- *Corresponding Author:
- J. D. Raja
Chemistry Research Centre, Mohamed Sathak Engineering College, Kilakarai-623 806, India
E-mail: jdrajapriya@gmail.com
| Date of Submission | 15 December 2016 |
| Date of Revision | 02 December 2017 |
| Date of Acceptance | 30 June 2018 |
| Indian J Pharm Sci 2018;80(4):727-738 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
A novel series of metal (II) complexes, [CuL(AcO)].H2O (1), [CoL(AcO)].4H2O (2), [MnL(AcO)].4H2O (3), [NiL(AcO)].4H2O (4) and [ZnL(AcO)].2H2O (5) have been synthesized from 2-(2-morpholinoethylimino)methyl)phenol Schiff base ligand in a 1:1 molar ratio. The resulting compounds were characterized using various physical, chemical and spectral studies. The spectral results indicated square planar geometry around the metal (II) ion and found to possess [ML] stoichiometry. Complexes (1), (2) and (4) exhibited potent antimicrobial, antioxidant and DNA cleavage activities as compared to complexes (3) and (5). In vitro DNA binding properties of complexes (1-5) have been carried out employing the electronic absorption technique. The calculated intrinsic binding constant (Kb) values for the complexes were in the order of 1.3512×105 (1) >2.9843×104 (2) >1.6432×104 (4) >4.2280×103 (5) >2.9100×103 M−1 (3) and the DNA binding affinity values of the complexes obtained by the viscosity titration measurements were in the order of (1)>(2)>(4)>(5)>(3). The observed results suggested that the complexes (1-5) bind to DNA via intercalation.
Keywords
Schiff base, square planar, morpholine, antimicrobial, antioxidant, DNA interactions
In coordination chemistry, Schiff bases are a paramount class of ligands that have widespread applications in various fields [1,2]. The interaction of these ligands with metal ions tends to the formation of complexes with different geometries, which are potentially biologically active [3]. The π system in a Schiff base often imposes a geometrical constriction and influences the electronic structure as well. Schiff base ligands and their metal complexes have been extensively studied over the past few decades and the relevant transition metal complexes are still found to be a great curiosity in inorganic chemistry due to various biological activities [4]. Das et al. studied the successful treatment of persistent bronchorrhea by gefitinib established remarkable cytotoxic potencies towards human HL- 60 leukemic cells [5]. Usually these derivatives inhibit the growth of tumor cells, but with a remarkably low effect on normal cells. Nowadays these complexes with a morpholine in the terminal base found use as antitumor agents. Morpholine as a substitution in many heterocyclic moieties has been reported to possess potential analgesic, antiinflammatory, antimicrobial, antimalarial and anticancer activities because of their specific structures [6,7]. Thermo chemical properties of Schiff bases have attracted the attention of many researchers in view of their ability to bind through NO or N2O2 donor atom sets [8]. These Schiff base metal derivatives involving bidentate or tridentate bonding of ligands in biological systems have considerable interest and also contributing to the knowledge of their structure and behaviour in various activities. Schiff bases offer a versatile and flexible series of ligands capable of binding with Cu(II), Co(II), Mn(II), Ni(II) and Zn(II) metal ions to give complexes with suitable properties for theoretical and practical applications. Since the publication of Schiff base complexes, a huge number of polydentate Schiff base complexes have been structurally characterized and extensively investigated due to their diverse chelating ability [9]. Copper(II)-salicylaldimine complexes play important roles in both synthetic and structural research because of their preparative accessibility and structural diversity [10]. The metal Schiff base complexes can serve as efficient models for the metal containing sites in metallo-proteins and enzymes due to the varied magnetic and catalytic properties and also they have been studied for their dioxygen up take and oxidative catalysis because of they are an excellent catalyst for the insertion of oxygen into an organic substrate [11]. Magnetic properties of Cu(II) and Ni(II) complexes bridged by the azo-2,2’-bispyridine ligand has been considered with special attention due to the ability of this ligand to mediate exchange interactions between paramagnetic metal centers [12]. The present study aimed to investigate the reaction of tridentate Schiff base ligand (HL) derived from the condensation of 4-(2-aminoethyl)-morpholine with salicylaldehyde. The complexes (1-5) were synthesized by the ligand (HL) coordinating to Cu(II), Co(II), Mn(II), Ni(II) and Zn(II) ions. The complexes (1-5) were more stable with good solubility in common solvents like water and methanol. These complexes (1-5) have also exhibited potent biological activities, which might contribute as drug in future.
Materials and Methods
The starting materials 4-(2-aminoethyl)-morpholine, salicylaldehyde, CuII(OAc)2.H2O, CoII(OAc)2.4H2O, MnII(OAc)2.4H2O, NiII(OAc)2.4H2O, ZnII(OAc)2.2H2O salts, Tris (hydroxyl methyl)aminomethane-HCl (Tris- HCl) and 2,2-diphenyl-1-(2,4,6-trinirophenyl)hydrazyl (DPPH) were procured from Sigma-Aldrich Chemical Repository (USA). Calf thymus DNA (CT DNA) was purchased from Bangalore Genei, Bangalore, India. Other reagents and solvents used were of analytical grade. Schiff base ligand (HL) and its complexes (1-5) were characterized by proton nuclear magnetic resonance (1H NMR; Bruker 300 MHz NMR spectrometer), Fourier-transform infrared spectroscopy (FTIR; Shimadzu FTIR spectrophotometer using KBr pellets technique in the range of 4000 to 400 cm-1), electronic absorption (Shimadzu UV/ Vis 1800 spectrophotometer in the range of 200 to 1100 nm), magnetic susceptibility (Guoy balance at room temperature), electron paramagnetic resonance (EPR, Varian E112 EPR spectrometer), mass (ESI-MS 3000 Bruker Daltonics instrument), elemental analysis (Elementar Vario EL III CHNS), molar conductivity (Systronics model 304 digital conductivity bridge using a dip type conductivity cell fitted with a platinum electrode), cyclic voltammogram (100 mVs-1 in the presence of 0.1 M n-Bu4NClO4 as supporting electrolyte in methanol) and melting points by Guna- BMQR Cintex apparatus in open glass capillaries.
Synthesis of complexes (1-5)
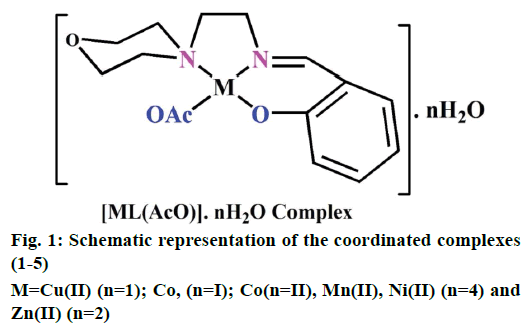
The preparation of 2-(2-morpholinoethylimino) methyl)phenol Schiff base ligand (HL) was previously reported [13]. A solution of the ligand HL (0.002 M) in methanol (40 ml) was added drop by drop to a solution of metal (II) acetates (0.002 M) in 30 ml of absolute methanol and stirred for 30 min. The resulting mixture was refluxed for 3 h. The formed solid product was separated out by filtration and purified by recrystallization from methanol and petroleum ether. Traces of water and solvents present were recovered using a vacuum desiccator consisting of anhydrous CaCl2. Similarly, complexes (1-5) have been successfully synthesized and the yield was found to be 77-81 % (Figure 1).
