- *Corresponding Author:
- K. R. Iyer
Department of Pharmaceutical Chemistry, Bombay College of Pharmacy, Kalina, Santacruz (E), Mumbai - 400098, India
E-mail: krishnaiye@gmail.com
| Date of Submission | 17 February 2005 |
| Date of Revision | 29 November 2006 |
| Date of Acceptance | 6 June 2007 |
| Indian J Pharm Sci, 2007, 69 (3): 448-451 |
Abstract
Coumarin has been reported to be a reliable probe for the determination of human microsomal CYP2A6 activity. Coumarin is converted to 7-hydroxycoumarin by CYP2A6. A high pressure liquid chromatographic assay for the estimation of CYP2A6 activity in microsomes was evaluated. A RP C-18 Novapak Waters (15 cm x 3.9 mm, 5 µm) column was used for the assay. The mobile phase composition was methanol : 1% glacial acetic acid (35:65 v/v) (pH~3.1), with a flow rate of 0.6 ml/min, injection volume of 100 µl and detection at 320 nm. The retention times for coumarin and 7-hydroxycoumarin were 8.7 min and 5.3 min, respectively. The limit of detection (LOD) was 0.05 µM, and the limit of quantitation (LOQ) was 0.1 µM, for 7-hydroxycoumarin. The percent coefficient of variation associated with 7-hydroxycoumarin determination after duplicate estimation was found to be in the range 0.03 to 3.6% for buffer matrix and 0.1 to 6.5% for microsomal matrix. The mean rate of 7-hydroxycoumarin formation in guinea pig liver microsomes was 0.084 nmol/min/nmol P450. Coumarin 7-hydroxylase activity was absent in rat liver microsomes. No interference was observed from incubation mixture components.
Keywords
HPLC assay, CYP2A6, coumarin, 7-hydroxycoumarin
Cytochrome P450 (CYP) comprises of a superfamily of heme containing monooxygenases that play an important role in biotransformation of numerous endogenous compounds and xenobiotics [1]. The major human CYP isoenzymes that are primarily responsible for hepatic drug metabolism are CYP1A2, CYP2A6, CYP2B6, CYP2C9, CYP2C19, CYP2D6, CYP2E1 and CYP3A4 [2,3]. The relative levels of these CYP isoenzymes in human liver microsomes can be determined by monitoring the biotransformation of suitable substrates (termed isoenzyme specific ‘probe’ substrates) that are selectively metabolized to specific metabolites by the individual CYP isoenzymes [4]. CYP2A6 is the predominant member of CYP2A subfamily. CYP2A6 metabolizes carcinogenic substrates including aflatoxin, N-nitrosodiethylamine, 6-aminochrysene [5]. CYP2A6 is inducible by barbiturates. Coumarin-7-hydroxylation is routinely used as an in vitro substrate probe to determine CYP2A6 activity [4].
HPLC assay for estimation of CYP2A6 activity in human liver sub-cellular fractions and during in vivo studies has been reported previously [6]. We have evaluated the reported procedure with minor modifications for the estimation of coumarin 7- hydroxylase activity under in-house conditions [6].
Coumarin AR and trichloroacetic acid were purchased from S. D. Fine Chemicals Ltd., Mumbai. 7- Hydroxycoumarin was synthesized in our laboratory [7]. Methanol (HPLC grade) was purchased from Qualigens Chemicals Ltd., Mumbai. The Novapak Waters RP C18 (15 cm×3.9 mm, 5 μm) column was a gift from Wockhardt Ltd., Aurangabad. All other chemicals were of analytical grade.
Microsomes were isolated from the livers of female Sprague-Dawley rats and male Albino guinea pigs. The liver microsomes were prepared by using the calcium aggregation method reported by Schenkman and Cinti [8] with minor modifications. CYP content of the microsomes was determined by the method of Omura and Sato [9]. Isolated rat liver microsomes and guinea pig liver microsomes showed a CYP content of 12.2 µM and 6.92 µM, respectively. The microsomes were stored at the temperature of –70° until further use.
Stock solution of coumarin (500 µM) and 7- hydroxycoumarin (1 mM) was prepared in methanol : double distilled water (1:9). The sub stock solutions of 7-hydroxycoumarin was prepared by suitable dilution of the stock solution with double distilled water to yield solutions with final concentrations of 0.5, 1, 2.5, 5, 10, 20, 50 and 100 µM. The stock solutions and sub-stock solutions of coumarin and 7-hydroxycoumarin were stored in refrigerator (0-4°) until use.
