- *Corresponding Author:
- V. B. Tatipamula
K L College of Pharmacy, Koneru Lakshmaiah Education Foundation, Vaddeswaram-522 502, India
E-mail: vinaybharadwajt@gmail.com
| Date of Submission | 22 January 2019 |
| Date of Revision | 03 April 2019 |
| Date of Acceptance | 19 July 2019 |
| Indian J Pharm Sci 2019;81(5):815-823 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
For the first time, 89 chemical constituents were identified by gas chromatography-mass spectrometry analysis of ethanol extract of Clathria baltica. The chemical examination of ethanol extract of Clathria baltica yielded three known metabolites, 1, 2 and 3, for first time from the marine algae Clathria baltica. All three metabolites and the ethanol extract of Clathria baltica were screened against cancer cell lines, MCF-7, DLD-1, HeLa, FADU, A549, and SKOV3, and one normal human cell line using sulforhodamine B assay and doxorubicin as the standard. Among all isolates, compound 3 showed significant growth inhibition of MCF-7, DLD-1 and FADU with IC50 values of 26.5, 15.5 and 16.5 µg/ml, respectively. Whereas, all the metabolites and the extract exerted lower growth inhibitory effect on the normal human cell line tested. This is the first in vitro gas chromatography-mass spectrometry analysis, as well as, cytotoxicity report on marine algae Clathria baltica.
Keywords
GC-MS analysis, isolation, characterization, anticancer activity, sulforhodamine B assay
Globally, cancer still remains an aggressive killer that severely effecting the human population[1]. According to the report of the International Agency for Research on Cancer (IARC), it was documented that within 5 y of diagnosis - 9.6 million cancer deaths from 33.7 million people living with cancer has taken place and 18.1 million new cancer cases were reported in 2018 across the world[2]. In addition, the IARC is also estimated that about 26 million new cancer cases and 17 million cancer deaths per year may be recorded by 2030[3]. In addition, the present therapies for cancer include chemotherapy and radiotherapy, which cause lot of strain to patients and ultimately damage their health[4]. Hence, researchers are focussing on developing new, safer and effective drugs to treat cancer.
From the dawn of ancient medicine, natural products have been used to treat many deadly diseases like cancer. In addition, from the past 30 y, natural products have received great attention in attaining potential as well as novel cancer therapeutic agents[5,6]. Moreover, the mechanism of action of inhibiting various stages of tumorigenesis by most of the natural products were also well-documented[6].
In folklore, marine algae (seaweeds) are used as food, feed, fuel and livelihood in Asian countries. Based on the pigmentation, marine algae are classified as red, brown and green algae. From the past few decades, marine algae have been highly acknowledged to possess noticeable pharmacological activities, which include antineoplastic, antimicrobial and antiviral activities due to their specific functional compounds (which are not available in other plants)[7-10]. Particularly, marine algae of Chara genus (family: Characeae- charophyte green algae) reported to possess and antioxidant enzyme, allelophatic activities and lipid peroxidation capabilities along with high levels of photosynthetic pigment content[11]. Earlier reports from our laboratory showed phytochemical analysis along with antioxidant activities of marine algae Chara baltica[12]. Till date, complete chemical and pharmacological evaluation has not been reported on C. baltica. Based on the aforementioned properties of Chara genus, as well as, C. baltica, C. baltica specimens (in July 2018) were subjected to gas chromatography-mass spectrometry (GC-MS) analysis, chemical and cytotoxicity evaluation, which are reported in this research communication.
Materials and Methods
The specimens of seaweed Clathira baltica was collected near Korangi coast, Kakinada, Andhra Pradesh, India at a depth of 8-10 ft (16°80’N and 82°08’E with 3 m elevation) on 3 July, 2018. The seaweed was authenticated in the Botany Department, Andhra University, Visakhapatnam, India. A voucher specimen was deposited in the Marine Organisms Museum in the University College of Pharmaceutical Sciences, Andhra University, Visakhapatnam, India (No. AU-SW-2018-947). All chemicals used in the present study were of analytical grade. Streptozotocin was from Himedia Laboratories Pvt. Ltd. (Mumbai, India); glibenclamide from the Aventis Pharma Ltd. (Mumbai, India); and rat feed from the Hindustan Lever Ltd. (Mumbai, India).