In vitro antimicrobial assay
Antimicrobial activities of the ligand (HL) and its complexes (1-5) were screened against the selected pathogenic bacteria, Escherichia coli, Salmonella typhi, Chromobacterim violaceum, Staphylococcus aureus, Bacillus cereus and fungi, Aspergillus flavus, Aspergillus niger and Candida albicans using the disc diffusion method. Amikacin and ketoconazole were used as standard drugs for antibacterial and antifungal studies, respectively. The test organisms were grown on nutrient agar medium in sterile Petri plates. The paper discs were saturated with 10 μl of the ligand (HL) and its complexes (1-5) solution. The saturated paper discs were placed aseptically in the Petri dishes containing Mueller-Hinton nutrient agar with 2 % of glucose media inoculated with the pathogenic bacteria and fungi separately. The inoculated culture plates were incubated at 37° for 24 h for the bacteria and at 30° for 48 h for the fungi. After incubation, the antimicrobial activity was evaluated by measuring the diameter (mm) of the inhibition zone formed around the discs.
DPPH radical scavenging studies
In vitro antioxidant activities were determined by DPPH free radical scavenging assay. The absorbance of control (DPPH) was measured at 517 nm without using antioxidant and special care was taken to minimize the loss of free radical activity of the DPPH stock solution [13]. The reducing abilities of the complexes (1-5) were determined by their interaction with free s PPH radical at different concentrations. The reduction of DPPH• radical form as indicated below is followed by monitoring the decrease in its absorbance at 517 nm but reduction by an antioxidant (AH) or a radical species (R•), the absorbance disappears (DPPH•+AH→DPPH-H+A• (or) DPPH•+R•→DPPH-R).
DNA interaction studies
The CT DNA cleavage studies of ligand (HL) and its complexes (1-5) were monitored by agarose gel electrophoresis method. The gel electrophoresis experiment was performed under aerobic conditions in a medium of 5 mM Tris-HCl/ 50 mM NaCl buffer solution at pH 7.2 in the presence of H2O2 and the incubation was maintained at 35° for 2 h [14]. During the experiments bromophenol blue is used as a photosensitizer, which is helpful to visibly analyze the migrated band lanes for the degree of DNA cleavage by comparing with standard CT DNA marker.
Electronic absorption spectral titration
The CT DNA stock solution was prepared by the buffer solution of 5 mM Tris-HCl/50 mM NaCl in deionized water at pH 7.2. The purity of CT DNA was ensured by monitoring the ratio of absorbance at 260 nm to that at 280 nm (A260/A280) the value was in the range of 1.8- 1.9 indicating that the DNA is sufficiently free from protein and the initial concentration of the CT DNA was confirmed from its absorption intensity at 260 nm with a molar extinction coefficient (ε) of 6600 M-1 cm-1 [15]. The electronic absorption titrations were performed by keeping a fixed complex concentration (10-5 M). In which 50 μl increments of the CT DNA stock solutions are continuous followed up to reaching the fully binding. The concentration of the solution was carefully noted from initial (1×10-5 M) to final state (12×10-5 M). The intrinsic binding constants (Kb) of metal (II) complexes (1-5) were calculated from the Wolfe-Shimmer Eqn. 1, [DNA]/(ɛa–ɛf) = [DNA]/(ɛb–ɛf))+1/(Kb(ɛb–ɛf)), where, [DNA] is the concentration of CT DNA in base pairs. The apparent extinction coefficient (ɛa) observed for the metal-to-ligand charge-transfer absorption band at the given CT DNA concentration was obtained by calculating Abs/ [complex], ɛf and ɛb correspond to the extinction coefficient of the complex free (unbound) and fully bound to DNA. The binding constant (Kb) values can be obtained from the ratio of the slope to the intercept of the plots of [DNA]/(ɛa–ɛf) versus [DNA]. The standard Gibb’s free energy change (ΔGb°) for DNA binding can be calculated by van ’t Hoff Eqn. 2, ΔGb° = RT ln Kb.
Viscosity titration measurements
These experiments were carried out using an Oswald viscometer, immersed in a thermostatic water bath maintained at 25°±0.1°, which helped to further clarify the nature of the interaction between the complexes and CT DNA. The results observed as the least ambiguous and the most critical test of binding mode in solution [16]. The titrations were performed for complexes (1-5) and the control ethidium bromide (0.2–1.0×10-5 M). Each compound was introduced into the CT DNA solution and the average flow time was observed for each resulting mixture during the viscosity titration [17]. The viscosity of CT DNA increased with rising ratio of complexes (1-5) to DNA, further suggesting a binding ability of the complexes (1-5) with CT DNA. The specific viscosity values of CT DNA (η) either in the presence or absence of complexes were calculated from the Eqn. 3. (η/η0)1/3 = {(tcomplex–t0)/t0}/ {(tDNA–t0)/t0}.
Data were analyzed as (η/ηo)1/3 versus binding ratio R= [complex]/ [DNA], where η is the specific viscosity of DNA in the presence of the complex and η0 is the specific viscosity of DNA alone; tcomplex, tDNA and t0 are the average flow time for the DNA in the presence of the complex, DNA alone and Tris-HCl buffer, respectively [18].
Results and Discussion
The synthesized ligand (HL) and its complexes (1-5) were found to be intensely colored and they were slightly hygroscopic nature at room temperature and they are easily soluble in water, methanol, ethanol, CHCl3 and DMSO. The analytical data and physical properties of the ligand (HL) and its complexes (1-5) were listed in Table 1.
| Compounds (formula weight and empirical formula) |
Colour | Yield (%) |
MP (°) |
Found (Calcd.) (%) | Molar conductance (Ohm-1cm2 mol-1; ∧m) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | H | N | M | ||||||||||||
| Ligand (HL) (234.31) (C13H18N2O2) |
Yellow liquid |
87.48 | -- | 66.90 (66.64) |
07.60 (07.74) |
11.79 (11.95) |
---- | ---- | |||||||
| [CuL(AcO)].H2O (1) (373.54) (C15H22N2O5)Cu |
Green | 81.62 | 108 | 48.46 (48.18) |
05.96 (05.88) |
07.61 (07.49) |
17.12 (17.01) |
38.48 | |||||||
| [CoL(AcO)].4H2O (2) (422.93) (C15H28N2O8)Co |
Dark-brown | 78.23 | 275 | 42.68 (42.56) |
06.70 (06.62) |
06.77 (06.62) |
13.98 (13.93) |
37.54 | |||||||
| [MnL(AcO)].4H2O (3) (418.94) (C15H28N2O8)Mn |
Black | 77.54 | 112 | 43.03 (42.96) |
06.76 (06.68) |
06.83 (06.68) |
13.35 (13.11) |
35.77 | |||||||
| [NiL(AcO)].4H2O (4) (422.70) (C15H28N2O8)Ni |
Dark-green | 78.92 | 310 | 42.83 (42.58) |
06.73 (06.62) |
06.78 (06.62) |
13.96 (13.88) |
36.23 | |||||||
| [ZnL(AcO)].2H2O (5) (393.37) (C15H24N2O6)Zn |
Yellow | 77.65 | 140 | 45.91 (45.75) |
06.28 (06.10) |
07.28 (07.11) |
16.76 (16.61) |
35.22 | |||||||
Table 1: Analytical and physical data of the Schiff base ligand (HL) and complexes (1-5)
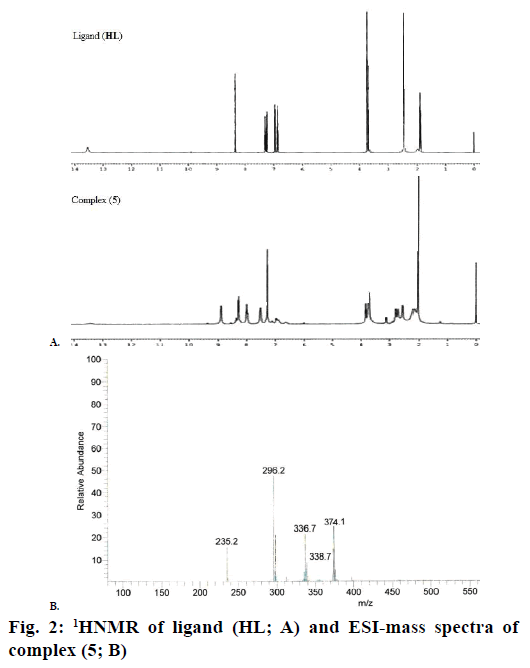
The 1H NMR spectra of the Schiff base ligand (HL) and complex (5) showed the following signals (Figure 2A): δ values of ligand (HL): aromatic protons (m, 4H) at 6.84-7.32 ppm; azomethine (-HC=N-) proton (s, 1H) at 8.35 ppm; morpholinic-OCH2 protons (t, 4H) at 3.72 ppm; morpholinic-N-CH2 (t, 4H) at 2.44 ppm; phenolic-OH proton (s, 1H) at 13.52 ppm [19]. Complex (5): aromatic protons (m, 4H) at 6.85- 7.95 ppm; azomethine proton (-HC=N-; s, 1H) at 8.88 ppm; morpholinic-OCH2 (t, 4H) at 3.72 ppm; morpholinic-N-CH2 (t, 4H) at 2.78 ppm; acetate protons (CH3COO-; s, 3H) at 2.038 ppm; The low intensity singlet peak at nearly 6.0 ppm attributable water protons [20] (s, 1H). The absence of singlet peak at the rage of 13 ppm is noted in the complex (5) indicates the loss of the –OH proton due to complexation [21] and there is no appreciable change in other signals in the complex (5).
