- *Corresponding Author:
- H. F. Li
Shanghai Key Laboratory of Psychotic Disorders, Shanghai Mental Health Center, Shanghai Jiao Tong University School of Medicine, Shanghai, PR China
Y. F. Shen
Department of Psychiatry, Shanghai Mental Health Center, Shanghai Jiao Tong University School of Medicine, Shanghai, PR China
E-mail: shenyifeng@yahoo.com
| Date of Submission | 22 November 2018 |
| Date of Revision | 18 March 2019 |
| Date of Acceptance | 14 June 2019 |
| Indian J Pharm Sci 2019;81(4):690-698 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The aim of this study was to investigate the association between two brain-derived neurotrophic factor polymorphisms (Val66Met and C270T) and response to citalopram/escitalopram treatment. Chinese patients who met diagnostic and Statistical Manual of Mental Disorders criteria for major depressive disorder (n=180) were prescribed citalopram/escitalopram for 6 weeks. Depression severity was evaluated using the 17-item Hamilton rating scale for depression at baseline and week 2, 4, and 6 of treatment. The expression quantitative trait loci analysis was performed to investigate the functional effect of Val66Met and C270T on brain-derived neurotrophic factor expression in the brain and blood. A significant difference was found in genotype (χ2=6.979, p=0.031, correction p=0.062) but not allele (p>0.05) frequencies of Val66Met between responders and non-responders. Homozygous GG showed a higher response than other two genotypes (χ2=5.218, p=0.022, correction p=0.044). In addition, the C allele and CC genotype of C270T were significantly related to higher remission over 6 weeks among females (C/T, p=0.027, correction p=0.054; CC/TT, p=0.023, correction p=0.046) and first-episode major depressive disorder patients (C/T, p=0.023, correction p=0.046; CC/TT, χ2= 6.870, p=0.009, correction p=0.018). Further expression quantitative trait loci analysis showed that Val66Met and C270T have functional effect on brain-derived neurotrophic factor expression in the brain. Brain-derived neurotrophic factor Val66Met and C270T polymorphisms were associated with therapeutic response to citalopram/escitalopram in Chinese major depressive disorder patients.
Keywords
Brain-derived neurotrophic factor, Val66Met, C270T, polymorphism, depression, antidepressant
Antidepressant drugs such as selective serotonin reuptake inhibitors (SSRIs), serotonin and noradrenergic reuptake inhibitors, noradrenergic and specific serotonergic antidepressants are a primary treatment for major depression disorder (MDD). However, only about two thirds of MDD patients achieve remission after undergoing up to several cycles of antidepressant treatment, with one-third of patients showing no response[1]. Genetic variations are thought to contribute to the variable responses to antidepressants[2], which was also supported by a genome-wide association study[3]. As such, identifying single nucleotide polymorphisms (SNPs) associated with treatment resistance can potentially predict patient treatment outcome.
Brain-derived neurotrophic factor (BDNF) is a neurotrophin associated with response to antidepressants in MDD[4]. It is widely expressed in the brain, especially in the hippocampus and cortex[5]. Not only can BDNF promote differentiation and survival in peripheral and central neurons, but also regulate brain maturation, synaptic plasticity, and axon outgrowth[6]. Evidence from animal models[7], neuronal cultures[8], and clinical[9,10] and post-mortem[11] studies have shown that BDNF expression is dysregulated in MDD patients, which can be reversed by antidepressant treatment. It is believed that BDNF interacts with the 5-hydroxytryptamine neurotransmitter system to modulate the response to antidepressants[12,13].
The BDNF gene, with 11 exons and located on chromosome 11p31, is a vital candidate for the mechanism of action of antidepressants[14]. Val66Met (rs6265, G>A) is a most frequently investigated polymorphism of BDNF and the G to A exchange leads to the valine-to-methionine change at position 66. Evidence has shown that this polymorphism is implicated in activity-dependent secretion[15], blood levels of BDNF[16] and antidepressant response[17]. Citalopram and escitalopram are widely prescribed SSRIs, highly efficacious to treat depression[18]. However, the relationship between Val66Met and citalopram/escitalopram response is inconsistent. For example, two findings showed Met allele carriers of Val66Met responded better to citalopram and escitalopram[19,20]; whereas, another study found no association between Val66Met and treatment response to escitalopram[21]. Thus, whether Val66Met is associated with citalopram/escitalopram response hasn’t been confirmed yet.
The C270T polymorphism (rs56164415, C>T) is located in the 5ꞌ untranslated region of BDNF and may affect transcription efficiency and protein expression[22]. C270T has been linked to increased susceptibility to schizophrenia[23], Parkinson’s disease[24], and autism[25]. Huuhka et al. found no association between C270T polymorphism and response to electroconvulsive therapy (ECT) in Caucasians; however, the CC genotype of C270T predicted good response in psychotic and late onset subtypes of depression[26]. To date, there have been no studies investigating the association between C270T and response to antidepressant drug treatment. Therefore, the purpose of this study was to explore the association between C270T and Val66Met of the BDNF gene and response to citalopram/escitalopram in Chinese population with MDD.
