- Corresponding Author:
- N. M. KAMAT
Department of Botany Goa University, Taleigao Plateau Goa-403 206, India
E-mail: nandkamat@gmail.com
| Date of Submission | 31 August 2010 |
| Date of Revision | 31 January 2011 |
| Date of Acceptance | 8 March 2011 |
| Indian J. Pharm. Sci., 2011, 73 (1): 223-228 |
Abstract
Out of the 30 actinobacterial cultures screened for antimicrobial activity, 28 cultures were found to produce active products against various pathogenic microorganisms such as Gram-negative, Gram-positive bacteria and yeast, using a modified cross streak method. The modified method helped in easy quantification of results and also in ruling out probable mutual antibiosis. The actinobacterial strains that showed the ability to produce antimicrobial compounds belonged to Streptomyces (53%), Micromonospora (13%) and Actinomadura (10%) genera. Streptomyces sp. strain MMA-5 showed the highest multispecific antibiosis efficiency score value. Broad antibiotic spectrum activity was exhibited by Streptomyces sp. strain MMA-2 and Micromonospora sp. strain MMA-8. The
Keywords
Actinobacteria, antibacterial and antifungal activity, modified cross streak method
As multidrug resistance is rising, there is a steep decline in antibiotic discovery and development [1]. Hence, there is a continuing and cyclical need of new antibiotics to combat antibiotic resistant strains of pathogenic bacteria [2]. The actinobacteria are a group of Gram-positive bacteria, majority of which are found in the soil playing a significant role in decomposition and humus formation. The discovery of new molecules from actinobacteria have marked an epoch in antibiotic research as most of its antibiotics are too complex to be synthesized by combinatorial chemistry [3,4]. Actinobacteria are the most prolific producer of antibiotics [5]. Out of more than eight thousand antimicrobial products described in antibiotic literature database (ABL database), 45.6% are produced by Streptomycetes and 16% by strains belonging to rare genera of actinobacteria[3], which forms an important component of the soil microflora [6]. The rate of discovery of new compounds from terrestrial actinobacteria has decreased, whereas the rate of reisolation of known compounds has increased [7].
Search of underexplored ecological niches have revealed new molecules which stresses the need to explore new groups of actinobacteria from such habitats as source of novel bioactive secondary metabolites supplemented by novel selected media and/or improvement of isolation methods [6-8]. Baltz has estimated that less than one part in 1012 of the earth’s soil surface has been screened for actinobacteria [1]. Only 10% of natural products from screened strains and just 1% of molecules from the global consortium of microbial producers may have been discovered by researchers [2].
There are many techniques for detecting antimicrobial activity; most of them are based on methods involving diffusion through solid or semi-solid culture media to inhibit the growth of sensitive microorganisms [9]. The cross-streaking is a easy and relatively rapid method for screening cultures in search for new antibiotics and thus establish a spectrum of inhibiting properties of any bacterium, mold, or actinobacteria which will grow discretely on an agar plate [10]. Studies have revealed that the ‘cross streak method’ resulted in higher inhibition zones on indicator bacteria than those obtained by agar well diffusion method [9] but the major drawback of the ‘cross streak method’ was difficulty in obtaining quantitative data, since the margins of the zone of inhibition were usually very fuzzy and indistinct [10]. Hence, in the present study, we reported the isolation as well as the inhibitory effects of local actinobacterial isolates on various human pathogenic bacteria and yeast using a modified cross streak method.
For screening, the soil samples were collected from various locations in Goa (Table 1) at a depth of 10-20 cm in clean polythene bags. Several diverse habitats in different areas were selected for isolation of actinobacterial strains, which included forest, biogenic, market, orchard and sediment soils. Samples were transported to the laboratory and stored at 4º until use. The bacterial strains used were procured from the Department of Microbiology, Goa Medical College, Goa while the yeast strain was procured from Microbial Type Culture Collections, Chandigarh (MTCC). The bacteria strains were maintained on slants of Nutrient Agar (Himedia, India), whereas the yeast strain was maintained on slants of Potato Dextrose Agar (Himedia, India). Other chemicals used were of analytical grade and were purchased from Sigma, Germany.