Extraction and isolation:
The seaweed was cleansed from extraneous substance and stored in ethanol-water (9:1) at the site of collection. The crude plant material (100 g) was extracted at least thrice with ethanol-water and concentrated at a reduced pressure and the combined extracts were lyophilized to obtain ethanol extract of C. baltica; as a reddish brown gummy residue (3.25 g). About 2 g of ethanol extract was subjected to column chromatography, afforded three fractions. Fraction I (510 mg) and fraction II (800 mg) were subjected to repeated column chromatography using hexane and ethyl acetate as a solvent system yielded 1 (240 mg) as a colourless liquid and 2 (330 mg) as a pale greenish liquid, respectively. Fraction III (400 mg) were purified by recrystallization technique using hexane and acetone as solvents yielded 3 (200 mg) as a greenish crystals.
Compound 1 (dihydrofuran-2,5-dione), colorless crystals, molecular formula: C4H4O3; yield: 240 mg; Rf: 0.7 (hexane:ethyl acetate, 1:9); 1H NMR (CDCl3, 400 MHz) δ 4.00 (4H, s, 2,3-CH2); 13C NMR (CDCl3, 400 MHz) δ 22.16 (C-2/C-3), 170.24 (C-1/C-4); LCMS m/z: 100.
Compound 2 (3-hydroxy-4,4-dimethyldihydrofuran- 2(3H)-one), pale greenish liquid, C6H10O3; yield: 330 mg; Rf: 0.6 (hexane:ethyl acetate, 1:4); 1H NMR (CDCl3, 400 MHz) δ 1.00 (6H, s, 5,6-CH3), 2.31 (1H, s, 2-CH), 3.86-3.88 (1H, d, J= 8 Hz, 4a-CH2), 4.04- 4.06 (1H, d, J= 8 Hz, 4a-CH2); 4.11 (1H, s, 2-OH); 13C NMR (CDCl3, 400 MHz) δ 23.15 (C-5/C-6), 43.40 (C-3), 80.44 (C-2), 87.23 (C-4), 182.11 (C-1); LC-MS m/z: 130.
Compound 3 (2,4-dihydroxy-2,5-dimethylfuran- 3(2H)-one), pale greenish crystals, molecular formula: C6H8O4; yield: 200 mg; Rf: 0.4 (hexane:ethyl acetate, 1:4); melting point: 163-164°; 1H NMR (CDCl3, 400 MHz) δ 1.87 (3H, s, 6-CH3), 2.05 (3H, s, 5-CH3), 3.62 (1H, s, 3-OH), 3.92 (1H, s, 1-OH); 13C NMR (CDCl3, 400 MHz) δ 12.14 (C-6), 22.41 (C-5), 99.46 (C-1), 139.32 (C-3), 159.75 (C-4), 192.74 (C-2); CHNS analysis: found C-50.00, H-5.60 (%), calcd. C, 50.46, H, 5.54 (%); LC-MS m/z: 144.
Gas chromatography-mass spectrometry (GC-MS) analysis:
The phytochemical investigation of ethanol extract of C. baltica was performed on a GC-MS system (GCMSQP2010 Plus, Shimadzu, Europe). Experimental parameters set for the GC-MS were, column oven temperature- 50°; injection temperature- 250°; injection mode- split; flow control mode- linear velocity; pressure- 29.7 kPa; total flow- 7.3 ml/min; column flow- 0.72 ml/min; linear velocity- 30.7 cm/s; purge flow- 3.0 ml/min; split ratio- 5.0; oven temperature program- 50° with rate of increase of 10°/min, 50-300° with rate of increase of 7.5°/min, hold time of 2.0 min; equilibrium time- 1.0 min; ion source temperature- 210°; interface temperature- 250°; solvent cut time- 3.0 min; detector gain- 0.99+0.20 kV; threshold- 1000; start time- 4.0 min; end time- 37.30 min; ACQ modescan; event time- 0.50 s; scan speep- 2500; start m/z- 34.0 and end m/z- 1090.00.
Cytotoxicity assay:
MCF-7 (breast), DLD-1 (colon), HeLa (cervical), FADU (head and neck), A549 (lung), SKOV3 (ovary) and normal human mammary epithelial (NHME) were kindly provided by National Centre for Cell Science, Pune. The cancer cells were maintained in minimum essential medium (MEM), containing 10 % fetal calf serum (FBS), 5 % mixture of 100 units penicillin and 100 μg/ml streptomycin in presence of 5 % carbon dioxide in an incubator with 90 % humidity at 37° for 72 h.