Materials and Methods
Subjects:
Unrelated Chinese patients (n=180) with MDD were recruited at the Shanghai Mental Health Center, Shanghai Jiao Tong University School of Medicine, China. For pharmacogenetic analyses, only patients with a treatment cycle of at least 4 w from baseline were included (n=147). All patients were assessed by trained psychiatrists using the structured clinical interview for DSM-IV Axis I disorders. The inclusion criteria were as follows: 1) age between 18 and 65 y; 2) at least moderately severe depression and a total score on the 17-item Hamilton rating scale for depression (HAMD-17) of 18 or higher; 3) physical, laboratory, and electrocardiogram examinations showing no other clinically significant abnormalities. Exclusion criteria were as follows: 1) patients with bipolar, psychotic, or anxiety disorder, mental retardation, or disease related to alcohol or drug abuse; 2) patients with neurological or any other general disorders; 3) patients who were pregnant or breastfeeding; 4) at serious risk of suicide or with drug allergies; 5) patients taking monoamine oxidase inhibitors within the 4-w period prior to the start of the study. Patients were provided written, informed consent for their participation in the study. The ethics committee of Shanghai Mental Health Center and Huashan hospital (Shanghai, China) approved the protocol.
Treatment:
A total of 147 patients were treated with citalopram (n=58) and escitalopram (n=89) for 6 w. Drugs were given as monotherapy in patients. All patients had a medication washout for a minimum of 5 half-lives of all current psychotropic medications. Patients were treated with citalopram at a dose of 20 mg/d and escitalopram at a dose of 10 mg/d for the first 2 w. Psychiatrists adjusted the dosage according to the improvement of symptoms; if these had improved at w 2, then patients continued taking the initial dose of these drugs, but the dose was doubled for patients who showed no response to the drugs or exacerbation of depressive symptoms. Dosage modification was not allowed after 4 w. No other medications including antipsychotics, anxiolytics, or other antidepressants were allowed during the study, except for medication to treat insomnia (no more than 1 w) such as zolpidem, zopiclone, or clonazepam.
Clinical assessment:
Clinical symptoms were evaluated at baseline (w 0) and at w 2, 4, and 6 with the HAMD-17, Hamilton anxiety rating scale, and the Clinical Global Impression scale. Responders were defined as patients exhibiting a decrease in HAMD-17 total scores of at least 50 % at the end of w 6, and remitters were defined as those with scores less than 8. Patients who did not satisfy these criteria were defined as non-responders and nonremitters, respectively.
DNA sequencing:
Genomic DNA from peripheral blood samples was phenol-chloroform extracted and purified. Genetic variations in the BDNF gene were genotyped by DNA sequencing. Fragments containing polymorphisms were amplified with the following forward and reverse primer sets: Val66Met, 5ꞌ-GAGTGATGACCATCCTTTTCCT- 3ꞌ and 5ꞌ-CCTCATGGACATGTTTGCAG-3ꞌ; and C270T, 5ꞌ-ATTCCTGCAAAGGACCATGT-3ꞌ and 5ꞌ-TGTGGCCCATCTGATTGTAA-3ꞌ. PCR products were sequenced using a BigDye Terminator v1.1 Cycle Sequencing kit (Applied Biosystem) in a 5-μl reaction consisting of 1 μl purified PCR product (10 ng/μl), 0.5 μl of 2.5×BigDye, 0.15 μl of 5×BigDye buffer, 1 μl primer (3.2 pmol/μl), and 2.35 μl of sterilized deionized water. Reactions were run on an ABI 3730 sequencer (Applied Biosystems, Foster City, CA, USA). Twenty samples were randomly selected for duplicate detection to verify and quality of the results.
eQTLs datasets:
In order to explore the functional effects of Val66Met and C270T on the expression of the BDNF gene across different brain regions, a gene expression analyses was conducted using data from the BRAINEA (http:// braineac.org). This dataset includes 10 brain regions from 134 neuropathologically normal individuals of European descent[27]. In addition, blood eQTL browser (https://molgenis58.target.rug.nl/bloodeqtlbrowser/) was used to understand the effects of these two SNPs on the expression of the BDNF in whole blood[28]. This resource contains the results of an eQTL meta-analysis from 5311 peripheral blood samples from 7 studies.
Statistical analysis:
Categorical variables of demographic data are presented as percent (%) frequency and were analyzed with the χ2 test. Continuous variables of demographic data are expressed as mean±standard deviation and were evaluated with the Student’s t test. Hardy-Weinberg equilibrium for genotype frequencies in patients was determined with the χ2 test. Allele and genotype distributions for Val66Met and C270T were compared between remitters and non-remitters and between responders and non-responders with the χ2 test. The haplotype analysis was performed on the SHEsis online software (http://analysis2.bio-x.cn/myAnalysis. php). The pharmacogenetic analysis of Val66Met and C270T variant effects on HAMD-17 total and change scores over the 6 w of antidepressant treatment was carried out with an overall repeated-measures analysis of variance (with genotype as the fixed factor and weeks as repeated measure). Age, gender, first episode or recurrence, recent episode (month), total course (month), BMI and drug categories were included as covariates. All statistical analyses were performed using SPSS v.19.0 software (SPSS Inc., Chicago, IL, USA). The level of statistical significance was assumed at p<0.05 (two-sided test). Bonferroni correction was performed by multiplying the p values by the number of SNPs.