A modified slide technique for isolation of soil actinobacteria in a natural tropical community was used, which employs slides coated with Arginine Vitamin Agar (AVA) medium to specifically capture ex situ actinobacterial diversity. The composition of AVA medium [11] is given in Table 2. The basal medium and the stock trace element solution were sterilized by autoclaving at 121º for 20 min and pH of the medium was adjusted to 6.8 by using 0.1N NaOH/ HCl. The stock vitamin and antibiotic solution was membrane sterilized and added later to the presterilised basal medium. The soil samples were filled in clean polyvinylchloride pots and pair of test slides (with antibiotics) was then inserted so that the coated surface was in contact with the surrounding soil horizon. The sets of slides were sampled at predetermined time points on 4, 7, 12, 15, 22, 26 and 28 days without disturbing the colonized coated surface and processed immediately. The slides were then washed with 1 ml of sterile distilled water and 0.1 ml of the suspension was then plated on antibiotic based AVA medium (the same used for coating the slides) and incubated at room temperature of 28-30º till visible colonies were seen on the plates. Incubated plates were observed under Olympus SZ51 stereomicroscope (Tokyo, Japan) and the actinobacterial colonies were then transferred to AVA slants and incubated at the ambient temperature of 28º for 4-7 days.
| Sampling location | Latitude and | Total no. of | No. of actinobacterial isolates grouped in different taxa | |||
|---|---|---|---|---|---|---|
| longitude | actinobacterial isolates | Streptomyces | Actinomadura | Micromonospora | Unidentified Genera | |
| Forest soils | ||||||
| Forest Karmali, Tiswadi (KFA) | 15°29’16”N | 7 | 3 | 0 | 2 | 2 |
| 73°54’40”E | ||||||
| Forest Canacona (CFA) | 14°59’45.76”N | 4 | 2 | 0 | 0 | 2 |
| 74°03’02.17”E | ||||||
| Biogenic soils | ||||||
| Termite mound, Karmali (TMK) | 15°29’16”N | 1 | 0 | 1 | 0 | 0 |
| 73°54’40”E | ||||||
| Hollow of Terminalia sp. tree, | 15°41’02.28”N | 6 | 5 | 1 | 0 | 0 |
| Arambol (MAS) | 73°43’05.17”E | |||||
| Market soils | ||||||
| Mapusa market, Bardez (MMA) | 15°35’16.73”N | 6 | 3 | 1 | 2 | 0 |
| 73°48’32.66”E | ||||||
| Panjim market, Tiswadi (PM) | 15°29’55.64”N | 1 | 1 | 0 | 0 | 0 |
| 73°49’18.40”E | ||||||
| Orchard soils | ||||||
| Orchard, Bicholim (BOA) | 15o35’13.96”N | 1 | 0 | 0 | 0 | 1 |
| 73°56’11.06”E | ||||||
| Sediment soil | ||||||
| Siolim, Bardez (SIA) | 15°37’39.35”N | 4 | 2 | 0 | 0 | 2 |
| 73°48’38.79”E | ||||||
| Total | 30 | 16 | 3 | 4 | 7 |
Table 1: Various Sites Selected For Soil Sample Collection
| Glycerol | 0.08 ml |
| Glucose | 0.1 g |
| K2HPO4 | 0.03 g |
| L-arginine | 0.03 g |
| NaOH | 0.03 g |
| MgSO4.7H2O | 0.02 g |
| Agar | 1.8 g |
| Distilled water | 100 ml |
| Stock trace element solution (Add 0.1 ml to the | |
| basal medium) | |
| Fe2 (SO4)3 | 50 mg |
| CuSO4.5H2O | 50 mg |
| MgSO4.4H2O | 50 mg |
| ZnSO4.7H2O | 50 mg |
| Distilled water | 50 ml |
| Stock vitamin solution (Add 0.1 ml to the basal | |
| medium) | |
| Thiamine | 100 mg |
| Calcium panthothenate | 25 mg |
| Meso-inositol | 25 mg |
| Nicotinic acid | 25 mg |
| P-aminobenzoic acid | 25 mg |
| Pyridoxine | 25 mg |
| Riboflavin | 25 mg |
| Biotin | 12.5 mg |
| Distilled water | 50 ml |
| Antibacterials (Added to basal medium) | |
| Nalidixic acid | 0.0176 mg/ml |
| Neomycin and polymyxin b sulphate | 0.01 mg/ml |
| Antifungals (Added to basal medium) | |
| Nystatin | 0.004 mg/ml |
| Actidione | 0.00192 mg/ml |
Table 2: Ava Media Compositiona
The isolates were phenotypically grouped according to the traditional criteria of classification including growth, aerial spore-mass colour, texture, elevation, substrate mycelium colour, margin, pigment production and presence or absence of exudates. For micromorphological details air dried smears stained in 1% w/v crystal violet were used and morphology of the spore bearing hyphae with entire spore chain was studied along with other microstructures [11,12]. The criteria used to select the actinobacterial strains were based on ex situ slide observations. The strains, which were recovered from slides having less or no fungal bacterial colonization, were selected as it was assumed that these strains could be potential antibiotics procedures.