Cancer cell lines were maintained in MEM (adjusted to 10 % FBS, 1.5 g/ml NaHCO3, 0.1 mM MEM non-essential amino acids and 1 mM sodium pyruvate). Three days prior to performing assay, the cells were washed with sterilized phosphate-buffered saline and grown using MEM media (supplemented with 0.25 % trypsin in versene-EDTA and 10 % FBS) and mixed to obtain homogeneous suspension of cells. The suspension was taken in a sterilized polypropylene tube and the cell count in each well was determined in a hemacytometer chamber under a microscope using 0.4 % trypan blue solution. The minimal seed density must be 1.9×104 cells per well. All crude extracts and the standard were dissolved in dimethyl sulfoxide (DMSO) to 100 mg/ml and 10 μg/ml, respectively. Doxorubicin and DMSO were used as a standard and control, respectively.
The sulforhodamine B (SRB) assay[13] is based on the estimation of cellular protein content. The prepared samples were taken in 96-well tissue-culture plate and added 190 μl screened ideal cell suspension and mixed occasionally and incubate at 37° with 5 % CO2 and 90 % relative humidity for 3 h. Then 100 μl cold TCA was added to each well and incubated at 4° for 1 h. After that the plates were gently washed using water, dried using blow dryer and air-dried at room temperature. To each completely dried well, 100 μl of 0.057 % SRB solution was added, kept aside for 30 min and quickly rinse with 1 % acetic acid. To the dried plate, 200 μl of 10 mM Tris base (pH 10.5) solution was added, shake for 5 min and measure the OD at 510 nm. The blank contains only medium while the control has only cancer cells with no test samples. Percent growth inhibition was calculated using the formula, growth inhibition (%) = 100–[(S–B)/(C–B)]×100, where S is mean OD value of sample, B is mean OD value of blank, C is mean OD value of control.
Results and Discussion
After the successful extraction of the whole marine algae material with ethanol-water, the dried ethanol extract was subjected to GC-MS analysis. The GCMS chromatogram revealed the presence of various chemical components with different retention times, whereas the MS analyzes the compounds eluted at different times to detect its nature and structure of the compounds. The results pertaining to GC-MS analysis of the ethanol extract lead to the identification of a number of compounds. A total of 86 compounds were identified by GC-MS analysis of ethanol extract, which were tabulated in Table 1.
| S. No | Compound name | Chemical structure | Molecular formula |
|---|---|---|---|
| 1 | (2Z)-1,3-dimethyl-6-oxo-2-hexenyl acetate | 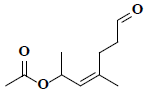 |
C10H16O3 |
| 2 | (2Z)-5-methyl-2-decene | 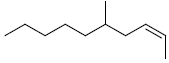 |
C11H12 |
| 3 | (9E)-9-octadecenal | 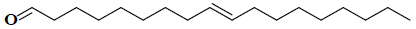 |
C18H34O |
| 4 | (9Z)-9-octadecenamide | 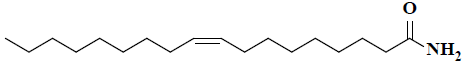 |
C18H35NO |
| 5 | 1-((E)-[(2-methylphenyl)imino]methyl)-2-naphthol | 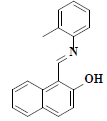 |
C18H15NO |
| 6 | 1,2,5-trimethyl-4-piperidinone thiosemicarbazone | 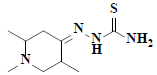 |
C9H18N4S |
| 7 | 1,3-cyclohexanedione | 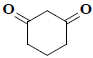 |
C6H8O2 |
| 8 | 1,4-dimethyl-5-oxabicyclo[2.1.0]pentane | 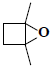 |
C6H10O |
| 9 | 1,5-anhydro-6-deoxyhexo-2,3-diulose | 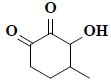 |
C7H10O3 |