The standard curve for determination of coumarin 7-hydroxylase activity was developed in duplicate, in guinea pig liver microsomal matrix and in 0.1 M phosphate buffer containing 0.001 M EDTA, pH 7.4. The sample preparation protocol for development of standard curve in microsomal matrix and buffer matrix is given in Tables 1 and 2, respectively. The sample work-up was done by using the following procedure. To each of the samples tubes, 250 µl of 50% trichloroacetic acid was added (for the precipitation of microsomal proteins) and the samples were transferred into 1.5 ml capacity Eppendorff tubes. The samples were centrifuged at 13000×g for 10 min at 4°. The supernatant was separated and stored at –70° till HPLC analysis.
| Final conc. | Concentration(µM)of sub stock7HC solutionadded (50 µl) | Vol. of 500 µM CMN solution (µl) | Vol. of buffer (µl) | Vol. of micro-somes (µl) |
|---|---|---|---|---|
| Blank | - | - | 400 | 100 |
| 10 | 100 µM | 50 | 300 | 100 |
| 5 | 50 µM | 50 | 300 | 100 |
| 2.5 | 25 µM | 50 | 300 | 100 |
| 1 | 10 µM | 50 | 300 | 100 |
| 0.5 | 5 µM | 50 | 300 | 100 |
| 0.25 | 2.5 µM | 50 | 300 | 100 |
| 0.05 | 0.5 µM | 50 | 300 | 100 |
The abbreviations used are 7HC= 7-hydroxycoumarin and CMN = coumarin. The buffer used was 0.1 M sodium phosphate buffer containing 0.001 M EDTA, pH 7.4. All samples were prepared in duplicate.
Table 1: Protocol For Standardcurve Preparation In Microsomal Matrix
| Conc.(µM) | Concentrationof sub stock7HC solution. added (50 µl) | Vol. of 500 µM(µl)CMN solution | Vol. of Buffer(µl) |
|---|---|---|---|
| Blank | - | 50 | 500 |
| 10 | 100 µM | 50 | 400 |
| 5 | 50 µM | 50 | 400 |
| 2.5 | 25 µM | 50 | 400 |
| 1 | 10 µM | 50 | 400 |
| 0.5 | 5 µM | 50 | 400 |
| 0.25 | 2.5 µM | 50 | 400 |
| 0.1 | 1 µM | 50 | 400 |
| 0.05 | 0.5 µM | 50 | 400 |
The abbreviations used are 7HC= 7-hydroxycoumarin and CMN = coumarin. The buffer used was 0.1 M phosphate buffer containing 0.001 M EDTA (pH 7.4). All samples were prepared in duplicate.
Table 2: Protocol For Standard Curve
The microsomal incubation conditions for coumarin 7-hydroxylase activity determination were adapted from reported procedures [6,10]. The incubations were conducted in a total volume of 0.5 ml of 0.1 M sodium phosphate buffer containing 0.001 M EDTA, pH 7.4 and contained 100 μl microsomes, coumarin at a final concentration of 50 μM, and 0.6 mM NADPH. Incubations were initiated by addition of NADPH and were performed at 37° for 15 min. After incubation, the reactions were stopped with 250 µl of 50% trichloroacetic acid and samples processed as mentioned for the standard curve samples. Control incubations were also performed either without microsomes or without coumarin, in order to evaluate possible interference from incubation components.
HPLC analysis was done with minor modifications of the method reported previously [6]. Analysis was performed with a Jasco HPLC with PU-980 intelligent pump, equipped with a UV-975 detector and Jasco- Borwin software (version 1.5) with Hercule-2000 interface. HPLC analysis was done on A RP C-18 Novapak Waters (15 cm x 3.9 mm, 5 μm) column. The mobile phase composition was methanol:1% glacial acetic acid (35 : 65 v/v) (pH~3.1), with a flow rate of 0.6 ml/min and detection at 320 nm. One hundred microlitres sample was injected onto the HPLC column for analysis. The standard (calibration) curve was plotted using area of 7-hydroxycoumarin on the ordinate and concentration of 7-hydroxycoumarin on the abscissa.