Determination of antimicrobial activities of 30 pure actinobacterial cultures was performed by modified cross-streak method (MCSM). Muller–Hinton agar (Himedia, India) plates [8] were prepared and inoculated with actinobacterial cultures by a single streak in the centre of the Petri dish and incubated at 28º for 7 days. This was done to provide enough time for the active organism to produce the antibiotic substance, which will diffuse into the agar medium [13]. The crossstreak method (CSM) [14] was modified by keeping the distance between the test pathogen streak fixed i.e. 1 cm; length of the test pathogen streak line, 3 cm; streak width, 0.5 cm; length of actinobacterial streak, 7 cm and width of the actinobacterial streak, 0.5 cm. The markings were drawn on a transparency sheet, which acted as a template and was attached below the plates to facilitate streaking and thereafter in quantifying the results.
The plates were seeded with test organisms by streaking perpendicular to the line of actinobacterial growth. The plates containing active organisms were kept for 14-28 days to determine whether the test pathogens made any further growth towards the master streak, or whether they remain stationary, or whether lysis of the test pathogens occurred [14]. Antagonism was observed based on the inhibitory interaction between the actinobacteria and test strains. The potential antagonistic producer was selected by screening the actinobacterial isolates against a group of pathogens which included human pathogenic Gram negative bacteria such as Salmonella typhi, Shigella flexneri, Proteus sp., Enterobacter aerogenes, Serratia marcescens, Gram-positive bacteria such as Staphylococcus aureus and Bacillus subtilis and dimorphic yeast such as Candida albicans (MTCC183).
Since the area of each test streak was fixed at 150 mm2 it was possible to quantify regions of growth and measure the clear/inhibited areas with a graph paper template. The clear areas for different test species are indicators of antibiotic activity and are proportional to antibiotic concentration and diffusion. To express these results quantitatively and to prepare quantitative antibiotic activity score matrices, the following Eqns. were used, Percent area specific differential antibiotic activity score (PASDAAS)= (AWG/TSA)×100….(1), where, AWG is the area on the plate without growth for each test panel streak, TSA is the total streak area (150 mm2) scored for each test panel streak.
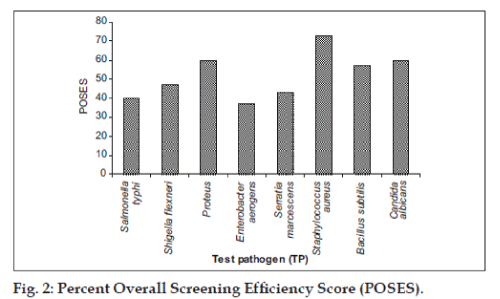
Percent multispecific antibiosis efficiency score (PMSAES), gives the efficiency of tested actinobacteria strain to inhibit more than a single test pathogen. The ideal score would be 100. This is computed using the Eqn. PMSAES=(ΣPASDAASTP1-8/ TPS)×100…….(2), where, ΣPASDAASTP1-8 is the percent area specific differential antibiotic activity score of test pathogens 1-8 and TPS is total possible score for all test pathogens (i.e. 100×8=800). Scoring total or percent overall inhibition efficiency score (POIES), since multiple microbial species were used in the test panel it was possible to calculate a total score for each actinobacteria using the following Eqn, POIES = (TNIS/TNTS)×100…..(3), where, TNIS is total number of inhibited species and TNTS is total number of test species. The ideal score for multispecific inhibition would be 100. Having calculated the total no of positive results of each test pathogen against 30 actinobacterial strains, it was possible calculate percent overall screening efficiency score (POSES). This is computed by the equation, POSES= (TPR/TAS)×100…..(4), where TPR is total no of positive results of each test pathogen and TAS is total no of actinobacterial strains.