| 10 | 10-undecen-1-ylester | 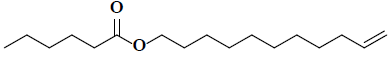 |
C17H32O2 |
| 11 | 1-chloro-4-decyne | 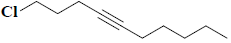 |
C10H17Cl |
| 12 | 1-hexadecylpyridinium chloride monohydrate | 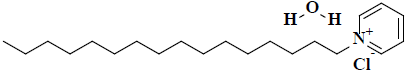 |
C21H40ClNO |
| 13 | 1-Tetradecanol, acetate | 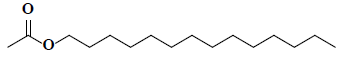 |
C16H32O2 |
| 14 | 1-undecene | 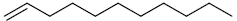 |
C11H22 |
| 15 | 3-hydroxy-4,4-dimethyldihydrofuran-2(3H)-one | 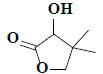 |
C6H10O3 |
| 16 | 2-(benzyloxy)-4-bromo-1,3-butanediol | 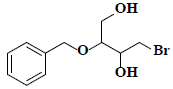 |
C11H15BrO3 |
| 17 | 2,10-dimethylundecane | 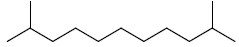 |
C13H28 |
| 18 | 2,2,4-trimethyl-1-pentanol |  |
C8H18O |
| 19 | 2,2-dimethyl-1-phenyl-1-propanol | 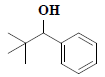 |
C11H16O |
| 20 | 2,2-dioctyl-1-oxohydrazine-1-oxide | 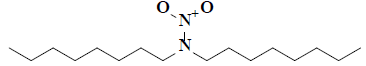 |
C16H34H2O2 |
| 21 | 9,12-Octadecadienoic acid (Z,Z)-, 2,3-bis(acetyloxy)propyl ester |  |
C25H42O6 |
| 22 | 2,3-dihexyl-2-cyclopropene-1-carboxylic acid |  |
C16H28O2 |
| 23 | 2,4,6-trimethyldecane |  |
C13H28 |
| 24 | 2,4-Dihydroxy-2,5-dimethyl-3(2H)-furanone |  |
C6H8O4 |
| 25 | 2,5-furandione | 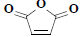 |
C4H2O3 |
| 26 | 2-acetoxytetradecane | 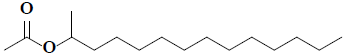 |
C16H32O2 |
| 27 | 2-acetoxytridecane |  |
C15H30O2 |
| 28 | 2-cyclohexen-1-one | 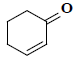 |
C6H8O |
| 29 | 2-cyclohexylethyl acetate | 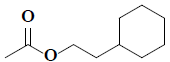 |
C10H18O2 |
| 30 | 2-hydroxy-3-methylbutanoic acid | 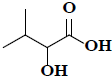 |
C5H10O3 |
| 31 | 2-propylheptan-1-ol | 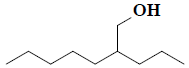 |
C10H22O |
| 32 | 3-(methoxymethoxy)-butanoic acid | 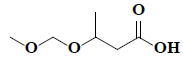 |
C6H12O4 |
| 33 | 3,5,5-trimethyl-4-(3-oxo-1-butenyl)-2-cyclohexen-1-one | 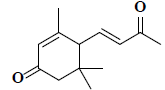 |
C13H18O2 |
| 34 | 3,5-bis(1,1-dimethylethyl)-phenol | 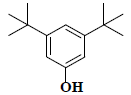 |
C14H22O |
| 35 | 3,6-dimethyl-2-octanone | 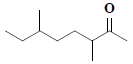 |
C10H20O |
| 36 | 3-acetoxydodecane | 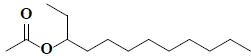 |
C14H28O2 |
| 37 | 3-ethyl-2,5-dimethylhexane | 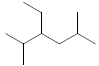 |
C10H22 |
| 38 | 3-hexene-2,5-diol | 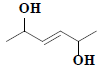 |
C6H12O2 |
| 39 | 3-hydroxy-2,2,6-trimethylcyclohexyl-methyl acetate | 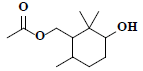 |
C12H22O3 |
| 40 | 3-hydroxydodecanoic acid | 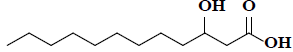 |
C12H24O3 |
| 41 | 3-nonyn-1-ol |  |
C9H16O |
| 42 | 4-acetoxypentadecane | 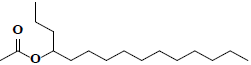 |
C17H34O2 |
| 43 | 4-heptanol | 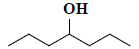 |
C7H16O |
| 44 | 5-(1-benzyl-1h-indol-3-ylmethylene)-1-(2-ethyl-phenyl)-pyrimidine-2,4,6-trione | 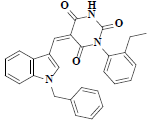 |
C28H23N3O3 |
| 45 | 5-bromo-3-methylpentyl acetate | 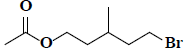 |
C8H15BrO2 |
| 46 | 5-cyclopropylidene-1-pentanol |  |
C8H14O |
| 47 | 7,9-ditert-butyl-1-oxaspiro[4.5]deca-6,9-diene-2,8-dione | 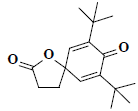 |
C17H24O3 |
| 48 | 9-octadecenamide | 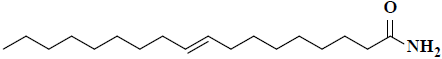 |
C18H35NO |
| 49 | 9-octadecenoic acid | 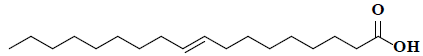 |
C18H34O2 |
