inferred that the 3-D QSAR model generated can be successfully expanded to predict the activity of structurally diverse compounds, which will be used in designing new chemical entities and predicting their activity.
| Compd. No. | CTC50 µg/ml | CTC50 µM/ml | pCTC50 Experimental | pCTC50 Predicted | Residual | % Residual |
|---|---|---|---|---|---|---|
| D 1 | 190 | 0.571 | 0.243 | 0.285 | -0.042 | 14.7 |
| D 2 | 165 | 0.448 | 0.349 | 0.361 | -0.012 | 3.3 |
| D 3 | 160 | 0.461 | 0.336 | 0.373 | -0.037 | 9.9 |
| D 4 | 165 | 0.437 | 0.360 | 0.393 | -0.033 | 8.4 |
| D 5 | 200 | 0.599 | 0.223 | 0.292 | -0.069 | 23.6 |
| D 6 | 215 | 0.583 | 0.234 | 0.317 | -0.083 | 26.2 |
| D 7 | 180 | 0.517 | 0.287 | 0.317 | -0.030 | 9.5 |
| D 8 | 195 | 0.557 | 0.254 | 0.238 | 0.016 | 6.7 |
Table 3: Comparative Variation In The Experimental And Predicted Anticancer Activity Of Compounds In Test Set
Acknowledgements
The authors wish to thank His Holiness Jagadguru Sri Sri Shivarathri Deshikendra Mahaswamigalavaru of Suttur Mutt, Mysore, for providing the facilities to carry out the research work.
References
- Cramer, R.D.(III), Patterson, D.E. and Bunce, J.D., J. Amer.Chem. Soc., 1988, 11, 5959.
- Clark, M., Cramer, R.D.(III), Jones, D.M., Patterson, D.E. andSimeroth, P.E., Tetrahedron Comput. Method, 1990, 3, 47.
- Pohjala, E., Hanhijarvi, H., Lauren, L., Hannuniemi, R., Juhakosi, A.,Kippo, K. and Kleimola, T. J. Med. Chem., 1991, 34, 2338.
- Hocart, S.J., Vik Reddy, Murphy, W.A. and Coy, D.H. J. Med.Chem., 1995, 38, 1974.
- Peter Hecht, Kroemer, T. and Peter Ettmayer, J. Med. Chem., 1995, 38, 4917.
- Kulkarni, S.S. and Kulkarni, V. M. J. Med. Chem., 1999, 42, 373.
- Bantley, R.K. Hunt, T.T., Stein, P.D. and Stouch, T.R. J. Med.Chem., 1995, 38, 659.
- Tong, W., Collantes, E.R., Chen, Y. and Welsh, W.J., J. Med.Chem., 1996, 39, 380.
- Ramsden, C.A., Eds., In; Quantitative Drug Design, Vol. 4, Pergamon, Oxford, 1990, 20.
- Kubinyi, H., QSAR: Hancsh Analysis and Related Approaches, VCH, Weinheim, 1993.
- SYBYL molecular modeling software, version 6.7, Tripos Associates Inc., 1699 Hanley Rd, St. Louis, MO 63144-2913.
- Clark, M., Cramer, R.D.(III) and Van Opdenbosh, J. Comput.Chem., 1989, 10, 982.
- Gasteiger, J. and Marsili, M., Tetrahedron, 1980, 36, 32192.
- Geladi, P., J. Chemom., 1988, 2, 231.