Results and Discussion
According to HAMD-17 scores at 6 w, 147 patients were divided into remitter (n=81, HAMD<8) and non-remitter (n=66, HAMD≥8) groups, and responder (n=123, decreased rate of HAMD-17≥50 %) and non-responder (n=24, decreased rate of HAMD-17< 50 %) groups. The demographic data are summarized in Table 1. There were no differences in age, duration of the current episode of depression, duration of illness, BMI, first/recurrent episode, and dosage of citalopram/ escitalopram. There were no sex differences between remitters and non-remitters, but a difference was observed between responders and non-responders (p=0.047), with the former comprising mostly females.
| Characteristics | Overall (n=147) | Remitter (n=81) | Non-remitter (n=66) | p | Responder (n=123) | Non-responder (n=24) | p |
|---|---|---|---|---|---|---|---|
| Sex: male | 59 (40.1 %) | 28 (34.6 %) | 31 (47.0 %) | 0.127 | 45 (36.6 %) | 14 (58.3 %) | 0.047* |
| Female | 88 (59.9 %) | 53 (65.4 %) | 35 (53.0%) | 78 (63.4 %) | 10 (41.7 %) | ||
| Age (year) | 38.41±13.39 | 36.78±13.60 | 40.41±12.94 | 0.102 | 38.54±13.58 | 37.75±12.58 | 0.793 |
| Duration of current episode (month) | 5.97±8.75 | 6.35±9.29 | 5.49±8.08 | 0.552 | 5.99±9.14 | 5.83±6.57 | 0.936 |
| Duration of illness (month) | 47.21±72.88 | 49.88±66.60 | 43.93±80.32 | 0.624 | 44.10±61.08 | 63.13±116.66 | 0.243 |
| BMI | 22.21±2.98 | 21.83±2.72 | 22.68±3.23 | 0.083 | 22.12±2.88 | 22.68±3.45 | 0.402 |
| First episode | 63 (42.9 %) | 34 (42.0 %) | 29 (43.9 %) | 0.811 | 53 (43.1%) | 10 (41.7 %) | 0.897 |
| Recurrence | 84 (57.1 %) | 47 (58.0) | 37 (56.1 %) | 70 (56.9 %) | 14 (58.3 %) | ||
| Dose (mg/day, 2-6 w) | |||||||
| Citalopram | 27.41±9.84 | 26.45±9.50 | 0.709 | 26.80±9.57 | 27.50±10.35 | 0.850 | |
| Escitalopram | 14.81±5.04 | 14.57±5.05 | 0.825 | 15.07±5.03 | 13.13±4.79 | 0.162 | |
*p<0.05, the difference is statistically significant
Table 1: Demographic and clinical characteristics of patients
The two SNPs were in Hardy-Weinberg equilibrium. The allele and genotype frequencies of the two polymorphisms are shown in Table 2. There was a significant association in genotype frequencies (χ2=6.979, p=0.031, correction p=0.062) not in allele frequencies (p>0.05) of Val66Met between responders and non-responders. For further analysis, homozygous GG exhibited a higher response to genotypes with A (GA and AA; χ2=5.218, p=0.022, correction p=0.044). There were no differences in allele or genotype frequencies of Val66Met between remitters and nonremitters. For C270T, there were no differences in allele and genotype frequencies between remitters and non-remitters or between responders and nonresponders (p>0.05).
| Genotype | χ2 | p | Allele | χ2 | p | ||||
|---|---|---|---|---|---|---|---|---|---|
| Val66Met | GG | GA | AA | G | A | ||||
| Remitter (n=81) | 20 | 49 | 12 | 1.549 | 0.461 | 89 | 73 | 1.214 | 0.271 |
| Non-remitter (n=66) | 12 | 40 | 14 | 64 | 68 | ||||
| Response (n=123) | 31 | 69 | 23 | 6.979 | 0.031* | 131 | 115 | 0.886 | 0.347 |
| Non-response (n=24) | 1 | 20 | 3 | 22 | 26 | ||||
| C270T | CC | CT | C | T | |||||
| Remitter (n=81) | 71 | 10 | 2.093 | 0.148 | 152 | 10 | 1.907 | 0.167 | |
| Non-remitter (n=66) | 52 | 14 | 118 | 14 | |||||
| Response (n=123) | 102 | 21 | 0.064 | 0.801 | 225 | 21 | 0.058 | 0.809 | |
| Non-response (n=24) | 21 | 3 | 45 | 3 | |||||
*p<0.05, the difference is statistically significant
Table 2: The genotype and allele analysis of SNPs with response and remission status
With frequencies >0.03 in our population, the two SNPs generated 4 common haplotypes with remission status and 3 haplotypes with response status. However, no haplotypes showed significant associations with remission and response status after antidepressant treatment in patients with MDD (all p>0.05).