| 50 | Acetic acid | 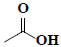 |
C2H4O2 |
| 52 | Araldite | 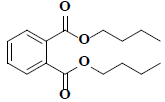 |
C16H22O4 |
| 53 | Benzenemethanol |  |
C7H8O |
| 54 | Benzoic acid |  |
C7H6O2 |
| 55 | Butanoic acid |  |
C4H8O2 |
| 56 | Cis-oleic acid |  |
C18H34O2 |
| 57 | Cyclopentaneundecanoic acid | 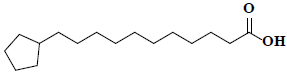 |
C16H30O2 |
| 58 | Dimethyl(ethenyl)propoxysilane | 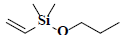 |
C7H16OSi |
| 59 | Di-n-butyl sulphite | 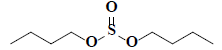 |
C8H18O3S |
| 60 | Di-n-octyl phthalate | 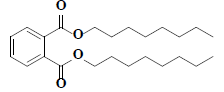 |
C24H38O4 |
| 61 | DL-alanine | 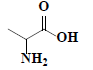 |
C3H7NO2 |
| 62 | E-2-undecen-1-ol |  |
C11H22O |
| 63 | Ethyl-(4E)-2-acetyl-2-methyl-4-hexenoate | 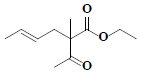 |
C11H18O3 |
| 64 | Ethyl docosanoate |  |
C24H48O2 |
| 65 | Ethyl nonadecanoate | 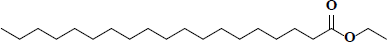 |
C21H42O2 |
| 66 | Hexadecyl acetate | 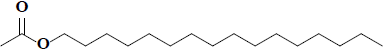 |
C18H36O2 |
| 67 | Methyl-(E)-docos-13-enoate |  |
C23H44O2 |
| 68 | Methyl-(9E)-9-octadecenoate |  |
C19H36O2 |
| 69 | Methyl-3-(3,5-di-tert-butyl-4-hydroxyphenyl)propionate | 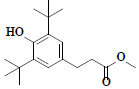 |
C18H28O3 |
| 70 | Methyl-3-isopropyl-6-oxoheptanoate | 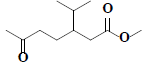 |
C11H20O3 |
| 71 | Methyl-4-hydroxy-3-methyl-2-butenoate | 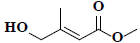 |
C6H10O3 |
| 72 | Methyl heptanoate | 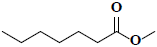 |
C8H16O2 |
| 73 | n-cyclohexyl methacrylamide | 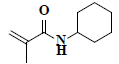 |
C10H17NO |
| 74 | n-decyl acetate | 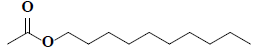 |
C12H24O2 |
| 75 | n-heptadecane |  |
C17H36 |
| 76 | Octadecane | 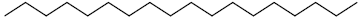 |
C18H38 |
| 77 | Octadecanoic acid | 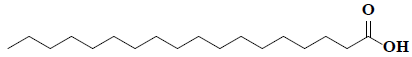 |
C18H38O2 |
| 78 | Octanal dimethyl acetal | 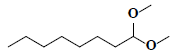 |
C10H22O2 |
| 79 | Palmitic acid | 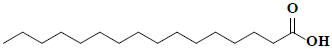 |
C16H32O2 |
| 80 | Palmitic acid ethyl ester |  |
C18H36O2 |
| 81 | Pelargonic acid methyl ester | 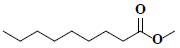 |
C10H20O2 |
| 82 | Pentanoic acid | 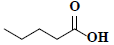 |
C5H10O2 |
| 83 | Phthalic acid | 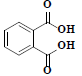 |
C8H6O4 |
| 84 | Tert-nonanethiol |  |
C9H20S |
| 85 | Tetradecane | 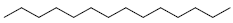 |
C14H30 |
| 86 | Xanthosine | 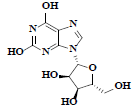 |
C10H12N4O6 |
Table 1: Chemical constitutents identified by GC-MS analysis of ethanol extracts of Chara baltica
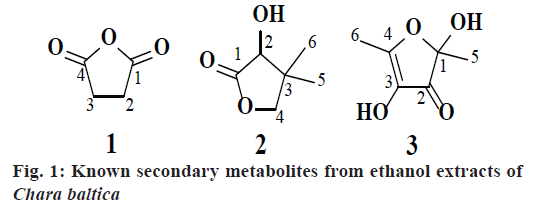
The collected seaweed was extracted with ethanol water at room temperature. The obtained ethanol extract C. baltica was subjected to chromatographic purification led to isolation of three metabolites (1-3, fig. 1). Compound 1 was obtained as a colorless crystals with Rf value 0.7 (hexane:ethyl acetate, 1:9) and the LC-MS ion peak at m/z 100 confirmed the molecular formula as C4H4O3. The 1H and 13C NMR data showed the presence of two methane groups at δH 4.00 (s, 4H) with corresponding carbon signal at δC 22.16 (C-2 and C-3). In addition, the 13C NMR also revealed the presence of dione groups at δC 170.24 (C-1 and C-4). Hence, compound 1 was confirmed after corroboration with the existing literature[14] as a dihydrofuran-2,5-dione (fig. 1).