The IR spectra of the complexes (1-5) were compared with the free ligand (HL) for the frequency changes during the complexation and their results are summarized in Table 2. IR spectrum of ligand (HL) exhibited a strong sharp band at 1635 cm-1 region is assigned to the -HC=N-mode of the azomethine group, which was shifted to lower frequencies in complexes (1-5), which indicates that imino nitrogen (-HC=N-) of the ligand (HL) is involved in coordination with the central metal ion [22]. Morpholinic-C-N-C bands found at 1342 cm-1 in the ligand (HL). The bands also shifted to lower frequencies in complexes (1-5) indicating the involvement of C-N-C nitrogen in coordination to the metal ion. The peak of ligand [HL] belonging to -OH group was identified at 3676 cm-1, the peak disappeared in all spectra of the complexes (1-5), which indicated deprotonation of -OH group upon complexation [23] and also the peak due to the presence of phenolic C-O at 1278 cm-1 in ligand (HL) was shifted to higher frequencies in complexes (1-5) indicating confirmed deprotonation of the phenolic–OH on chelation [24]. In the spectra of complexes (1-5) a broad band was identified (stretching) in the range of 3500-3400 cm-1, followed by another week band in-plane bending (rocking) at 831-845 cm-1 that suggests the presence of water molecules in the metal complexes (1-5) [25]. In the complexes (1-5), carboxylate of the acetate group absorbs strongly in the range of 1664-1670 cm-1 (γasymmetry) and more weakly at 1398-1402 cm-1 (γsymmetry). It suggested that they were involved in unidentate coordination with the metal ion because of the difference values between asymmetry and symmetry stretching frequencies were greater than 200 cm-1 [26]. The far IR spectra of the complexes (1-5) showed medium bands in the region 462-466 and 494-520 cm-1 corresponding to M-N and M-O vibrations, respectively and no appreciable change was observed in the other absorption bands of the ligand (HL) and complexes (1-5), Table 2.
| Compounds (cm-1) |
-HC=N | Phen- (C-O) |
Morp- (C-N-C) |
Morp- (C-O-C) |
Ar- (C-H) |
Ali- (C-H) |
Acetate (CH3COO) |
Ph-(OH) | H2O | M-N | M-O |
|---|---|---|---|---|---|---|---|---|---|---|---|
| HL | 1635 | 1278 | 1342 | 1114 (s) | 3056 | 2974 | --- | 3676 | --- | --- | --- |
| 1197 (as) | |||||||||||
| -1 | 1595 | 1284 | 1322 | 1112 (s) | 3063 | 2970 | 1398 (s) | --- | 831(b) | 466 | 494 |
| 1193 (as) | 1670 (as) | ||||||||||
| -2 | 1595 | 1284 | 1330 | 1112 (s) | 3068 | 2972 | 1398 (s) | --- | 831(b) | 464 | 506 |
| 1191 (as) | 1670 (as) | ||||||||||
| -3 | 1600 | 1288 | 1333 | 1112 (s) | 3065 | 2970 | 1400 (s) | --- | 811(b) | 466 | 514 |
| 1195 (as) | 1666 (as) | ||||||||||
| -4 | 1593 | 1309 | 1336 | 1114 (s) | 3065 | 2970 | 1402 (s) | --- | 844(b) | 464 | 510 |
| 1195 (as) | 1668 (as) | ||||||||||
| -5 | 1627 | 1286 | 1332 | 1112 (s) | 3052 | 2972 | 1402 (s) | --- | 842(b) | 462 | 518 |
| 1193 (as) | 1662 (as) |
s: Symmetry, as: asymmetry, b: in-plane bending (rocking), Phen-C-O: phenolic C-O, Morp-C-N-C: morpholinic C-N-C, Morp-C-O-C: morpholinic C-O-C, Ar-C-H: aromatic C-H, Ali-C-H: aliphatic C-H, Ph-OH: phenolic OH
Table 2: Infrared spectral data of the ligand (HL) and complexes (1-5)
The electronic absorption spectral data of the ligand (HL) and its complexes (1-5) were recorded in methanol. The absorption maxima and magnetic moment values are depicted in Table 3. The electronic spectra of the free ligand (HL) displayed two bands at 258 nm (38 760 cm-1) and 319 nm (31 348 cm-1) are intra ligand charge transfer, which indicates π→π* and n→π* transitions for phenyl ring and the azomethine chromophore, respectively [27]. The bands in the complexes (1-5) were shifted to a higher wavelength, which might be attributed to the donation of a lone pair electron in a sp2-hybridized orbital of the imino nitrogen atom of the ligand (HL) to the metal.Complex (1) in methanol revealed a broad band at 526 nm (19 011 cm-1) assignable to 2B1g→ 2A1g transition and the observed magnetic susceptibility value (μeff) of the complex (1) is slightly higher (1.86 Bohr magnetons, BM) than the spin-only value (1.73 BM) for one unpaired electron [28]. Complex (2) showed d-d bands at 572 nm (17 482 cm-1) and 322 nm (31 055 cm-1) 1A1g→1B1g and n→π*, respectively. The observed μeff value of the complex (2) is 3.12 BM, which indicated the square planar geometry for the Co(II) ion [29]. The absorption spectrum of the complex (4) showed two d-d bands at 566 nm (17 667 cm-1) and 438 nm (22 831 cm-1), which were assigned as 1A1g (D)→1A2g (G) and 1A1g (D)→1B2g (G) transitions. The magnetic susceptibility of the complex (4) indicated that μeff=0.00 BM, which was due to diamagnetic behaviour [30]. Complexes (3) and (5) presented no absorption in the visible domain, which was in agreement with the electronic structure of the central metallic ion with d5 and d10 electronic configurations, respectively. The crystal field theory did not predict the d-d transitions of these complexes (3) and (5) due to the absence of absorption bands in the visible region. The two absorption bands at 292 nm (34 246 cm-1) and 392 nm (25 510 cm-1) for the complex (3) and two absorption bands at 274 nm (36 496 cm-1) and 364 nm (27 472 cm-1) for the complex (5) were found in the UV region. These were considered to be characteristic of the ligand. In the metal complexes (3) and (5), the first bands responsible for the π→π* transitions and the second bands due to presence of ligand to metal charge transfer transitions were shifted to higher wavelength, which could be attributed to the binding of these coordination centers to the central metal ions [31]. The obtained all spectral data results suggested a square planar environment around the metal (II) ion.
| Compounds | Band position λmax nm (γ-cm-1) |
Assignment | μeff (B.M) |
Suggested geometry |
|---|---|---|---|---|
| HL | 319 (31 348) 258 (38 760) |
n→π* π→π* |
-- | -- |
| (1) | 526 (19 011) 372 (26 881) 271 (36 900) |
2B1g→2A1g INCT INCT |
1.86 | Square planar |
| (2) | 572 (17 482) 322 (31 055) |
1A1g→1B1g n→π* |
3.12 | Square planar |
| (4) | 566 (17 667) 438 (22 831) |
1A1g (D)→1A2g (G) 1A1g (D)→1B2g (G) |
-- | Square planar |
Table 3: Electronic spectral data and magnetic susceptibility values of (HL) and complexes (1-5).