For the subgroup analysis, the 147 patients were divided according to sex (male, n=59 and female, n=88). The results are shown in Table 3. Among females, the C allele and CC genotype of C270T were significantly related to higher remission over 6 w (C/T, p=0.027, correction p=0.054; CC/TT, p=0.023, correction p=0.046). A significant association for Val66Met genotypes in females but not in males was noted between responders and non-responders (χ2=7.057, p=0.029, correction p=0.058), and females who were homozygous (GG+AA) showed a better response than those who were heterozygous (GA; p=0.040, correction p=0.080).
| Val66Met | C270T | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GG | GA | AA | χ2 | p | CC | CT | χ2 | p | ||
| Male (n=59) | remitters | 8 | 18 | 2 | 2.744 | 0.254 | 21 | 7 | 0.273 | 0.601 |
| non-remitters | 5 | 20 | 6 | 25 | 6 | |||||
| responders | 12 | 27 | 6 | 2.880 | 0.237 | 34 | 11 | - | 0.713a | |
| non-responders | 1 | 11 | 2 | 12 | 2 | |||||
| Female (n=88) | remitters | 12 | 31 | 10 | 0.239 | 0.887 | 50 | 3 | - | 0.023*,a |
| non-remitters | 7 | 20 | 8 | 27 | 8 | |||||
| responders | 19 | 42 | 17 | 7.057 | 0.029* | 68 | 10 | - | 1.000a | |
| non-responders | 0 | 9 | 1 | 9 | 1 | |||||
| First episode (n=63) | remitters | 9 | 22 | 3 | 2.804 | 0.246 | 32 | 2 | 6.870 | 0.009* |
| non-remitters | 6 | 16 | 7 | 20 | 9 | |||||
| responders | 14 | 31 | 8 | 1.470 | 0.480 | 43 | 10 | - | 0.676a | |
| non-responders | 1 | 7 | 2 | 9 | 1 | |||||
| Recurrence (n=84) | remitters | 11 | 27 | 9 | 0.717 | 0.699 | 39 | 8 | 0.195 | 0.659 |
| non-remitters | 6 | 24 | 7 | 32 | 5 | |||||
| responders | 17 | 38 | 15 | 10.312 | 0.006* | 59 | 11 | - | 1.000a | |
| non-responders | 0 | 13 | 1 | 12 | 2 | |||||
aFisher's exact test; *p<0.05, the difference is statistically significant
Table 3: Subgroup analysis of SNPs with response and remission status
Patients were divided into first-episode (n=63) and recurrent-episode (n=84) groups according to the number of depression episodes (Table 3). The C-allele and CC genotype of C270T were related to higher remission over 6 w in first-episode patients (C/T, p=0.023, correction p=0.046; CC/TT, χ2=6.870, p=0.009, correction p=0.018). Among recurrentepisode patients, a significant association was found across the three genotypes of Val66Met between responders and non-responders (χ2=10.312, p=0.006, correction p=0.012), and recurrent patients who were homozygous (GG+AA) showed a better response than those who were heterozygous (GA; χ2=7.277, p=0.007, correction p=0.014). There were no differences in other subgroups for Val66Met and C270T between remitters and non-remitters or between responders and non-responders.
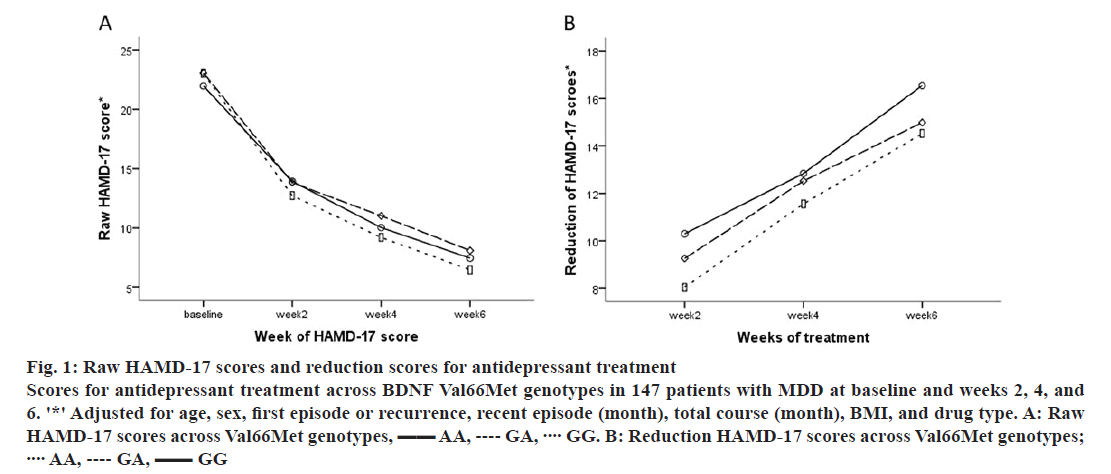
There were no differences in HAMD-17 raw scores over 6 w across Val66Met genotypes (all p>0.05; fig. 1A). However, a trend in HAMD-17 change scores over 6 w was observed across Val66Met genotypes (GG>GA>AA; fig. 1B). In addition, there were no differences across C270T genotypes in raw or change HAMD-17 scores over time (p>0.05). In analyses stratified by gender and depression status, there were no differences in HAMD-17 total or change scores across genotypes of Val66Met and C270T (all p>0.05).