Compound 2 was obtained as a pale greenish liquid. Based on the LC-MS ion peak at m/z 130 confirmed the molecular formula as C6H10O3. The 1H NMR showed the presence of two singlets for two methyl groups at δH 1.00 (s, 6H) with corresponding carbon signal at δC 23.15 (C-5 and C-6), including one methine group at δH 2.31 (s, 1H) with corresponding carbon signal at δC 80.44 (C-2), and one hydroxyl group at δH 4.11 (s, 1H), additionally, two doublets at δH 3.86-3.88 (d, J=8 Hz, 1H) and 4.04-4.06 (d, J=8 Hz, 1H), which confirmed the presence of methane group at δC 87.23. In addition, the 13C NMR also revealed the presence of dione group at δC 182.11 (C-1). By corroboration with the existing literature compound 2 was found to be 3-hydroxy-4,4- dimethyldihydrofuran-2(3H)-one (fig. 1)[15].
Compound 3 was obtained as a pale greenish crystals, based on the LC-MS ion peak at m/z 144 confirmed the molecular formula as C6H8O4. The 1H NMR showed the presence of four singlets for two methyl groups at δH 1.87 (s, 3H) and 2.05 (s, 3H), and for two hydroxyl groups at δH 3.62 (s, 1H) and 3.92 (s, 1H). The 13C NMR revealed the presence of two methyl group at δC 12.14 (C-6) and 22.41 (C-5), two hydroxyl groups at δC 99.46 (C-1) and 139.32 (C-3), dione group at δC 192.74 (C-2), and one methene carbon signal at δC 159.75 (C-4). Based on the aforementioned data, compound 3 was compared with the existing literature on lichen metabolites and it was confirmed as a 2,4-dihydroxy- 2,5-dimethylfuran-3(2H)-one (fig. 1). Compound 3 was earlier reported from Sysepalum dulcificum[16].
Earlier reports of C. baltica indicated free radical scavenging activity[13]. Based on the report, the isolated metabolites (1-3) were screened along with ethanol extract against six different cancer cell lines using SRB assay with doxorubicin as standard. Initially, compounds (1-3) at 30 μg/ml concentration, extract at 100 μg/ml concentration and doxorubicin at 10 μg/ml concentration were screened against the cell lines tested and percent inhibition of cell growth was tabulated in Table 2.
| Sample | Percent growth inhibition at 30 µg/ml | ||||||
|---|---|---|---|---|---|---|---|
| MCF-7 | DLD-1 | HeLa | FADU | A549 | SKOV3 | NHMEc | |
| 1 | 1.46±0.18 | 5.64±0.65 | 3.97±0.70 | 13.80±1.06 | 21.37±1.94 | 5.69±0.87 | 0.52±0.01 |
| 2 | 7.17±0.24 | 11.91±0.50 | 5.12±0.18 | 16.41±1.26 | 14.57±0.39 | 8.14±1.05 | 1.04±0.16 |
| 3 | 56.86±4.78 | 76.37±5.34 | 22.27±6.23 | 64.08±3.92 | 34.07±4.84 | 46.11±4.74 | 9.40±0.33 |
| Ethanol extract* | 71.53±4.71 | 87.11±3.58 | 40.90±2.57 | 70.46±4.96 | 57.62±5.58 | 48.24±4.44 | 2.91±0.31 |
| Doxorubicin# | 84.40±0.80 | 66.71±0.71 | 90.71±0.13 | 88.34±0.27 | 65.40±0.60 | 77.05±0.22 | 10.08±0.95 |
Mean±SEM values (n=3); #10 µg/ml; *100 µg/ml; cnormal human mammary epithelialium
Table 2: Cytotoxicity studies of compounds 1-3 and ethanol extract of Chara baltica
Further, the samples that caused 50 % or more of cell death were further screened at 5, 10, 20 and 30 μg/ml concentrations for pure compounds; 25, 50, 75 and 100 μg/ml concentrations for extracts; 2.5, 5.0, 7.5 and 10 μg/ml concentrations for doxorubicin. The results were plotted to obtain IC50 values. The lower IC50 value indicated better inhibitory profile against cancer cell lines.
