- *Corresponding Author:
- R. Ghosh
Department of Pharmacy, Tripura University (A Central University), Suryamaninagar, West Tripura-799 022, India
E-mail: rajatghosh@tripurauniv.in
| Date of Submission | 12 April 2017 |
| Date of Revision | 28 January 2019 |
| Date of Acceptance | 02 July 2019 |
| Indian J Pharm Sci 2019;81(5):797-806 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The term quorum sensing in bacteria is used to describe the communication among them through signalling molecules, known as autoinducers or pheromones. This type of communication usually triggers at a time when the bacteria reach their threshold population. This in turn leads to expression of various genes, resulting in co-operative behaviour in bacteria. Biofilm formation, virulence expression are the examples of such kind of quorum sensing. In Gram-negative bacteria such as Pseudomonas aeruginosa, quorum sensing is achieved through acyl homoserine lactone class of signalling molecules, two such molecules are 3-oxo-dodecanoylhomoserine lactone and butylhomoserine lactone. The productions of these molecules are controlled by LasI and RhlI genes through the acylhomoserine lactone synthetases. At high concentration, those signal molecules bind to cytoplasmic transcription factors LasR and RhlR and activate target genes leading to expression of virulence factors namely elastase, protease, rhamnolipids, pyocyanin, siderophores. In Pseudomonas aeruginosa, the Las and Rhl systems function in tandem with the Las system at the top and these two systems are interconnected by Pseudomonas quinoline signal. Quorum sensing in bacteria can be inhibited by targeting either signal generator, the signal molecule or the signal receptor. Quorum sensing inhibitors can be obtained from natural sources including traditional medicines or can be synthesized in the laboratory.
Keywords
Autoinducers, biofilm, Pseudomonas aeruginosa, quorum sensing, quorum sensing inhibitors
Pseudomonas aeruginosa is a Gram-negative, ubiquitous, and opportunistic bacteria[1,2], which causes a large number of infectious diseases including chronic lung infection[3,4], soft tissue infection, dermatitis[5], chronic wounds[6], urinary tract infections[7] and respiratory tract infection[8] affecting especially the immune compromised subjects and those with cystic fibrosis[9,10]. It can be isolated from soil and water and it regularly contaminates medical devices like catheters, implants like prosthetic heart valves, cardiac pacemakers, and total joint prostheses[11,12]. Moreover, P. aeruginosa is the major cause of morbidity and mortality in patients with cystic fibrosis, a genetic disease, which affects about 1 out of 3000 newborn in the Caucasian population[13,14]. Apart from having intrinsic resistance to several antibiotics it can acquire new resistance factors with ease. Like many other host-assisted bacteria, P. aeruginosa can control its population density and gene expression by release of certain chemicals, sometimes referred to as autoinducers (AIs) or pheromones, a phenomenon known as quorum-sensing, which is essentially a cell to cell communication system, which ultimately leads to expression of specific genes[15]. P. aeruginosa uses N-acylhomoserine lactones (AHLs) as the chemical signal to communicate among them, the signalling compound being synthesised by LuxI type of synthetase. The result is the expression of specific genes leading to virulence expression and formation of biofilms, which are highly structured protective layers embedding the bacteria[16,17].
Quorum sensing (QS):
Bacteria are social organisms and they interact with each other, which according to some researchers are similar to those performed by insects, vertebrates and humans. The term QS is used to describe bacterial cell to cell communication process, which involves the production, detection and response to extra cellular signalling molecules called AIs. The increase in concentration of these signalling molecules or AIs are directly proportional to the bacterial population as a result bacteria in turn use this information to alter a number of factors, such as gene expression, virulence[18,19].