The X-band EPR spectra of the complex (1) in powder state was recorded at room temperature and liquid nitrogen temperature under 9.10 GHz microwave field modulation using tetracyanoethylene (ge=2.00277). The complex (1) at 300 K is exhibited only one intense absorption band in the high field region, which is isotropic because of the tumbling motion of the molecules and the complex (1) at 77 K is also exhibited anisotropic pattern with well-resolved hyperfine lines. The spin-Hamiltonian parameters have been calculated by Kivelson’s method [32]. In square planar complexes, the unpaired electron lies in the dx 2 –y 2 orbitals giving 2B1g as the ground state, which leads to g||>g⊥>ge and the unpaired electron lies in the dz2 orbital giving 2A1g as the ground state, which leads to g⊥>g||>ge . The observed g-values were in the order g|| (2.22)>g⊥ (2.04)>ge (2.00277), which indicated that g|| values for the complex were less than 2.3 in agreement with the covalent environment character of the M-L bond [33] and the covalent nature of the M-L bond in the complex (1) was further supported by the geff = [(g|| + g⊥)/3] value less than 2.00277 [34]. The observed value of geff (1.42) was less than ge (2.00277) and hyperfine constant parameters were in the order A|| (153 G)>Aav (86 G)>A⊥(52 G). It was very clear that the EPR parameters of the complex (1) coincided well with the related system, which suggested that the complex had square planar geometry and the system was axially symmetrical [35]. The values were obtained from the following Eqn. 4-8, g⊥ = (3gav–g||)/2; K⊥ = (3Aav–A||)/2; gav = (g||–2g⊥)/3; Aav = (A||–2A⊥)/3; G = (g||–2.00277)/ (g⊥–2.00277).
The observed value of G (5.834) was greater than 4 in the complex (1) suggesting that there was no interaction between Cu-Cu centers in the solid state complex (1) and the absence of half field signal at 1600 G corresponding to the ΔMs=±2 transition rules out a Cu-Cu interaction [36,37]. The values of molecular orbital coefficient parameters α2, β2 and γ2 were calculated by Kivelson and Neimann Eqn. 9 and 10, α2 = A||/P+(g||– 2.00277)+3/7(g⊥–2.00277)+0.04; β2 = (g||–2.00277) (Ed–d/–8λ0a2) and γ2 = (g⊥–2.00277)(Ed–d/–2λ0a2).
In-plane σ-bonding parameter α2=1.0 indicated the pure ionic character whereas, α2=0.5 indicated pure covalent bonding. The observed value of α2= was 0.658, which indicated that the complex has covalent character and also the observed β2= (in-plane π-bonding) and γ2 (out-plane π-bonding) values were less than 1.0, which indicated π-bonding was completely of covalent character. Hathaway [38] has pointed out that for the pure σ-bonding in case of orbital reduction factor K|| and K⊥ are equal. In case of K|| value is less than K⊥, which represents in-plane π-bonding and if the K|| value is greater than K⊥, which leads to out-of-plane π-bonding. The observed value of K║ (0.623) was greater than K⊥ (0.427) for the complex (1), which indicated the presence of out-plane π-bonding in metal ligand π-bonding. The values were calculated from the Eqn. 11-14, K||2 = (g||–2.00277)(Ed–d/–8λ0); K⊥ 2 = (g⊥–2.00277)(Ed–d/–2λ0); Kfermi = Aav/Pβ2+(gav–2.00277)/ β2=; P = 2 ϒCu β0BN (γ–3), where, the calculated value of free ion dipolar term (P)=0.036 cm-1, ϒCu= magnetic moment value for copper, β0= Bohr magneton, BN= nuclear magneton, γ= the distance from the central nucleus to the electron. The Co-factors of degree of geometrical distortion f||=145.098 cm-1 indicating square planar geometry around the Cu(II) ion. Fermi contact hyperfine interaction term (Kfermi=0.356) is a dimensionless quantity, which is measure of the polarization produced by the uneven distribution of d-electron density on the inner core s-electron was expressed by dipolar term (P), which is calculated from the Eqn. 14.
Electrospray ionisation mass spectrometry (ESI/MS) is used to confirm the stoichiometry composition of compounds. The mass spectrum of ligand (HL) shows the molecular ion peak at m/z 235 corresponding to C13H18N2O2 and the complex (1) molecular ion peak at m/z 374 (Figure 2B) corresponding to (C15H22N2O5) Cu, which confirms the formation of [ML] stoichiometry. The molecular ion peaks of other complexes were observed at m/z with relative abundance 423 (2), 419 (3), 422 (4) and 393 (5) and they are in good agreement with the formula weight (Table 1). The composition and purity of the coordinative compounds were determined by the elemental analysis (C, H, N and metal contents) and the result presented in Table 1 indicated good agreement with the proposed structure shown in Figure 1.
The molar conductivity values of complexes (1-5) were in the range of 35.22-38.48 ohm-1 cm2/mol (Table 1). The low values indicated that these were non-electrolytic in nature due to lack of dissociation and ligand (HL) was also non-electrolytic nature due to very least molar conductivity value as compared to complexes (1-5). The electrochemical techniques are the most effective and important methods available for the mechanistic study of redox systems of the complexes.
The results were summarized in Table 4. The cyclic voltammogram for complexes (1-5) revealed that one electron reduction peak (Epc) corresponding to M(II) to M(I) and oxidation peak (Epa) corresponding to M(I) to M(II) [39]. The observed value of peak-to-peak separation (ΔEP) for complexes (1-5) were in the range of 262-829 mV, which was larger than the Nernstian value (EPa–EPc) (or) ΔEP=2.303RT/nF = 59/n mV, for the M(II)/M(I) redox couple revealed that this process was the best quasi-reversible. If the peak current ratio was greater or less than unity (ipa/ipc> or <1), which represented quasi-reversible one electron transfer process [40]. The observed values of peak current ratio of complexes (1-5) were less than unity. In quasi-reversible the peak current (ip) is proportional to the root of the scan rate (√ν) and separation in peak potential (ΔEP) [41]. In the case of reversible process, since the peak current was proportional to the scan rate is independent to scan rate and concentration. In reversible process current is controlled purely by mass transport only. But, in quasi reversible process current is controlled by mass transport as well as charge transfer kinetics. These observed systems are truly intermediate state between reversible and irreversible process and also they exhibits a large peak-to-peak potential separation. The observed values of formal electrode potential (E°) were in the ranges –0.4537 to –0.7836 V, which indicate that all couple corresponds to one electron transfer process. Antimicrobial assays revealed that most of the complexes (1-5) exhibited good antimicrobial activity than the free ligand [42]. The observed higher activity of the complexes (1-5) could be explained on the basis of Overtone’s concept and Tweedy’s chelation theory [43,44]. The polarity of the metal ion in a chelated complex is reduced to a greater extent due to the overlap of the ligand orbital and partial sharing of the positive charge of the metal ion. It increases the delocalization of π and d-electrons over the whole chelated ring and enhances the lipophilicity of the metal complexes. The increased lipophilicity of complexes enhances the cell permeability into lipid membranes, which leads to breakdown of the barrier of the cell and thus retards the normal cell processes. Generally, it is suggested that the chelated complexes deactivate various cellular enzymes vital to various metabolic pathways of these microorganisms and also solubility, conductivity and dipole moment factors affected by the presence of metal ions, which might also be possible reasons for increasing the biological activity of the complexes as compared to the ligand [45]. The observed results were recorded in the form of inhibition zone (mm). Although the complexes (1-5) exhibited better antimicrobial activities than the free ligand (HL), the complexes (1), (2) and (4) showed significant antimicrobial activity as compared to complexes (3) and (5), Table 5. The observed zone of inhibition diameter values for the complexes (1), (2) and (4) were in the range 11-16 mm. However, these were less active than the standard drugs amikacin and ketoconazole. In addition, the activities of the complexes (1-5) were also confirmed by calculating the activity index according to the following Eqn. 15, activity index (A) % = inhibition zone of compounds (mm)/inhibition zone of standard drug (mm)×100.