During the initial screening, the isolate 3 and ethanol extract depicted prominent degree of specificity against MCF-7, DLD-1, FADU and A549. Besides, all the samples exhibited very low toxicity towards NHME cell lines (Table 2). From the outcomes of the SRB assay, it is interesting to note that compound 3 (30 μg/ml) and ethanol extract (100 μg/ml) showed prominent degree of specificity against DLD-1 with 76.37±5.34 and 87.11±3.58 % growth inhibition which was found to be better than activity of 66.71±0.71 % shown by the standard at 10 μg/ml (Table 2).
From the Table 3, the IC50 values of 3 and ethanol extract against MCF-7 were 26.5 and 75.0 μg/ml, respectively, whereas standard value was 5.5 μg/ml. In addition, the concentration of 3 and ethanol extract needed for 50 % inhibition of DLD-1 were found to be 15.5 and 61.0 μg/ml, respectively, while for doxorubicin it was 5.4 μg/ml (Table 3).
| Cell line sample | Concentration (µg/ml) | IC50 values (µg/ml) | ||||
|---|---|---|---|---|---|---|
| 5 | 10 | 20 | 30 | |||
| MCF-7 | ||||||
| Compound 3 | 18.09±2.87 | 28.17±2.87 | 38.59±1.74 | 56.86±4.78 | 26.5 | |
| Ethanol extracts** | 25.06±1.78 | 36.58±2.68 | 50.05±3.57 | 71.53±4.71 | 75.0 | |
| Doxorubicin* | 30.87±0.61 | 45.45±0.18 | 67.08±0.11 | 81.25±0.27 | 5.5 | |
| DLD-1 | ||||||
| Compound 3 | 22.55±4.84 | 39.77±2.83 | 57.92±7.78 | 76.37±5.34 | 15.5 | |
| Ethanol extract ** | 29.75±1.77 | 42.57±2.49 | 59.22±1.77 | 87.11±3.58 | 61.0 | |
| Doxorubicin* | 19.75±4.61 | 31.58±6.85 | 44.75±2.84 | 52.57±7.97 | 5.4 | |
| FADU | ||||||
| Compound 3 | 29.68±2.68 | 40.88±4.06 | 54.87±4.54 | 64.08±3.92 | 16.5 | |
| Ethanol extract ** | 26.47±2.97 | 37.18±2.52 | 50.17±3.58 | 70.46±4.96 | 74.5 | |
| Doxorubicin* | 41.01±3.85 | 52.14±2.84 | 68.88±1.77 | 85.55±6.24 | 4.5 | |
| A549 | ||||||
| Ethanol extract ** | 26.47±2.97 | 37.18±2.52 | 50.17±3.58 | 57.62±5.58 | 74.5 | |
| Doxorubicin* | 41.01±3.85 | 52.14±2.84 | 68.88±1.77 | 85.55±6.24 | 4.5 | |
Each value is a mean+SD of % growth inhibition of the cell line tested with n=3; *2.5, 5.0, 7.5 and 10 µg/ml concentrations; **25, 50, 75 and 100 µg/ml concentrations
Table 3: Growth inhibition and IC50 values of compound 3 andethanol extracts of Chara baltica
From the SRB assay of FADU (Table 3) it can be calculated that the IC50 values of 3 and ethanol extract against FADU were 16.5 and 74.5 μg/ml, respectively, whereas standard value was 4.5 μg/ml. Additionally, the concentration of ethanol extract needed for 50 % inhibition of A549 was found to be 74.5 μg/ml, while standard (doxorubicin) was 4.5 μg/ml (Table 3).