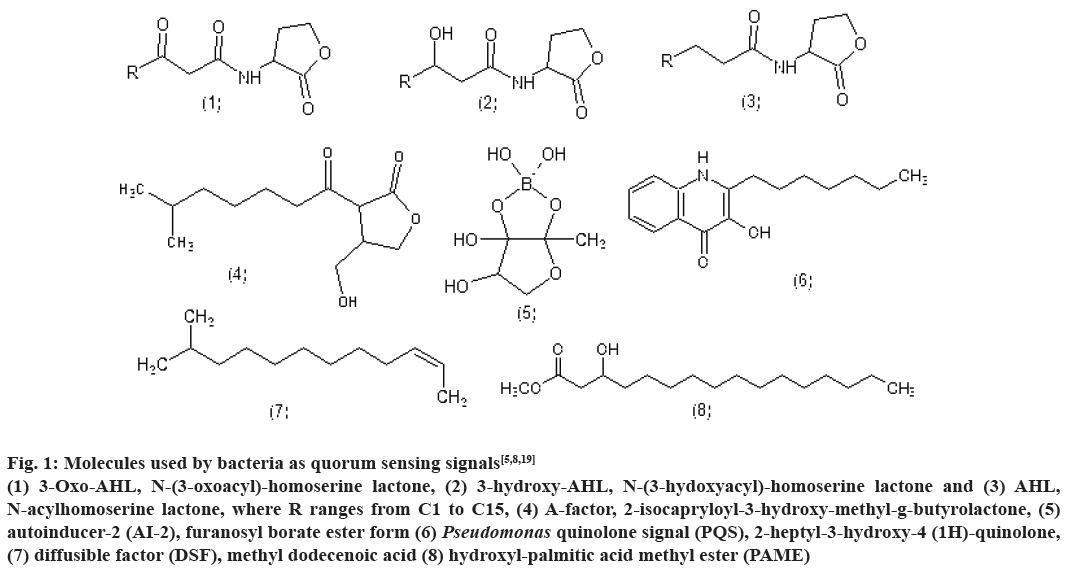
Bioluminescence, competence, antibiotic production, biofilm formation are some of the processes controlled by QS. Till date, several classes of such signalling molecules have been described in bacteria including AHLs, 2-alkyl-4(1H)-quinolones, cyclic dipeptides, AI-2 and small modified peptides. Fig. 1 represents the structure of few signalling molecules such as, 3-oxo- AHL, N-(3-oxoacyl)-homoserinelactone, 3-hydroxy- AHL, N-(3-hydoxyacyl)-homoserine lactone and AHL, N-acylhomoserine lactone, where R ranges from C1 to C15, A-factor, 2-isocapryloyl-3-hydroxymethyl- g-butyrolactone, AI-2, furanosyl borate ester form, Pseudomonas quinolone signal (PQS), 2-heptyl- 3-hydroxy-4-(1H)-quinolone, diffusible factor, methyl dodecenoic acid and hydroxyl palmitic acid methyl ester[5,8,19].
Figure 1: Molecules used by bacteria as quorum sensing signals[5,8,19]
(1) 3-Oxo-AHL, N-(3-oxoacyl)-homoserine lactone, (2) 3-hydroxy-AHL, N-(3-hydoxyacyl)-homoserine lactone and (3) AHL,
N-acylhomoserine lactone, where R ranges from C1 to C15, (4) A-factor, 2-isocapryloyl-3-hydroxy-methyl-g-butyrolactone, (5)
autoinducer-2 (AI-2), furanosyl borate ester form (6) Pseudomonas quinolone signal (PQS), 2-heptyl-3-hydroxy-4 (1H)-quinolone,
(7) diffusible factor (DSF), methyl dodecenoic acid (8) hydroxyl-palmitic acid methyl ester (PAME)
Generally, bacterial communication can be divided into the following three categories. The intra-species in which the communication between bacteria of the same species is known as intra-species communication. For example, communication between P. aeruginosa through signal molecules N-3- oxo-dodecanoyl homoserine lactone (3OC12HSL) and butyl homoserine lactone (C4HSL) to determine gene expression and virulence factor production[18,20]. Inter-species communication where the inter-species communication usually occurs in between two or more bacterial species, for example, communication between P. aeruginosa and Burkholderia cepacia in a cystic fibrosis lung. However, this communication seems to be a one way communication[21,22] and the inter-kingdom communication related to the communication between bacteria and higher species. Such kind of communication is observed in between marine red algae Delisea pulchra and Serratia liquefaciens, where halogenated furanones produced by the marine red algae Delisea pulchra disrupts QS and swarming motility of Serratia liquefaciens[23].