| Complexes | Couple | Epa (V) |
Epc (V) |
Δ EP (mV) | E° (V) |
ipa 10-6 (µA) |
ipc 10-6 (µA) |
ipa/ipc (µA) |
|---|---|---|---|---|---|---|---|---|
| (1) | Cu(II)/Cu(I) | –0.6103 | –0.9569 | 346 | –0.7836 | –0.9443 | 1.3764 | –0.6861 |
| (2) | Co(II)/Co(I) | –0.3308 | –1.1606 | 829 | –0.7457 | –1.2702 | 3.0056 | –0.4225 |
| (3) | Mn(II)/Mn(I) | –0.5427 | –0.8048 | 262 | –0.6737 | –2.1896 | 4.2920 | –0.5101 |
| (4) | Ni(II)/Ni(I) | –0.0670 | –0.8469 | 779 | –0.4569 | –2.6539 | 3.4382 | –0.7718 |
| (5) | Zn(II)/Zn(I) | –0.2548 | –0.6527 | 397 | –0.4537 | –1.1477 | 2.2275 | –0.5152 |
Formal electrode potential E° (or) E1/2 = ½ (EPa+EPc), scan rate: 100 mV s-1
Table 4: Electrochemical data of the synthesized complexes (1-5)
| Compounds | Antibacterial activity (mm, %) | Antifungal activity (mm, %) | ||||||
|---|---|---|---|---|---|---|---|---|
| E. coli | S. typhi | C. bacterim |
S. aureus |
B. cereus | A. flavus |
A. niger | C. albicans | |
| HL | 10 (62) | 09 (47) | 07 (39) | 09 (50) | 10 (59) | 08 (47) | 09 (60) | 11 (69) |
| (1) | 16 (100) | 15 (79) | 14 (78) | 16 (89) | 15 (88) | 13 (76) | 14 (93) | 15 (94) |
| (2) | 13 (81) | 13 (68) | 12 (66) | 14 (78) | 14 (82) | 12 (71) | 13 (87) | 13 (81) |
| (3) | 12 (75) | 12 (63) | 11 (61) | 13 (72) | 11 (65) | 10 (59) | 12 (80) | 12 (75) |
| (4) | 14 (88) | 13 (68) | 11 (61) | 12 (66) | 12 (71) | 11 (65) | 12 (80) | 14 (88) |
| (5) | 13 (81) | 14 (74) | 12 (66) | 15 (83) | 13 (76) | 13 (76) | 12 (80) | 13 (81) |
| Amikacin | 16 (100) | 19 (100) | 18 (100) | 18 (100) | 17 (100) | --- | --- | --- |
| Ketoconazole | --- | --- | --- | --- | --- | 17 (100) | 15 (100) | 16 (100) |
Table 5: Antimicrobial activities and activity index of HL and complexes (1-5)
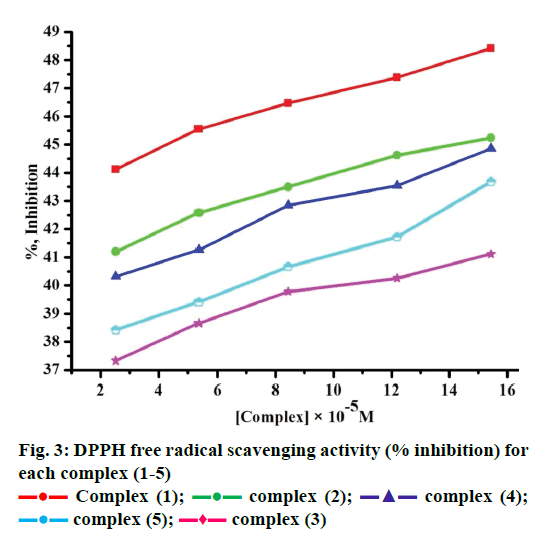
Free radicals produced by several redox reactions in the human body. Many natural antioxidants in plants play an important role in defence against free radicals [46-48]. Antioxidants are chemical substances that donate an electron to the free radical and convert into a harmless molecule. They may decrease the energy of the free radical formation or break chain propagation or repair damage. The DPPH assay provides a simple and rapid approach to evaluate antioxidants by electronic spectrophotometer and it can be useful to assess various products at a time. Free radical scavenging activity test is based on the ability of DPPH. The DPPH contains an odd electron, which is stable at room temperature and responsible for 515-517 nm and also for a visible deep purple colour. DPPH accepts an electron donated by an antioxidant compounds and reduced, giving rise to colourless in alcoholic medium [49]. They can be quantitatively measured from the changes in absorbance. The lower absorbance of the reaction mixture exhibited higher free radical activity. The percent of antioxidant activity of each substance was assessed by DPPH free radical assay. The annihilation activity of free radicals was determined by the following Eqn. 16. ADPPH is absorbance of DPPH (control) and AS is absorbance of sample. The observed radical scavenging activities were found as in the order (1)>(2)>(4)>(5)>(3), Figure 3. Eqn. 16, % RSA = ADPPH–AS/ADPPH×100.
The DNA cleavage efficiency of the complexes (1-5) was observed due to the different binding affinity of complexes (1-5) respect to rate of the conversion of open circular into linear [50]. In oxidative DNA cleavage mechanism, metal ions of the complexes (1-5) reacted with H2O2 to generate the hydroxyl radical, which attacked the C4 position of the sugar moiety, cleaved the DNA and inhibited the replication ability of the cancer gene. Lane 1 control (DNA+H2O2), which did not exhibit significant cleavage even on longer exposure time and lane 2, ligand (HL) alone was inactive in the presence and absence of external agents. Lane 3 complex (1), lane 4 complex (2), lane 5 complex (4), lane 6 complex (3) and lane 7 complex (5). During the end of experiment CT DNA was completely degraded resulting in the disappearance of bands on agarose gel, which indicated higher efficiency. The observed DNA cleavage efficiency for the complexes (1), (2) and (4) was higher than (3) and (5) in the presence of hydrogen peroxide. This was attributable to the formation of diffusible hydroxyl free radicals, which may damage the DNA.