Figure 1: Raw HAMD-17 scores and reduction scores for antidepressant treatment
Scores for antidepressant treatment across BDNF Val66Met genotypes in 147 patients with MDD at baseline and weeks 2, 4, and 6. '*' Adjusted for age, sex, first episode or recurrence, recent episode (month), total course (month), BMI, and drug type. A: Raw HAMD-17 scores across Val66Met genotypes,  AA,
AA,  GA,
GA, 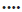 GG. B: Reduction HAMD-17 scores across Val66Met genotypes;
GG. B: Reduction HAMD-17 scores across Val66Met genotypes; 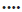 AA,
AA, 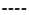 GA,
GA, 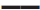 GG
GG
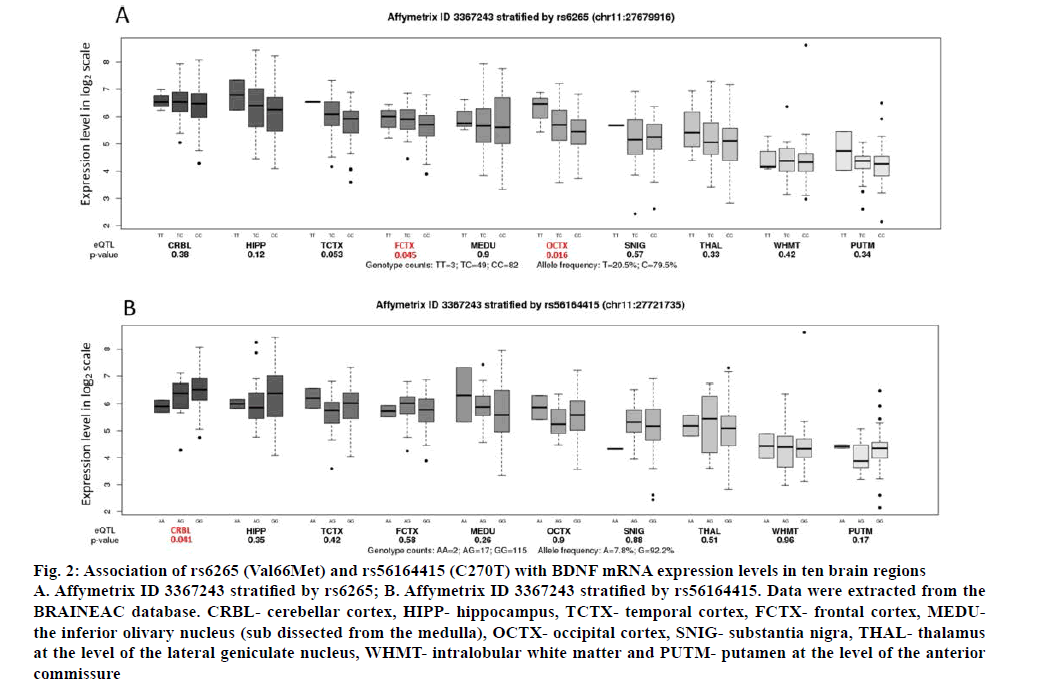
Using BRAINEAC data, we found that Val66Met genotypes differentially affect BDNF gene expression within the frontal cortex (p=0.045, correction p=0.090) and the occipital cortex (p=0.016, correction p=0.032; fig. 2A). In these regions, individuals with the GG genotype show significantly lower expression of BDNF. In addition, it was also found that C270T genotypes differentially affect BDNF gene expression within the cerebellar cortex (p=0.041, correction p=0.082). Individuals with the GG genotype show significantly higher expression of BDNF than other two genotypes (AA+GA; fig. 2B). The Val66Met was also identified as eQTLs for BDNF (p=0.0014, correction p=0.0028) using blood eQTL browser, however, the Z-score of –3.21 indicated that the G allele was associated with higher BDNF expression in the blood opposite to what we found using BRAINEAC (fig. 2B). The C270T across BDNF was not cis-eQTLs in this database.
Figure 2: Association of rs6265 (Val66Met) and rs56164415 (C270T) with BDNF mRNA expression levels in ten brain regions
A. Affymetrix ID 3367243 stratified by rs6265; B. Affymetrix ID 3367243 stratified by rs56164415. Data were extracted from the BRAINEAC database. CRBL- cerebellar cortex, HIPP- hippocampus, TCTX- temporal cortex, FCTX- frontal cortex, MEDUthe inferior olivary nucleus (sub dissected from the medulla), OCTX- occipital cortex, SNIG- substantia nigra, THAL- thalamus at the level of the lateral geniculate nucleus, WHMT- intralobular white matter and PUTM- putamen at the level of the anterior commissure
In this study, it was found that BDNF Val66Met and C270T polymorphisms were associated with the therapeutic response to citalopram/escitalopram in Chinese patients with MDD. To our knowledge, this is the first study to investigate the association between C270T and response to antidepressant drug treatment.