The occurrence and distribution of different genera of actinobacteria isolated from different soil samples are presented in Table 1. Despite the addition of antibiotics in the slide coating medium (SCM) to inhibit growth of unwanted microbes and allow actinobacteria to dominate, bacterial and fungal spores were observed, which shows their partial resistance towards the incorporated antibiotics. These slides acted as reference to the slides in which less or no fungal and bacterial colonization was observed. The strains recovered from such slides were thus assumed to be potential producers of antibiotics (antifungal and antibacterial) and were selected for screening.
Based on the spore morphology and mycelium, the isolated strains were classified into various genera. Out of the 30 actinobacterial isolates, 16 isolates were identified as genus Streptomyces (spore chain with coiling, spiral and looped), 3 as Actinomadura (spore chains are straight and open hooked), 4 as Micromonospora (clusters of single conidia on substrate mycelium) and 7 were unidentified actinobacterial strains. Among the genera recorded in the present study, Streptomyces was the most predominant as compared to others. The dominance of Streptomyces among the actinobacteria especially in soils has also been reported by many workers [15,16]. Preliminary tests for antimicrobial activity of isolated strains obtained from different locations in Goa, clearly demonstrated that 53% of actinobacterial strains belonging to Streptomyces genera, 13% belonging to Micromonospora and 10% belonging to Actinomadura genera have the ability of producing antimicrobial compounds. Results demonstrate the fertility of the sampled soils in possessing such microbes, which are valued for their unparalleled ability to produce bioactive molecules of biotechnological and pharmaceutical importance. The cross streak method was modified as the streak non uniformity caused a problem with respect to quantifying the results [10]. Besides mutual antibiosis due to use of multiple cultures in the test panel could not be ruled out. The distance between the test steak and the actinobacterial strain was kept constant to reduce the effect of probable mutual antibiosis, as multiple cultures were used in the test panel. Since the test pathogen population was delimited within a defined streaking zone, antibiosis would indicate its impact with respect to that zone and thus the antibiosis reaction could be easily quantified.
Out of 30 cultures screened for antimicrobial activity, 28 cultures showed positive activity. Fifty percent of all antimicrobial producing strains had inhibited five Gram positive bacteria, thirty percent strains had inhibited two Gram negative bacteria and twenty percent strains inhibited multidrug resistant human pathogenic strain of Candida albicans. The multidrug resistance is presently an urgent focus of research and new bioactive compounds are necessary to combat these multidrug resistance pathogens [8,17]. The results of the antimicrobial screening are represented in Table 3.
Streptomyces sp. strain MMA-5 showed high PASDAAS against Serratia marcescens (87), Salmonella typhi (80), Staphylococcus aureus (80), Bacillus subtilis (80), and Candida albicans (80). while Streptomyces sp. strain MAS-11 showed high PASDAAS against Serratia marcescens (87) and Shigella flexneri (80) whereas Streptomyces sp. strain SIA-2 and Streptomyces sp. strain MAS-30 were found to be active against Bacillus subtilis (80) and Candida albicans (80). Unidentified actinobacterial strain KFA-1Gb was found active against Shigella flexneri (80) and Candida albicans (80). Streptomyces sp. strain KFA-5 was found active against Staphylococcus aureus (80). Streptomyces sp. strain KFA-1 was found to be active against Shigella flexneri (80) where as Micromonospora sp. strain KFA-9 and Micromonospora sp. strain MMA-8 was found to be active against Shigella flexneri (80). Streptomyces sp. strain MMA-5 showed the highest PMSAES (59) value (fig. 1). Broad antibiotic spectrum activity against eight test pathogens was exhibited by Streptomyces sp. strain MMA-2 and Micromonospora sp. strain MMA-8 (Table 3). The highest POSES was exhibited against Staphylococcus aureus followed by Proteus sp. and Candida albicans (fig. 2).