Electronic absorption spectroscopy is one of the important techniques to study the binding mode of DNA with metal complexes. In this study, the binding constant (Kb) and standard Gibb’s free energy change (ΔGb°) values for complexes (1-5) have been evaluated by absorption titration (Table 6). The DNA binding abilities were characterized by monitoring the changes in the absorbance of π–π* bands and shift in wavelength on each addition of DNA solution to the complex. After intercalating the base pairs of DNA, the π* orbital of the intercalated ligand could couple with π orbital of base pairs, thus decreasing the π–π* transition energy. On other hand, the coupling π orbital with partially filled electrons thus decreases the transition probabilities, which led to hypochromism and further resulted in the bathochromism (red shift). The hypochromism results indicated that the contraction of DNA helix axes as well as the conformational changes on molecule of DNA. Since hypochromism due to π–π* strong stacking interactions between the chromophore and the base pairs of DNA might appear in the case of the intercalated binding mode when the DNA duplex is stabilized [51]. It was observed that complexes (1-5) have a shoulder peak at 366 nm (1), 365 nm (2), 392 nm (3), 371 nm (4) and 364 nm (5). While the concentration of CT DNA increases from 1×10-5 to 12×10-5 M, all peaks decreased gradually and wavelengths also slightly increased with range of 3-8 nm (Figure 4). Therefore, based on this view point, the observed results of hypochromism effect with a red shift revealed that the interaction between complexes (1-5) and CT DNA could be non-covalent intercalative binding. The binding constant (Kb) values are determined from the ratio of the slope to the intercept of the plot of [DNA]/(ɛa-ɛf)×10-7 verses [DNA]×10-5 M by Wolfe-Shimmer Eqn. 1 and ΔGb ° values for the complexes (1-5) were calculated by van’t Hoff Eqn. 2, which indicate that the complexes can interact with DNA in a spontaneous manner and the percent of hypochromicity for the complexes (1-5) were also determined from the Eqn. 17, % H = (ɛb–ɛf)/ ɛf×100.
Figure 4: Absorption spectra of complexes (1-5) in the presence of increasing amount of CT DNA
Absorption spectra of complexes (1-5) in buffer pH=7.2 at 25° in the presence of increasing amount of CT DNA. Arrow indicates
the changes in absorbance upon increasing the CT DNA concentration. Inset: plot of [DNA]/(ɛa-ɛf) vs [DNA] for the absorption
titrations of CT DNA with complexes (1-5)
| Complexes | λmax free (nm) |
λmax bound (nm) |
Δ λ (nm) |
Chromism (%) |
Binding constant Kb (M-1) |
ΔGb° (k j mol-1) |
|---|---|---|---|---|---|---|
| (1) | 366 | 374 | 08 | 42.16 | 1.3512×105 | –29.2751 |
| (2) | 365 | 371 | 06 | 36.45 | 2.9843×104 | –25.5327 |
| (3) | 392 | 396 | 04 | 28.06 | 2.9100×103 | –19.7644 |
| (4) | 371 | 376 | 05 | 32.26 | 1.6432×104 | –24.0540 |
| (5) | 364 | 367 | 03 | 30.12 | 4.2280×103 | –20.6901 |
Table 6: Spectral parameters for DNA interaction with complexes (1-5)
The Kb values of the synthesized complexes (1-5) were summarized in Table 6. From these results, it was clear that complex (1) has higher binding efficacy via intercalation than complexes (2-5). The length of the DNA double helix was increased upon binding of the intercalating agents. This elongation of the polymer led to increase in the viscosity of the solution [52]. The interaction between CT DNA and the complexes (1-5) were investigated by viscosity measurements. Although the experiments provided lots of information to elucidate the type and strength of the complex-DNA interaction, the hydrodynamic measurements were considered as the least ambiguous and the most critical test of a DNA binding model in solution. An intercalative metal complex causes a separation of DNA base pairs due to lengthening of the nucleic acids helix. The interaction is responsible for the DNA viscosity, which is strictly dependent on the DNA length changes. The viscosity also increases by lengthening the DNA double helix through classical intercalation. Ethidium bromide (EtBr) is used as a well-known DNA classical intercalator. Partial or non-classical intercalation (binding in the grooves or in the sugar phosphate backbone) involved insertion of a planar molecule between DNA base pairs [53], the complex can bend or kink the DNA double helix, causing either a less apparent change (an increase or a decrease) or no change at all in the DNA viscosity. When the concentration of the complexes (1-5) increased, the relative viscosity also increased. The raised degree of viscosity led to the binding affinity to DNA. Significant increase in viscosity of the complexes that the partial insertion of the ligand between the DNA base pairs leading to an increase in the separation of base pairs at intercalation locations, hence an increase in overall DNA contour length. The η/η0 ratio value of complex (1) obtained from Eqn. 3 was greater than other complexes (1-4) and there was lesser than control (EtBr). It is clear that the observed binding affinities were found as in the order EtBr>(1)>(2)>(4)>(5)>(3), Figure 5.
The elaborately described research work in this report involved synthesis and characterization of Schiff base ligand (HL) and its complexes (1-5). The spectral data of the complexes suggested a square planar geometry and [ML] stoichiometry. The lower electrical conductivity values revealed that these were non-electrolytes. The results of gel electrophoresis disclosed that the complexes (1), (2) and (4) possessed significant DNA cleaving property relative to the remaining complexes in the presence of H2O2. The DNA binding results from electronic absorption and viscosity titration measurements for these complexes (1), (2) and (4) were higher than the complexes (3) and (5) but lower than the control EtBr. These results indicated that the complexes (1-5) bind to DNA via intercalation and also that the complexes possessed significant radical scavenging activity in the presence of DPPH. Most of these complexes exhibited fairly good antimicrobial activity when compared to the ligand.
Acknowledgements
Authors acknowledge the Department of Science and Technology (DST) Science and Engineering Research Board (SERB), Government of India, New Delhi for the financial support granted under the Project Ref. No. SR/FT/CS–117/2011 and also express their deep gratitude to the Managing Board, Dean, Principal and Chemistry Research Centre MSEC, Kilakarai for providing research facilities.
Conflicts of interest
There are no conflicts of interest.
References
- Tarafder MH, Saravanan N, Crouse KA, Ali AM. Coordination chemistry and biological activity of nickel(II) and copper ion complexes with nitrogen-sulphur donor ligands derived from S-bezyldithiocarbazate (SBDTC). Transit Metal Chem 2001;26:613-8.
- Hughes MN. The inorganic chemistry of biological processes. 2nd ed. New York: John Wiley & Sons Inc.; 1981.
- Dharamraj N, Viswanathamurthi P, Natrajan K. Ruthenium(II) complexes containing bidentate Schiff bases and their antifungal activity. Transit Metal Chem 2001;26:105-9.
- Yimer AM, Baraki T, Upadhyay RK, Masood A. Spectromagnetic and antimicrobial studies on some 3d metal complexes with ethylenedianil of ortho-hydroxyphenylglyoxal. Am J Appl Chem 2014;2:15-8.
- Das U, Sakagami H, Chu Q, Wang Q, Kawase M, Selvakumar P. et al. 3,5-Bis(benzylidene)-1-[4-2-(morpholin-4-yl)ethoxyphenylcarbonyl]-4-piperidone hydrochloride: A lead tumor-specific cytotoxin which induces apoptosis and autophagy. Bioorg Med Chem Lett 2010;20:912-7.
- Benitez J, Becco L, Correia I, Leal SM, Guiset H, Pessoa JC, et al. Vanadium polypyridyl compounds as potential antiparasitic and antitumoral agents: new achievements. J Inorg Biochem 2011;105:303-12.
- Patel NB, Purohit AC, Rajani DP, Moo-Puc R, Rivera G. New 2-benzylsulfanyl-nicotinic acid based 1,3,4-oxadiazoles: Their synthesis and biological evaluation. Eur J Med Chem 2013;62:677-87.
- Costamagna J, Vargas J, Latorre R, Alvarado A, Mena G. Coordination compounds of copper, nickel and iron with Schiff bases derived from hydroxynaphthaldehydes and salicylaldehydes. Coord Chem Rev 1992;119:67-88.