Val66Met is a functional non-synonymous single nucleotide polymorphism that has been extensively investigated in the context of various antidepressants, including fluoxetine[14], mirtazapine[16], and venlafaxine[29]. Several studies have examined the association between Val66Met with citalopram/ escitalopram, but the results are inconsistent. An imaging study found that Met (A) allele carriers of BDNF Val66Met had lower fractional anisotropy values in the left uncinate fasciculus relative to Val/ Val (GG) individuals, reflecting a lower percent change of depression severity after 8 w of treatment with citalopram or quetiapine XR[30]. Lanctôt et al. found GG homozygotes of the BDNF polymorphism showed a greater percent change of HAMD-17 scores in response to citalopram treatment[31]. In this study, it was found that patients who were homozygous GG carriers of Val66Met showed a positive association with response as compared to genotypes with A (GA and AA). Thus, the results of our study are consistent with these previous findings. In addition, clinical reports[32] and animal experiments[33] have shown that the Met/Met (AA) genotype was associated with worse response to fluoxetine, which is an SSRI like citalopram/escitalopram. These findings confirm that the GG genotype is linked to better response to citalopram/escitalopram.
Some studies found no association between Val66Met and response to antidepressant treatment or even the opposite trends. For instance, Choi et al. found M allele carriers responded better to citalopram treatment in a Korean population[20], which may indicate ethnic differences. Moreover, Alexopoulos et al. found depressed older Caucasian BDNF Met carriers had a higher remission rate than Val/Val (GG) homozygotes after 12 w of treatment with escitalopram[19]. Similarly, Kang et al. found patients carrying the Met allele (A) who were treated with escitalopram had a higher rate of remission than non-carriers[34]. The reasons for these different findings may be that the former study population consisted of geriatric depression patients, whereas the latter included MDD patients with acute coronary syndrome. In addition, another study has reported that polymorphisms of the BDNF gene are not associated with the response to escitalopram[21]; this may be due to the relatively small sample size in this study.
In the present investigation it was found that homozygous (GG+AA) of Val66Met showed a better response to citalopram/escitalopram than those who were heterozygous (GA) among females and recurrent MDD patients. A meta-analysis has demonstrated that the BDNF gene influences MDD in a gender-specific way, and the level of gender differences in gene environment interaction, of sexual dimorphism of the hippocampus, or of gender-related differences in symptoms of depression may explain this phenomenon[35]. Other studies have showed that Val66Met polymorphism interacted with gender to influence the serum BDNF level[36], cortisol responses to mental stress[37], or bimanual motor control in healthy humans[38]. The reason why the Val66Met homozygous among females and recurrent MDD patients exhibited a better response to antidepressant than the heterozygous patients, need further exploration.
The association between C270T and antidepressant treatment response had not been previously reported. In our study, for the first time, we found that the C allele and CC genotypes of C270T were significantly associated with higher remission in females and in first-episode MDD patients. A previous study reported no relationship between C270T polymorphism and response to ECT in MDD patients, however, the CC genotype of C270T could predict good response within psychotic and late-onset depression subgroups[26]. It was consistent with our findings in a sense. In another study, Zhang et al. found no association between C270T genotypes and paroxetine treatment after 8 w in Alzheimer’s disease-related depression, a possible explanation for such discrepancy was disease characteristics[39]. Moreover, the frequency of the T allele was quite low and the minor allele frequency was 0.03 for Chinese, indicating the possibility of type II error due to a lack of statistical power[40]. Thus, further studies were required to verify the results.
We found that the Val66Met was identified as eQTLs for BDNF in both of the brain and blood databases. The conclusions, however, are not consistent. The G allele was associated with higher BDNF expression in the blood opposite to what we found using BRAINEAC. Therefore, this SNP is cis-eQTLs for BDNF, but there appears to be tissue-specific directionality of associations with the SNP. In addition, we found individuals with the CC genotype of C270T show significantly higher expression of BDNF than do either those with the CT genotype or those with the TT genotype. These results may support that these two SNPs of BDNF may influence its expression.
There were several limitations to our study. First, the sample size didn’t derive from calculation and was relatively small, especially for subgroup analyses, which limits the generalizability of the findings. Second, only Chinese participants were included into this study for ethnic homogeneity. Various frequencies of Val66Met and C270T polymorphisms in other ethnic populations should be taken into consideration when evaluating antidepressant efficacy. Third, several positive findings are modest and cannot survive after correction for multiple comparisons. Fourth, further studies were not performed to verify our findings. Thus, the datum is not replicated at any level of outcome and not graduated for the different genotypes, which makes it less clearly interpretable. Moreover, there were no available blood concentrations of citalopram/escitalopram in our study. Finally, some confounding factors were not considered in the study including dietary habits, lifestyle, social support, and economic conditions that may have influenced the response to antidepressant treatment.
In conclusion, the present study found that BDNF Val66Met and C270T polymorphisms were associated with the antidepressant treatment and may predict the response to citalopram/escitalopram in MDD patients in the Chinese population. These findings could help to devise a personalized treatment strategy prior to antidepressant treatment. Confirmation of these findings across other populations and exploration of the BDNF polymorphisms effect in pharmacogenetics may lead to novel strategies in the development of antidepressants.