Moreover, the phenomenon of cell to cell communication is also observed in the marine bioluminescent bacterium Vibrio fischeri. It was earlier observed in 1970 by Nealson and his colleagues that bioluminescence occurred only at a threshold bacterial population density, and spent culture supernatants induced luminescence at low bacterial population density[24]. It was attributed to the presence of AIs, which was later identified as N-(3-oxohexanoyl)- homoserine lactone (3-oxo-C6HSL). This belongs to the AHL family of molecules. Similar phenomenon was observed in the terrestrial plant pathogen Erwinia carotovora employed in the production of the β-lactam antibiotic 1-carbapen-2-em-3-carboxylic acid (carbapenem)[20].
In general, all known QS systems follow the below feed forward algorithm; production and concentration of AIs at high cell density→detection of AIs by receptors present in cytoplasm and cell membrane→activation of genes necessary for co-operative behaviour and further enhancement of production of AIs[25]. Virulence of P. aeruginosa is regulated by QS to overcome activation of the host’s immunity, by deleting the production of virulence factors until the bacteria population raises sufficient enough to cause infection[26].
QS allows bacteria to show a unified response that benefits the population. Bacterial QS approaches amplify access to nutrients and more convenient environmental slots, and they elevate action against competing bacteria and environmental strains. The cellular processes regulated by QS are symbiosis, transfer of conjugative plasmids, sporulation, antimicrobial peptide synthesis, regulation of virulence, and biofilm formation[27].
It has been well documented that in few bacteria, the biofilm formation is a cautiously engineered process managed by QS. The use of mutant gene of bacterial strains involved in the formation of signalling molecules and the perusal of differential gene expression in biofilms are unveiling information on the molecular mechanisms of biofilm development and the involvement of QS. Where it has been observed that most researchers support the role of QS in biofilm development, few studies oppose the role of QS involvement in the formation of biofilms. Additionally the understanding of chemical structures of various types of signalling molecules permits the spotting out of scaffolds that can be applied to regulate QSassociated processes, comprising biofilm formation. Further research is required to gain knowledge about mechanistic work frame of QS in biofilms and influence the virulence and antimicrobial resistance of biofilm communities by cell-to-cell signalling. The investigational data is important to identify possible targets and to design strategies and pharmacophores that may control biofilm formation[28].
In Gram-negative bacteria P. aeruginosa, two QS circuits LuxI/LuxR are arranged in tandem, along with the PQS controls biofilm formation and virulence expression[24].
The biofilm concept:
Biofilms are a highly organised and structured collection of bacterial cells belonging either to a single or mixed species enclosed in a polysaccharide matrix secreted by the bacteria[22,28,29]. These could be linked in nature to ant nests or beehives[18]. Oral cavities, wounds, burnt, damaged tissues, medical devices are among many places where biofilms can exist[30]. The protective polysaccharide matrix renders the biofilms highly resistant to host immune responses and other antimicrobial agents[31]. The existence of dormant cells, phenotypic variation, QS system, multidrug efflux pumps, limited agent penetration and increased production of matrix polysaccharide alginate are several hypotheses, which brought forward to explain for such resistance[24,31-33]. The direction of growth in a biofilm could be horizontal, vertical or both. Horizontal growth occurs when there is availability of substratum for growth or when the bacteria are forced to do so due to scarcity of nutrients or for new carbon or nutrient source. Vertical growth is the result of physical barriers or when the bacteria have utilised the available substratum. Differential combination of horizontal and vertical biofilm growth results in different structures such as mushroom, tower and channels[4,34]. Different gradients of chemicals, nutrients and oxygen in these biofilms forces the micro-organisms to undergo genotypic and phenotypic adaptations[22].