Drug metabolism studies (in vitro, in vivo and/or in silico) are very important in the assessment of safety of pharmaceuticals, individualization of drug therapy and prediction of drug-drug interactions [4,11-16]. The present study illustrates the evaluation of the suitable HPLC assay to determine coumarin 7-hydroxylase activity in liver microsomal preparations for drug metabolism studies. The conversion of coumarin to 7-hydroxycoumarin in humans is selectively mediated by CYP2A66. The concentration of the substrate i.e. coumarin, used for determination of CYP2A6 activity in liver microsomes is 50 µM [11]. Since the concentration generally used for determination of the Vmax is five times the Km, the projected Km would be 10 µM. Further, considering ≤ 5% conversion [for adherence to Michaelis-Menten steady state assumptions] in the microsomal incubations [6,10], the maximum expected concentration of 7- hydroxycoumarin at the end of incubation would be 2.5 µM or 0.5 µM, if incubations are performed at Vmaxor Km, conditions respectively. Thus, a standard curve with range from 0.05 to 10 µM (0.05, 0.1, 0.25, 0.5, 1, 2, 5 and 10 µM) should be sufficient for application of this assay for CYP2A6 activity determination and drug-drug interaction studies that are performed either at saturating and non-saturating substrate concentrations, respectively.
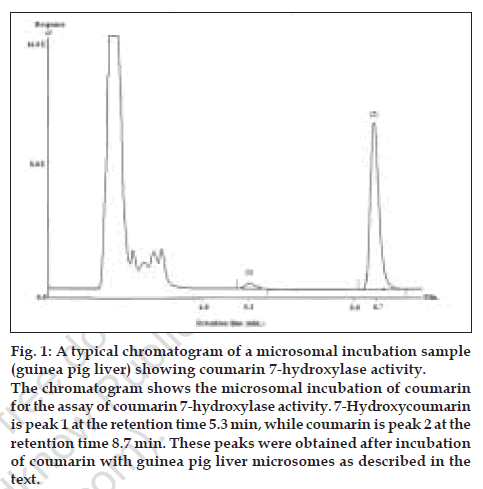
The mean retention times for 7-hydroxycoumarin and coumarin were found to be 5.3 min and 8.7 min, respectively under the stated HPLC conditions. A typical chromatogram in microsomal matrix is shown in fig. 1. Coumarin was included in the standard curve samples to evaluate potential effects on HPLC profile since it would normally be present in microsomal incubation at high concentrations. Linear regression (Microsoft Excel XP) of the standard curve data in microsomal matrix gave the equation Y=85001X with an r2 of 0.9998. Linear regression (Microsoft Excel XP) of the standard curve in buffer matrix followed the equation Y=134651X with r2 of 0.9997. The limit of detection (LOD) was 0.05 µM and the limit of quantitation (LOQ) was 0.1 µM, in both matrices. The mean coefficient of variation associated with two assay experiments, each done in duplicate was found to be in the range 0.03 to 3.6% for buffer matrix and 0.1 to 6.5% for microsomal matrix, respectively. The above results indicate that the coumarin 7-hydroxylase activity assay developed above is suitable for routine drug metabolism research work. The buffer matrix was evaluated in this study in order to avoid use of microsomes during preparation of standard curve while performing coumarin 7-hydroxylase activity assay on a regular basis. Liver tissues are difficult to obtain and microsomes preparation is tedious and as such use of microsomes just as a matrix for standard curve preparation is unwise.
Figure 1: A typical chromatogram of a microsomal incubation sample
(guinea pig liver) showing coumarin 7-hydroxylase activity.
The chromatogram shows the microsomal incubation of coumarin
for the assay of coumarin 7-hydroxylase activity. 7-Hydroxycoumarin
is peak 1 at the retention time 5.3 min, while coumarin is peak 2 at the
retention time 8.7 min. These peaks were obtained after incubation
of coumarin with guinea pig liver microsomes as described in the
text
The biotransformation of coumarin to 7- hydroxycoumarin was observed only in guinea pig liver microsomes, which shows the presence of coumarin 7-hydroxylase activity in the given liver preparation. Control incubations showed no production of 7-hydroxycoumarin. The HPLC profile on incubation of coumarin with guinea pig liver microsomes is shown in fig. 1. No interference was observed from the incubation matrix components. The mean rate of 7-hydroxycoumarin formation in guinea pig liver microsomes was 0.084 nmol/min/ nmol P450. In contrast, metabolism of coumarin to 7-hydroxycoumarin was not observed in rat liver microsomes indicating the absence of this activity. This is not a new finding and it has been reported previously that CYP2A6-like activity shows species differences [17]. Coumarin metabolism in monkey, mouse, hamster and rabbit seems to be most similar to humans [17]. In mouse, the isoform CYP2A5 is responsible for coumarin 7-hydroxylation while in rats, coumarin is mainly metabolized by opening of lactone ring via hydrolysis by esterase. It is suggested that this may be the reason for coumarin hepatotoxicity in rats (which is not observed in case of guinea pigs or humans) [17] where less than 1% of coumarin is metabolized to 7-hydroxycoumarin.