Conflict of interest:
The authors declare no conflict of interest.
Acknowledgements:
This work was supported by National Major Project for IND (2018ZX09734005), Collaborative Innovation Center for Translational Medicine at Shanghai Jiao Tong University School of Medicine (TM201506, TM201624) and Clinical Research Center, Shanghai Jiao Tong University School of Medicine (DLY201620).
References
- Rush AJ, Trivedi MH, Wisniewski SR, Nierenberg AA, Stewart JW, Warden D, et al. Acute and longer-term outcomes in depressed outpatients requiring one or several treatment steps: a STAR*D report. Am J Psychiatry 2006;163:1905-17.
- Franchini L, Serretti A, Gasperini M, Smeraldi E. Familial concordance of fluvoxamine response as a tool for differentiating mood disorder pedigrees. J Psychiatr Res 1998;32:255-9.
- Uher R, Perroud N, Ng MY, Hauser J, Henigsberg N, Maier W, et al. Genome-wide pharmacogenetics of antidepressant response in the GENDEP project. Am J Psychiatry 2010;167:555-64.
- Castrén E, Kojima M. Brain-derived neurotrophic factor in mood disorders and antidepressant treatments. Neurobiol Dis 2017;97:119-26.
- Binder DK, Scharfman HE. Brain-derived neurotrophic factor. Growth Factor 2004;22:123-31.
- Zagrebelsky M, Korte M. Form follows function: BDNF and its involvement in sculpting the function and structure of synapses. Neuropharmacology 2013;76:628-38.
- Ma M, Ren Q, Yang C, Zhang JC, Yao W, Dong C, et al. Adjunctive treatment of brexpiprazole with fluoxetine shows a rapid antidepressant effect in social defeat stress model: Role of BDNF-TrkB signaling. Sci Rep 2016;19;39209.
- Lepack AE, Bang E, Lee B, Dwyer JM, Duman RS. Fast-acting antidepressants rapidly stimulate ERK signaling and BDNF release in primary neuronal cultures. Neuropharmacology 2016;111:242-52.
- Chiou YJ, Huang TL. Serum Brain-Derived Neurotrophic Factors in Taiwanese Patients with Drug-Naïve First-Episode Major Depressive Disorder: Effects of Antidepressants. Int J Neuropsychopharmacol 2016;20(3):213-8.
- Hashimoto T, Shiina A, Hasegawa T, Kimura H, Oda Y, Niitsu T, et al. Effect of mirtazapine versus selective serotonin reuptake inhibitors on benzodiazepine use in patients with major depressive disorder: a pragmatic, multicenter, open-label, randomized, active-controlled, 24-week trial. Ann Gen Psychiatry 2016;19:15-27.
- Chen B, Dowlatshahi D, MacQueen GM, Wang JF, Young LT. Increased hippocampal BDNF immunoreactivity in subjects treated with antidepressant medication. Biol Psychiatry 2001;50:260-5.
- Mössner R, Daniel S, Albert D, Heils A, Okladnova O, Schmitt A, et al. Serotonin transporter function is modulated by brain-derived neurotrophic factor (BDNF) but not nerve growth factor (NGF). Neurochem Int 2000;36:197-202.
- Vaidya VA, Marek GJ, Aghajanian GK, Duman RS. 5-HT2A receptor-mediated regulation of brain-derived neurotrophic factor mRNA in the hippocampus and the neocortex. J Neurosci 1997;17:2785-95.
- Zou YF, Wang Y, Liu P, Feng XL, Wang BY, Zang TH, et al. Association of brain-derived neurotrophic factor genetic Val66Met polymorphism with severity of depression, efficacy of fluoxetine and its side effects in Chinese major depressive patients. Neuropsychobiology 2010;61:71-8.
- Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell 2003;112:257-69.
- Katsuki A, Yoshimura R, Kishi T, Hori H, Umene-Nakano W, Ikenouchi-Sugita A, et al. Serum levels of brain-derived neurotrophic factor (BDNF), BDNF gene Val66Met polymorphism, or plasma catecholamine metabolites, and response to mirtazapine in Japanese patients with major depressive disorder (MDD). CNS Spectr 2012;17:155-63.
- Xu G, Lin K, Rao D, Dang Y, Ouyang H, Mai G, et al. Brain-derived neurotrophic factor gene polymorphism (Val66Met) and the early response to antidepressant in Chinese Han population. Psychiatr Genet 2012;22:214-5.
- Brøsen K, Naranjo CA. Review of pharmacokinetic and pharmacodynamic interaction studies with citalopram. Eur Neuropsychopharmacol 2001;11:275-83.
- Alexopoulos GS, Glatt CE, Hoptman MJ, Kanellopoulos D, Murphy CF, Kelly RE Jr, et al. BDNF val66met polymorphism, white matter abnormalities and remission of geriatric depression. J Affect Disord 2010;125:262-8.