Biofilm formation is synchronized by various genetic and environmental factors. Genetic studies reveal that bacterial mobility, cell membrane proteins, extracellular polysaccharides and signalling molecules play remarkable roles in biofilm formation. Bacterial mobility is empowered by two types of protein growths on the cell surface, flagella and fimbriae. Flagella are long, spiral growths that empower bacteria to float in liquid phase, and fimbriae are short, straight growths that empower little, jerking movements of bacteria on substrate surface[26,35].
Detailed studies of Pseudomonas aeruginosa revealed that both kinds of bacterial mobility are required for biofilm formation. Bacterial mobility empowered by flagella is required for developing the connection between the bacteria and the surface, where the mobility empowered by fimbriae is required for the development of microcolonies. The stable connection between bacteria and substrate surface is regulated by specific cell membrane proteins, known as adhesions. It has been observed that in the inhibition of adhesion activity, biofilm formation is also inhibited. The extracellular polysaccharide matrix (EPS) has an important role in biofilm formation. Molecular studies on bacteria P. aeruginosa revealed that activation of genes required for EPS after establishing stable connection between bacteria and substrate surface. Whereas few studies conducted on other bacteria showed that the bacteria lose ability to form biofilm if the responsible genes for synthesis of EPS matrix are inactivated[36]. Interactive communication through signalling molecules empowers bacteria to organize into a community to functionalize the biofilm as a multicellular organism. Various signals from environment, like presence of certain nutrients, availability of oxygen, temperature and pH, take part in synchronizing of a biofilm formation. The EPS matrix provides protection to biofilm cells from several negative environmental stresses, like UV radiation, sudden changes in pH values. Between micro-colonies, there are passages through which water flows. These water passages function in a biofilm as a circulatory system diffusing nutrients to micro-colonies and collecting harmful metabolites. Moreover, studies reveal the effect of different glucose concentration on biofilm and show at high glucose concentration, micro colonies grow fast and accordingly biofilm thickness increases remarkably, whereas at decreased concentration of glucose, biofilm mass is diminished to restore former structure[37].
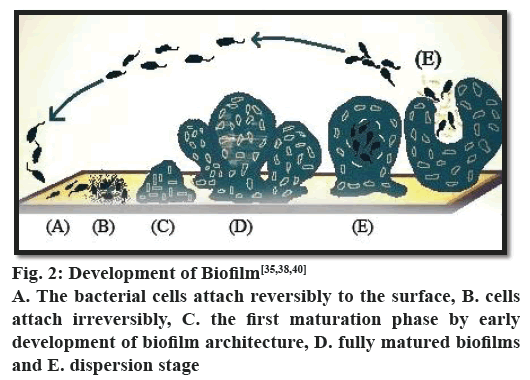
The formation of biofilms is a step-wise process (fig. 2). The process starts with the adhesion of microorganisms to a surface. This is followed by active division and colonialization. The next step is the co-adhesion of the microorganisms, which could be intra or inter species as a biofilm may contain several types of bacteria. It is then followed by growth and maturation. Finally, some of the bacteria are detached from the biofilm and the process continues. In each phase of biofilm formation, bacteria cells are physiologically different from cells of other phase. In the mature biofilm, all phases can exist concomitantly. It has been observed that in P. aeruginosa biofilm maturation involves various phases, which can be differentiated as adsorption, attachment, maturation and dispersion. The threshold of the event biofilm formation is interaction between substrate surface and planktonic bacteria, which is known as reversible adsorption due to bacterial attaching property to the substrate surface, but it stands only for a hasty period and then gets detached immediately. In the next phase, irreversible attachment takes place, where bacteria stick firmly to the surface of substrate and lose their mobility. Afterwards bacterial cells attach to each other and to the surface of substrate to start forming bacterial micro-colonies. Ones the attachment and formation of micocolonies is over, there is a next phase known as maturation of biofilm formation. In this phase EPS is synthesized[38,39]. Different studies on comparison of cells in maturation phase and planktonic cells reveal remarkable difference in protein structures, which further propel the knowledge in the domain of biofilm formation with the conclusion of great physiological difference between biofilm bacteria and planktonic bacteria. The last phase in the development of biofilm formation is dispersion. In this particular phase, the structure of microcolony changes from the bacteria cells located in their central part retrieve their mobility and detach from the formerly formed structure. The phase dispersion mostly takes place to permit bacterial cells better access to nutrients. During this particular phase, water passages form between micro-colonies. Protein expression in this phase is similar to protein expression in planktonic cells, which further proves that some bacteria return into planktonic stage[38,39].