From these results we concluded that, a total of 28 actinobacterial isolates associated with soil have shown the ability to produce antimicrobial compounds against 8 test pathogens, especially multiple antibiotic resistant Gram positive bacteria and yeast using ‘modified cross streak method’. Further studies on molecular taxonomical characterization of the potential organisms, purification, and characterization of antibacterial and anticandidial compounds are warranted.
| Actinobacterial strain | TPI | TP2 | TP3 | TP4 | TP5 | TP6 | TP7 | TP8 | POIES |
|---|---|---|---|---|---|---|---|---|---|
| Streptomyces sp. strain KFA-5 | 20 | 0 | 47 | 0 | 0 | 80 | 73 | 73 | 63 |
| Unidentified actinobacteria strain KFA-8 | 0 | 0 | 60 | 0 | 0 | 0 | 0 | 0 | 13 |
| Micromonosporasp.strain KFA-9 | 0 | 80 | 60 | 0 | 67 | 53 | 60 | 0 | 63 |
| Micromonosporasp. strain KFA-3 | 13 | 73 | 73 | 0 | 20 | 67 | 0 | 0 | 63 |
| Streptomyces sp. strain KFA-6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Unidentified actinobacteria strain KFA-1Gb | 0 | 80 | 0 | 27 | 20 | 60 | 0 | 80 | 63 |
| Streptomyces sp. strain KFA-1 | 0 | 80 | 67 | 0 | 0 | 67 | 20 | 53 | 63 |
| Streptomyces sp. strain CFA-1 | 0 | 0 | 0 | 0 | 0 | 60 | 0 | 0 | 13 |
| Unidentified actinobacteria strain CFA-2 | 0 | 53 | 73 | 73 | 60 | 60 | 40 | 0 | 75 |
| Unidentified actinobacteria strain CFA-3 | 0 | 27 | 47 | 0 | 0 | 0 | 53 | 53 | 50 |
| Streptomyces sp. strain CFA-2 | 0 | 73 | 0 | 60 | 0 | 73 | 0 | 0 | 38 |
| Actinomadurasp. strain TMK-33 | 0 | 60 | 0 | 0 | 0 | 53 | 67 | 27 | 50 |
| Streptomyces sp. strain MAS-2 | 0 | 0 | 40 | 20 | 27 | 60 | 60 | 67 | 75 |
| Actinomadurasp. strain MAS-4 | 27 | 0 | 0 | 20 | 0 | 0 | 0 | 73 | 38 |
| Streptomyces sp. strain MAS-11 | 0 | 80 | 0 | 0 | 87 | 20 | 0 | 0 | 38 |
| Streptomyces sp. strain MAS-14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Streptomyces sp.strain MAS-28 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 13 | 25 |
| Streptomyces sp. strain MAS-30 | 20 | 0 | 40 | 60 | 67 | 67 | 80 | 80 | 88 |
| Streptomyces sp. strain MMA-2 | 60 | 67 | 53 | 40 | 40 | 27 | 47 | 53 | 100 |
| Micromonosporasp. strain MMA-4 | 20 | 53 | 40 | 0 | 20 | 20 | 40 | 13 | 88 |
| Micromonosporasp. strain MMA-8 | 73 | 80 | 40 | 53 | 20 | 67 | 60 | 60 | 100 |
| Actinomadurasp. strain MMA-9 | 0 | 40 | 73 | 0 | 0 | 53 | 40 | 73 | 63 |
| Streptomyces sp. strain MMA-2 | 0 | 0 | 0 | 27 | 0 | 40 | 53 | 53 | 50 |
| Streptomyces sp. strain MMA-5 | 80 | 0 | 67 | 0 | 87 | 80 | 80 | 80 | 75 |
| Streptomyces rubrolavendulae | 0 | 7 | 67 | 0 | 40 | 73 | 0 | 0 | 50 |
| Unidentified actinobacteria strain BOA-10 | 0 | 0 | 0 | 0 | 0 | 20 | 0 | 0 | 13 |
| Streptomyces sp. strain SIA-2 | 0 | 0 | 27 | 53 | 0 | 67 | 80 | 80 | 63 |
| Streptomyces sp. strain SIA-2G | 40 | 0 | 40 | 0 | 60 | 67 | 47 | 0 | 63 |
| Unidentified actinobacteria strain SIA-2Gb | 7 | 0 | 20 | 0 | 40 | 0 | 0 | 53 | 50 |
| Unidentified actinobacteria strain SIA-12 | 20 | 0 | 0 | 27 | 0 | 73 | 73 | 73 | 63 |
TABLE 3: Pasdaas And Poies For Each Actinobacterial Strain
Acknowledgements
The Authors would like to acknowledge UGC-SAP for providing financial assistance to carry out the above work.