- Cotton AF, Wilkenson G. Advanced inorganic chemistry. 5th ed. New Yark: John Wiley & Sons; 1988.
- Jacobsen EN, Zhang W, Muci AR, Ecker JR, Deng L. Highly enantioselective epoxidation catalysts derived from 1,2-diaminocyclohexane. J Am Chem Soc 1991;113:7063-64.
- Campos-Fernandez CS, Galan-Mascaros JR, Smucker BW, Dunbar KR. Synthesis, X-ray studies and magnetic properties of dinuclear Ni(II) and Cu(II) complexes bridged by the azo-2,2-bipyridine ligand. Eur J Inorg Chem 2003;2003:988-94.
- Yimer AM. Chemical preparation, spectromagnetic and biocidal studies on some divalent transition metal complexes of Schiff’s base derived from 1-phenyl-2-(pyridin-2-yl)ethane-1,2-dione and ethylenediamine. Mod Chem Appl 2014;2:122.
- Raja JD, Sakthikumar K. Synthesis of water soluble transition metal(II) complexes from morpholine condensed tridentate Schiff base:structural elucidation, antimicrobial, antioxidant and DNA interaction studies. J Chem Pharm Res 2015;7:23-34.
- Rajesh J, Gubendran A, Rajagopal G, Athappan P. Synthesis, spectra and DNA interactions of certain mononuclear transition metal(II) complexes of macrocyclic tetraaza diacetyl curcumin ligand. J Mol Struct 2012;1010:169-78.
- Raman N, Jeyamurugan R, Sakthivel A, Mitu L. Novel metal-based pharmacologically dynamic agents of transition metal(II) complexes: designing, synthesis, structural elucidation, DNA binding and photo-induced DNA cleavage activity. Spectrochim Acta A Mol Biomol Spectrosc 2010;75:88-97.
- Charies JB, Dattagupta N, Crothers DM. Studies on interaction of anthracycline antibiotics and deoxyribonucleic acid: equilibrium binding studies on interaction of daunomycin with deoxyribonucleic acid. Biochemistry 1982;21:3933-40.
- Tan C, Liu J, Chen L, Shi S, Ji L. Synthesis, structural characteristics, DNA binding properties and cytotoxicity studies of a series of Ru(III) complexes. J Inorg Biochem 2008;102:1644-53.
- Muhammad S, Nooruddin, Saqib Ali, McKee V, Khan S, Khan M. Synthesis, spectroscopic characterization, crystal structure, DNA interaction study and in vitro biological screenings of 4-(5-chloro-2-hydroxyphenylamino)-4-oxobut-2-enoic acid. Spectrochim Acta A Mol Biomol Spectrosc 2015;134:244-50.
- Silverstein M, Webster X. Spectrometric Identification of Organic Compounds. 6th ed. New York: John Wiley & Sons Inc.; 1998. p.150.
- Tas E, Aslanoglu M, Kilic K, Kaplan O, Temel H. Preparation, characterisation and redox properties of four new tetradentate salicylaldimines with their Cu(II) complexes. J Chem Res 2006;4:242-45.
- Asadi M, Savaripoor N, Asadi Z, Ghatee MH, Moosavi F, Yousefi R, et al. Synthesis and characterization of some new Schiff base complexes of group 13 elements, ab initio studies, cytotoxicity and reaction with hydrogen peroxide. Spectrochim Acta A Mol Biomol Spectrosc 2013;101:394-9.
- Aazam ES, El Husseiny AF, Al-Amri HM. Synthesis and photoluminescent properties of a Schiff-base ligand and its mononuclear Zn(II), Cd(II), Cu(II), Ni(II) and Pd(II) metal complexes. Arab J Chem 2012;5:45-53.
- Raman N, Raja JD, Sakthivel A. Design, synthesis, spectroscopic Characterization, biological screening and DNA nuclease activity of transition metal complexes derived from a tridentate Schiff base. Russ J Coord Chem 2008;34:400-6.
- Li Y, Yang ZY, Liao ZC, Han ZC, Liu ZC. Synthesis, crystal structure, DNA binding properties and antioxidant activities of transition metal complexes with 3-carbaldehyde-chromone semicarbazone. Inorg Chem Commun 2010;13:1213-16.
- Masoud MS, Amira MF, Ramadan AM, El-Ashry GM. Synthesis and characterization of some pyrimidine, purine, amino acid and mixed ligand complexes. Spectrochim Acta A Mol Biomol Spectrosc 2008;69:230-38.
- Gupta LK, Bansal U, Chandra S. Spectroscopic and physicochemical studies on nickel(II) complexes of isatin-3,2’-quinolyl-hydrazones and their adducts. Spectrochim Acta A Mol Biomol Spectrosc 2007;66:972-75.
- Shakir M, Abbasi A, Azam M, Khan AU. Synthesis, spectroscopic studies and crystal structure of the Schiff base ligand derived from condensation of 2-thiophenecarboxaldehyde and 3,3’-diaminobenzidine and its complexes with Co(II), Ni(II), Cu(II), Cd(II) and Hg(II): Comparative DNA binding studies of L and its Co(II), Ni(II) and Cu(II) complexes. Spectrochim Acta A Mol Biomol Spectrosc 2011;79:1866-75.
- Chandra S, Gupta LK. EPR, mass, IR, electronic and magnetic studies on copper(II) complexes of semicarbazones and thiosemicarbazones. Spectrochim Acta A Mol Biomol Spectrosc 2005;61:269-75.
- Raman N, Sakthivel A, Rajasekaran K. Design, structural elucidation, DNA interaction and antimicrobial activities of metal complexes containing tetraazamacrocyclic Schiff bases. J Coord Chem 2009;62:1661-76.
- Kianfar AH, Keramat L, Dostani M, Shamsipur M, Roushani M, Nikpour F. Synthesis, spectroscopy, electrochemistry and thermal study of Ni(II) and Cu(II) unsymmetrical N2O2 Schiff base complexes. Spectrochim Acta A Mol Biomol Spectrosc 2010;77:424-29.
- Abdallah SM, Mohamed GG, Zayed MA, Abou El-Ela MS. Spectroscopic study of molecular structures of novel Schiff base derived from o-phthaldehyde and 2-aminophenol and its coordination compounds together with their biological activity. Spectrochim Acta A Mol Biomol Spectrosc 2009;73:833-40.
- Kivelson D, Neeman R. ESR studies on the bonding in copper complexes. J Chem Phys 1961;35:149-55.
- Dutta RL, Syamal A. Elements of Magnetochemistry. 2nd ed. New Delhi: East-west Press Pvt. Ltd.;1993. p. 106.
- Syamal A. Some improper terms in coordination chemistry. J Chem Edu 1985;62:143.
- Ray RK, George K. EPR Spectra and covalency of Bis(amidinourea/O-alkyl-1-amidinourea) copper(II) Complexes Part II. Properties of the CuN42- chromophore. Inorganica Chim Acta 1990;173:207-14.
- Hathaway BJ, Billing DE. The electronic properties and stereochemistry of mononuclear compounds of the copper(II) ion. Coord Chem Rev 1970;5:143-207.
- Sharma AL, Singh IO, Singh MA, Singh HR, Kadam RM, Bhide MK, et al. EPR studies on dichloromono(1-phenylamidino-o-alkylurea) copper(II) complexes: Evidence for field induced partial ordering in the solid state and some unusual features in solution. Transit Metal Chem 2001;26:532-37.
- Hathaway BJ, Wilkinson G, Gillard RD, McCleverty JA. Comprehensive Coordination Chemistry. Oxford: Pergamon Press; 1987.