There is emerging evidence that the expression of genes required in the various stages is well regulated[20] and can be represented as follows, stage 1- the bacterial cells attach reversibly to the surface; stage 2- the cells attach irreversibly; stage 3- the first maturation phase is reached, as indicated by early development of biofilm architecture; stage 4- fully mature biofilms and stage 5- dispersion stage[40].
The infection caused by P. aeruginosa is hard to eradicate due to development of strong resistance to the traditional antibiotics. Which is further intensified by the potentiality of the pathogen to develop biofilm architecture and provides bacterial cells a shielded environment resist diverse stresses including conventional antibiotics. QS plays an important role in synchronizing of the bacterial virulence and biofilm development. The QS network is decorated in a multilayered hierarchy of approx. four interconnected signalling mechanisms. The genes those are being involved in integrated quorum sensing signal (IQS) synthesis, are non-ribosomal peptide synthase gene cluster ambBCDE, and it has been observed that when it is interrupted, it caused a reduction in the production of PQS and N-butyryl homoserine lactone signals, in addition the virulence factors such as pyocyanin, rhamnolipids and elastase. Distortion of either LasR or LasI completely dissolves the expression of ambBCDE along with the production of IQS. Literature survey in this particular field is providing knowledge that the QS regulatory network not only reacts to bacterial population changes but also could respond to environmental stress signals. This QS mechanism could be a favourable target for flourishing new approaches against P. aeruginosa infection in the development of novel antiQS therapeutics[41,42].