References
- Fishbach AM, Walsh TC. Antibiotics for emerging pathogens. Science 2009;325:1089-93.
- Clardy J, FishbachAM, Walsh TC. New antibiotics from bacterial natural products. Nature Biotechnol 2006;24:1541-50.
- Lazzarini A, Cavaletti L, Toppo G, Marinelli F. Rare genera of actinomycetes as potential producers of new antibiotics. Antonie van Leeuwenhoek 2000;78:399-405.
- Baltz HR. Antimicrobials from Actinomycetes: Back to the future. Microbe 2007;2:125-31.
- Berdy J. Bioactive microbial metabolites. J Antibiot 2005;58:1-26.
- Singh LS, Indra B, Bora TC. Actinomycetes of Loktak Habitat: Isolation and Screening for Antimicrobial Activities. Biotech 2006;5:217-21.
- Sanasam S, Ningthoujam SD. Screening of local actinomycetes isolates in Manipur for anticandidial activity. Asian J Biotechnol 2010;2:139-45.
- Joseph S, Shanmughapriya S, Gandhimathi R, Kiran SG, Ravji RT, Natarajaseenivasan K, et al. Optimization and production of novel antimicrobial agents from sponge associated marine actinomycetesNocardiopsisdassonvilleiMAD08. ApplMicrobiolBiotechnol2009;83:435-45.
- Lertcanawanichakul M, Sawangnop S. A comparison of Two Methods used for measuring the antagonistic activity of Bacillus Species. Walailak J Sci Tech 2008;5:161-71.
- Williston EH, Zia-Walrath P, Youmans GP. Plate methods for testing antibiotic activity of actinomycetes against Virulent human type Tubercle Bacilli. J Bacteriol 1947;54:563-8.
- Locci R. Streptomycetes and Related Genera. In: Goodfellow M, Williams ST, Mordarski M editors. Bergey s Manual of Systematic Bacteriology. New York: Academic Press; 1989. p. 1-32.
- Cross T, Goodfellow M. Taxonomy and classification of the actinomycetes. In: Sykes G, Skinner FA, editors. Actinomycetales: Characteristics and Practical Importance. New York: Academic Press; 1973. p. 11-91.
- Waksman S, TheActinomycetes. Vol. 3. Baltimore: The Williams and Wilkins Company; 1962. p. 20-30.
- Fernando C. Screening Tests for Antibiotics. Mycologia 1947;39:128-30.
- Vijayakumar R, Muthukumar C, Thajuddin N, Panneerselvam A, Saravanamuthu R. Studies on the diversity of actinomycetes in the Palk Strait region of Bay of Bengal, India. Actinomycetologica 2007;21: 59-65.
- Ceylan O, Okmen G, Ugur A. Isolation of soil Streptomyces as source antibiotics active against antibiotic-resistant bacteria. Eur Asia J BioSci 2008;2:73-82.
- Selvameenal L, Radhakrishnan M, Balagurunathan R. Antibiotic pigment from desert soil actinomycetes. Indian J Pharm Sci 2009;71:499-504.