- Jeyasubramanian K, Samath SA, Thambidurai S, Murugesan R, Ramalingam SK. Cyclic voltammetric and ESR studies of a tetraaza 14-membered macrocyclic copper(II) complex derived from 3-salicylideneacetyl-acetone and o-phenlyenediamine: Stabilization and activation of unusual oxidation states. Transit Metal Chem 1995;20:76-80.
- Chakravarty AR, Shyamala A. Synthesis, structure and electrochemical properties of complexes with a (µ-oxo)bis-(µ-carboxylato)diruthenium(III) core. Polyhedron 1993;12:1545-52.
- Bard AJ, Izatt Eds LR. Electrochemical methods fundamentals and applications. 2nd ed. New York: John. Wiley & Sons Inc.; 2001.
- Panneerselvam P, Nair RR, Vijayalakshmi G, Subramanian EH, Sridhar SK. Synthesis of Schiff bases of 4-(4-aminophenyl)-morpholine as potential antimicrobial agents. Eur J Med Chem 2005;40:225-29.
- Thimmaiah KN, Lloyd WD, Chandrappa GT. Stereochemistry and fungitoxicity of complexes of p-anisaldehydethiosemicarbazone with Mn(II), Fe( II), Co(II) and Ni(II). Inorganica Chim Acta 1985;106:81-3.
- Raman N, Syed Ali Fathima S, Raja JD. Designing, synthesis and spectral characterization of Schiff base transition metal complexes: DNA cleavage and antimicrobial activity studies. J Serb Chem Soc 2008;73:1063-71.
- Venkatadri B, Arunagirinathan N, Rameshkumar MR, Ramesh L, Dhanasezhian A, Agastian P. In vitro antibacterial activity of aqueous and ethanol extracts of Aristolochia indica and Toddalia asiatica against multidrug resistant bacteria. Indian J Pharm Sci 2015;77:788-91.
- Hyun TK, Kim HC, Kim JS. In vitro Screening for Antioxidant, Antimicrobial and Antidiabetic Properties of Some Korean Native Plants on Mt. Halla, Jeju Island. Indian J Pharm Sci 2015;77:668-74.
- Peng LX, Zou L, Wang JB, Zhao JL, Xiang DB, Zhao G. Flavonoids, antioxidant activity and aroma compounds analysis from different kinds of tartary buckwheat tea. Indian J Pharm Sci 2015;77:661-7.
- Sindhu T, Rajamanikandan S, Srinivasan P. In vitro antioxidant and antibacterial activities of methanol extract of Kyllinga nemoralis. Indian J Pharm Sci 2014;76:170-4.
- Meng DE, Tian YC, Yang Y, Shi J. Evaluation of DPPH free radical scavenging activity of various extracts of Ligularia fischeri in vitro: A case study of Shaanxi region. Indian J Pharm Sci 2016;78:436-42.
- Detmer CA, Pamatong FV, Bocarsly JR. Nonrandom double strand cleavage of DNA by a monofunctional metal complex: mechanistic studies. Inorg Chem 1996;35:6292-98.
- Uma V, Kanthimathi M, Weyhermuller T, Nair BU. Oxidative DNA cleavage mediated by a new copper(II) terpyridine complex:crystal structure and DNA binding studies. J Inorg Biochem 2005;99:2299-307.
- Mussardo P, Corda E, Gonzalez-Ruiz V, Rajesh J, Girotti S, Martin MA, et al. Study of non-covalent interactions of luotonin: A derivatives and the DNA minor groove as a first step in the study of their analytical potential as DNA probes. Anal Bioanal Chem 2011;400:321-27.
- Parveen S, Arjmand F. Design, synthesis and spectroscopic characterization of chiral benzimidazole-derived amino acid Zn(II) complexes: Development of tryptophan-derived specific hydrolytic DNA artificial nuclease agent. Spectrochim Acta A Mol Biomol Spectrosc 2012;85:53-60.
 Complex (1);
Complex (1);  complex (2);
complex (2);  complex (4);
complex (4); complex (5);
complex (5);  complex (3)
complex (3)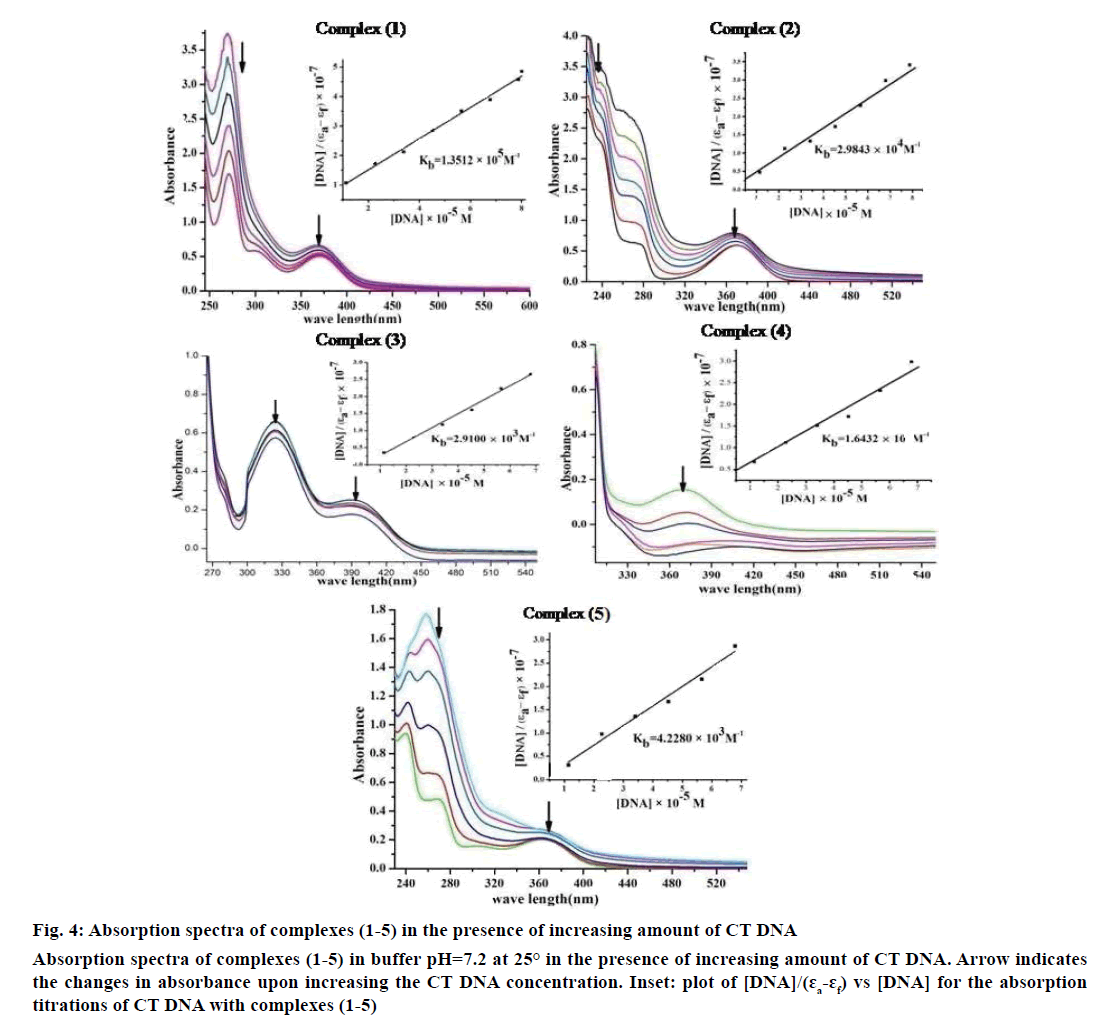
 Control (EtBr);
Control (EtBr);  complex (1);
complex (1); 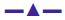 complex (2);
complex (2);  complex (4);
complex (4);  complex (5);
complex (5); complex (3)
complex (3)