Hence, from the overall SRB assay it can be concluded that the key agent responsible for anticancer activity is compound 3. In addition, the growth inhibitory properties of compound 3 is mainly due to the higher levels of oxygenated substituents present in its chemical structure, this higher levels of oxygenated content help in irradiating free radicals that are usually formed during massive cell division of cancer cells, which ultimately lead to the cell death[13,17].
In conclusion, this is the first report of GC-MS analysis, chemical and cytotoxicity profile of marine algae C. baltica. The GC-MS analysis of ethanol extract C. baltica revealed presence of 89 chemical constituents. In addition, the chemical examination of ethanol extract showed the presence of three metabolites 1-3. Additionally, the cytotoxicity studies of ethanol extract indicated specificity towards MCF-7, DLD-1, FADU and A549 without affecting NHME. The SRB assay screening proved that compound 3 has greater cytotoxicity. The results of the current study could be useful for further research on cancer to identify potential bioactive molecules from different aquatic fauna, such as marine algae like C. baltica.
Acknowledgement:
The authors thank the authorities of CDRI-Lucknow, India for providing the cytotoxicity testing facilities to complete present work. The authors wish to thank Dr. G. Mohan Narasimha Rao, Professor of Botany Department, Andhra University, Visakhapatnam, for authenticating the seaweed.
Conflict of interest:
No conflict of interest between any of the authors.
References
- Ochwang’I DO, Kimwele CN, Oduma JA, Gathumbi PK, Mbaria JM, Kiama SG. Medicinal plants used in treatment and management of cancer in Kakamega county Kenya. J Ethnopharmacol 2014;151:1040-55.
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424.
- Solowey E, Lichtenstein M, Sallon S, Paavilainen H, Solowey E, Lorberboum-Galski H. Evaluating Medicinal Plants for Anticancer Activity. Scientific World J 2014;2014:721402.
- Greenwell M, Rahman PK. Medicinal plants: their use in anticancer treatment. Int J Pharm Sci Res 2015;6:4103-12.
- Newman DJ. Natural products as leads to potential drugs: an old process or the new hope for drug discovery. J Med Chem 2008;51(9):2589-99.
- Newman DJ, Cragg GM, Snader KM. Natural products as sources of new drugs over the period 1981-2002. J Nat Prod 2003;66:1022-37.
- Val A, Platas G, Basilio A, Cabello A, Gorrochategui J, Suay I, et al. Screening of antimicrobial activities in red, green and brown macroalgae from Gran Canaria (Canary Islands, Spain). Int Microbiol 2001;4(1):35.
- Patterson GML, Larsen LK, Moore RE. Bioactive natural products from blue-green algae. J Appl Phycol1994;6:151.
- Pietra F. Secondary metabolites from marine microorganisms: bacteria, protozoa, algae and fungi. Achievements and prospects. Nat Prod Rep 1997;14(5):453-64.
- Cabrita MT, Vale C, Rauter AP. Halogenated compounds from marine algae. Mar Drugs 2010;8(8):2301-17.
- Berger J, Schagerl M. Allelopathic activity of Characeae. Hydrobiologia 2003;501(1-3):109.
- Naidu KK, Bharadwaj TV, Alekya K, Lakshmi YB, Sastry VG. Qualitative analysis and free radicals scavenging ability of marine algae, Chara baltica. J Integr Sci 2018;1:1-5.
- Tatipamula VB, Vedula GS, Sastry AVS. Antarvediside A-B from manglicolous lichen Dirinaria consimilis (Stirton) D.D. Awasthi and their pharmacological profile. Asian J Chem 2019;31:805-12.
- Buechi G, Demole E, Thomas AF. Syntheses of 2,5-dimethyl-4-hydroxy-2,3-dihydrofuran-3-one (furaneol), a flavor principle of pineapple and strawberry. J Org Chem 1973;38:123-5.
- Pansare SV, Shinkre BA, Bhattacharyya A. Enantioselective synthesis of α-hydroxy γ-butyrolactones from an ephedrine-derived morpholine-dione. Tetrahedron 2002;58:8985-1.
- Chukwu CJ, Omaka ON, Aja PM. Characterization of 2,5-dimethyl-2,4-dihydroxy-3(2H) furanone, a flavourant principle from Sysepalum dulcificum. Nat Prod Chem Res 2017;5:8.
- Shoemaker M, Hamilton B, Dairkee SH, Cohen I, Campbell MJ. In vitro anticancer activity of twelve Chinese medicinal herbs. Phytother Res 2005;19:649-51.