The QS circuit in P. aeruginosa and virulence expression:
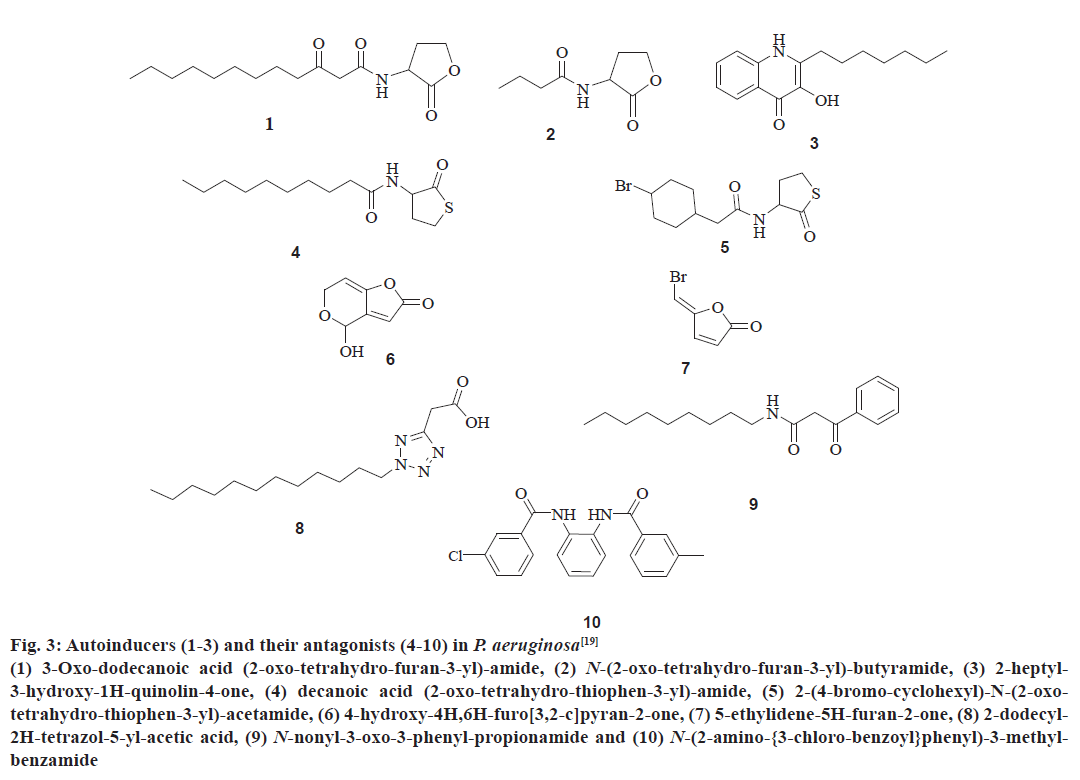
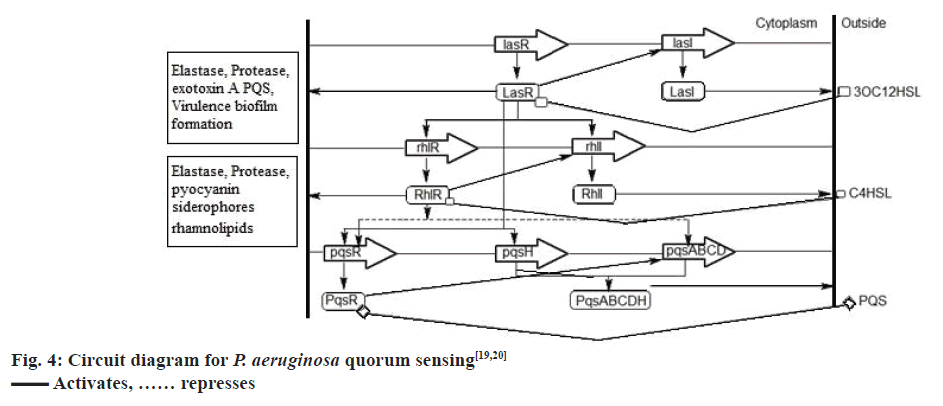
The QS communication system in P. aeruginosa consists of the Las and Rhl systems[27,28]. Both the systems essentially consist of two pairs of genes, LasI, LasR, and RhlI, RhlR[29]. Both the ‘I’ genes encode for the synthetases LasI and RhlI, respectively[43,44]. These synthetases direct the synthesis of freely diffusible acyl homoserine lactones (AHL’s) by catalysing a reaction between S-adenosyl methionine (SAM) and an acyl carrier protein (ACP)[19,45]. In the las system LasI directs the synthesis of 3-oxo-dodecanoyl-homoserine lactone (3OC12HSL) while in the Rhl system RhlI directs the synthesis of butyl homoserine lactone (C4HSL)[46-48]. On the other hand both the R genes viz. LasR and RhlR encode for the synthesis of cytoplasmic transcription factors LasR and RhlR[43,44]. At high concentrations, AHLs bind to their respective cytoplasmic transcription factors, thus stabilising them. This allows to fold, bind DNA and activate the target genes[19,43,44]. This leads to the expression of virulence factors including elastase, protease, rhamnolipids, Las A protease, exotoxin A, hydrogen cyanide, pyocyanin, and siderophores[3,21,43] another activity shown by AHL bound LasR type transcription factor is the activation of genes encoding the synthesis of synthetases synthesizing AHL’s. This forms a feed forward auto induction loop as mentioned earlier[19]. In P. aeruginosa the two QS systems are arranged in tandem with the Las QS system on top, thus exerting transcriptional control over Rhl QS system. Fig. 3 represents few AIs and antagonists of P. aeruginosa.