The above studies indicate the utility of this assay for determination of coumarin 7-hydroxylase activity is animal liver microsomes and potentially, CYP2A6 activity in human liver microsomal preparations.
Acknowledgements
The authors would like to extend special thanks to Dr. M.L. Kubal, Wockhardt Ltd., Aurangabad for providing HPLC columns used in this work. This work was conducted with the support of AICTE grant - 8019/RDII/R&D/PHA(213)/2001-02.
References
- Mannering GJ. Microsomal enzyme systems which catalyze drug metabolism. In: La Du BN, Mandal HG, Way LE, editors. Fundamentals of drug metabolism and disposition. 1 st ed. Baltimore; Waverly Press Inc: 1972. p. 206-45.
- Parkinson A. Biotransformation of xenobiotics. In: Klassen CD, Amdur MO, Doull J, editors. Toxicology - The basic science of poisons. 5 th ed. New York; McGraw Hill: 1996. p. 139-63.
- Wrighton SA, Stevens JC. The human hepatic cytochrome P450s involved in drug metabolism. Crit Rev Toxicol 1992;22:1-21.
- Ortiz de Montellano PR. The cytochrome P450 oxidative system. In: Woolf TW, editor. Handbook of drug metabolism. 1 sted. New York; Marcel Dekker Inc: 1999. p. 109-30.
- Williams DA. Drug metabolism. In: Foye WO, Lemke TL, Williams DA, editors. Principles of Medicinal Chemistry. 4 th ed. New Delhi; BI Waverly Pvt. Ltd: 1999. p. 83-140.
- Iyer KR, Sinz, MW. Characterization of Phase I and Phase II hepatic drug metabolism activities in a panel of human liver preparations.ChemBiol Interact 1999;118:151-69.
- Bhopale AK. Synthesis of metabolites of probe substrates of CYP2A6 and CYP2C9.M Pharm Sci Thesis. University of Mumbai: October 2001.
- Schenkman J, Cinti DT. Preparation of microsomes with calcium. Meth Enzymol 1978;52:84-7.
- Omura T, Sato R. The carbon monoxide binding pigment of liver microsomes. J BiolChem 1964;239:2370-7.
- Tassaneeyakul W, Birkett DJ, Veronese ME, McManus ME, Tukey RH, Quttroachi LC, et al . Specificity of substrates and inhibitor probes for human cytochromes. J PharmacolExpTher 1993;265:401-7.
- Chauret N, Gauthier A, Martin J, Nicoll Griffith DA. In vitro comparison of cytochrome P450 mediated metabolic activities in human, dog, cat, and horse. Drug MetabDispos 1997;25:1130-43.
- Rodrigues AD, Lin JH. In vitro models for early studies of drug metabolism. In: Testa B, Van de Waterbeemd H, Folkers G, Guy R, editors. Pharmacokinetic optimization in drug research-biological, physicochemical and computational strategies. 1 st ed. Germany; Wiley VCH: 2001. p. 217-44.
- Gillette JR. Techniques for studying drug metabolism in vitro . In: La Du BN, Mandel HG, Way LE, editors. Fundamentals of drug metabolism and disposition. 1 st ed. Baltimore; Waverly Press Inc: 1997. p. 400-16.
- Hayes RN, Pool WF, Sinz MW, Woolf TF. Recent developments in drug metabolism methodology. In: Welling PG, Tse FL, editors. Pharmacokinetics. 2 nded. New York; Marcel Dekker Inc: 1995. p. 201-34.
- Sinz MW, Iyer KR. The role and practice of drug metabolism in drug discovery and development. Asian ChemLett 2001;5:93-104.
- Clarke SE, Jones BC. Human cytochrome P450s and their role in metabolism based drug-drug interactions. In : Rodrigues AD, editor. Drug-drug interactions. 1 sted. New York; Marcel Dekker Inc: 2000. p. 55-88.
- Spatzenegger M, Born SL, Halpert JR. Cytochrome P450 in laboratory animal species. In : Lee JS, Obach RS, Fisher MB, editors. Drug metabolizing enzymes - Cytochrome P450 and other enzymes in drug discovery and development. Netherlands; Fontis Media SA: 2003. p. 179-209.