- Choi MJ, Kang RH, Lim SW, Oh KS, Lee MS. Brain-derived neurotrophic factor gene polymorphism (Val66Met) and citalopram response in major depressive disorder. Brain Res 2006;1118:176-82.
- Rajewska-Rager A, Skibińska M, Szczepankiewicz A, Kapelski P, Dmitrzak-Weglarz M, Leszczyńska-Rodziewicz A, et al. Association between polymorphisms of Val66Met in the BDNF gene and the response to escitalopram and nortriptyline treatment in the light of the neurodevelopmental hypothesis of depression. Psychiatr Pol 2008;42:915-23.
- >Guo XJ, Hang WH, Guo LY. A family based association study of BDNF C270T polymorphism with schizophrenia. J Nerv Ment Dis 2011;37:568-70.
- Watanabe Y, Nunokawa A, Someya T. Association of the BDNF C270T polymorphism with schizophrenia: updated meta-analysis. Psychiatry Clin Neurosci 2013;67:123-5.
- Ji H, Dai D, Wang Y, Jiang D, Zhou X, Lin P, et al.Association of BDNF and BCHE with Alzheimer's disease: Meta-analysis based on 56 genetic case-control studies of 12 563 cases and 12 622 controls. Exp Ther Med 2015;9:1831-40.
- Cheng L, Ge Q, Sun B, Yu P, Ke X, Lu Z. Polyacrylamide gel-based microarray: a novel method applied to the association Study between the polymorphisms of BDNF gene and autism. J Biomed Nanotechnol 2009;5:542-50.
- Huuhka K, Anttila S, Huuhka M, Leinonen E, Rontu R, Mattila K, et al. Brain-derived neurotrophic factor (BDNF) polymorphisms G196A and C270T are not associated with response to electroconvulsive therapy in major depressive disorder. Eur Arch Psychiatry Clin Neurosci 2007;257:31-5.
- Ramasamy A, Trabzuni D, Guelfi S, Varghese V, Smith C, Walker R, et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci 2014;17:1418-28.
- Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet 2013;45:1238-43.
- Shen X, Qian M, Yuan Y, Sun J, Zhong H, Yang J, et al. Research on association of BDNF gene Val66Met polymorphism with efficacy of antidepressants and plasma BDNF level. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2014;31:196-200.
- Tatham EL, Hall GB, Clark D, Foster J, Ramasubbu R. The 5-HTTLPR and BDNF polymorphisms moderate the association between uncinate fasciculus connectivity and antidepressants treatment response in major depression. Eur Arch Psychiatry Clin Neurosci 2017;267:135-47.
- Lanctôt KL, Rapoport MJ, Chan F, Rajaram RD, Strauss J, Sicard T, et al. Genetic predictors of response to treatment with citalopram in depression secondary to traumatic brain injury. Brain Inj 2010;24:959-69.
- Chi MH, Chang HH, Lee SY, Lee IH, Gean PW, Yang YK, et al. Brain derived neurotrophic factor gene polymorphism (Val66Met) and short-term antidepressant response in major depressive disorder. J Affect Disord 2010;126:430-5.
- Bath KG, Jing DQ, Dincheva I, Neeb CC, Pattwell SS, Chao MV, et al. BDNF Val66Met impairs fluoxetine-induced enhancement of adult hippocampus plasticity. Neuropsychopharmacology 2012;37:1297-304.
- Kang RH, Chang HS, Wong ML, Choi MJ, Park JY, Lee HY, et al. Brain-derived neurotrophic factor gene polymorphisms and mirtazapine responses in Koreans with major depression. J Psychopharmacol 2010; 24:1755-63.
- Verhagen M, van der Meij A, van Deurzen PA, Janzing JG, Arias-Vásquez A, Buitelaar JK, et al. Meta-analysis of the BDNF Val66Met polymorphism in major depressive disorder: effects of gender and ethnicity. Mol Psychiatry 2010;15:260-71.
- Elfving B, Buttenschøn HN, Foldager L, Poulsen PH, Andersen JH, Grynderup MB, et al.Depression, the Val66Met polymorphism, age, and gender influence the serum BDNF level. J Psychiatr Res 2012;46:1118-25.
- Jiang R, Babyak MA, Brummett BH, Siegler IC, Kuhn CM, Williams RB. Brain-derived neurotrophic factor (BDNF) Val66Met polymorphism interacts with gender to influence cortisol responses to mental stress. Psychoneuroendocrinology 2017;79:13-9.
- Smolders R, Rijpkema M, Franke B, Fernández G. BDNF Val66Met polymorphism interacts with sex to influence bimanual motor control in healthy humans. Brain Behav 2012;2:726-31.
- Zhang L, Fang Y, Zeng Z, Lian Y, Wei J, Zhu H, et al. BDNF gene polymorphisms are associated with Alzheimer's disease-related depression and antidepressant response. J Alzheimer’s Dis 2011;26:523-30.
- Kunugi H, Ueki A, Otsuka M, Isse K, Hirasawa H, Kato N, et al. A novel polymorphism of the brain-derived neurotrophic factor (BDNF) gene associated with late-onset alzheimer's disease. Mol Psychiatry 2001;6:83-6.