Figure 3: Autoinducers (1-3) and their antagonists (4-10) in P. aeruginosa[19]
(1) 3-Oxo-dodecanoic acid (2-oxo-tetrahydro-furan-3-yl)-amide, (2) N-(2-oxo-tetrahydro-furan-3-yl)-butyramide, (3) 2-heptyl-
3-hydroxy-1H-quinolin-4-one, (4) decanoic acid (2-oxo-tetrahydro-thiophen-3-yl)-amide, (5) 2-(4-bromo-cyclohexyl)-N-(2-oxotetrahydro-
thiophen-3-yl)-acetamide, (6) 4-hydroxy-4H,6H-furo[3,2-c]pyran-2-one, (7) 5-ethylidene-5H-furan-2-one, (8) 2-dodecyl-
2H-tetrazol-5-yl-acetic acid, (9) N-nonyl-3-oxo-3-phenyl-propionamide and (10) N-(2-amino-{3-chloro-benzoyl}phenyl)-3-methylbenzamide
P. aeruginosa has a third QS system called the PQS, which is intertwined between the first two QS systems[49]. Chemically, PQS is 2-heptyl-3-hydroxy-4- quinolone, the synthesis of which is directed by PqsA, PqsB, PqsC, PqsD, and PqsH through the synthetases PqsA, PqsB, PqsC, PqsD, and PqsH. This signal is detected by the regulator PqsR, whose synthesis is directed by pqsR[19]. Now, LasR-3OC12HSL activates the expression of PqsR and PqsH, while RhlR-C4HSL represses the expression of PqsR and PqsA, PqsB, PqsC, PqsD. The PQS, when binds to the cytoplasmic transcription factor PqsR causes autoinduction of PQS synthesis and expression of RhlI and RhlR genes[19,20], fig. 4 represents the QS circuitry in P. aeruginosa.
AI synthetases LasI, RhlI, and PqsABCDH synthesize the AIs 3OC12HSL, C4HSL and PQS, respectively. These SIs are detected and bound by the cytoplasmic transcription factors LasR, RhlR, PqsR respectively, which in turn regulates the expression of genes responsible for the production of virulence factors and further synthesis of AIs[50].
Quorum sensing inhibitors (QSIs):
QSI are the agents which can act at different levels of the QS circuit and interfere with bacterial QS. In designing a QSI, following three components of the QS pathway can be targeted[38,48,51,52].
The signal generator (the LuxI homologue) and blockage of signal generation:
This is the least investigated strategy of QSI. A few substrates namely, butyl-SAM, S-adenosylhomocystine, holo-ACP, and sinefungin have been found to block the production of AHL’s in vitro. However, these compounds have not been investigated in vivo in bacteria and their mode of action is not known at present[52].
The signal molecule:
Inhibition of QS circuit can be achieved by identifying and modulating quorum-quenching enzymes which can degrade bacterial QS molecules. Till date, three such classes of enzymes have been reported- lactonases, acylases, and oxido-reductases. Lactonases belong to the family of metallo β-lactamases or phosphotriesterases, which causes lactonolysis of the AHL lactone ring leading to an inactive product[38]. However, the reaction is reversible under low pH conditions. Acylases are the members of the N-terminal nucleophile hydrolase super family, which breakdown the amide bond of the AHLs releasing the homoserine lactone and an acyl moiety. The oxido-reductases as the name suggests are enzymes that causes either oxidation or reduction of the signal molecules. For example, reduction of 3-oxo substituted AHLs to 3-hydroxy substituted AHLs. Several bacteria including Pseudomonas sp. Variovarax sp. Comamonas sp. Arthrobacter sp. Klebsiella pneumonia, Rhodococcus erythropolis have enzymes which could degrade AHLs[52,53]. These are more recent development targeting bacterial communication system. These antibodies bind to the signal molecules thus eliciting immune response, which renders the signal inactive[51].
The signal receptor:
This is one of the attractive ways to inhibit the bacterial QS system which can be achieved by using natural and synthetic signal mimics. These bind to the transcriptional promoters thus interfering with the bacterial signalling system. One of the best studied example of signal mimicry are the halogenated furanone compounds from the Australian marine macro alga Delisea pulchra[53]. These compounds interfere with the binding of AHL to the LuxR homologue causing rapid turnover of these proteins[53]. Thus, little amount of LuxR is left to interact with the signal molecules and act as a transcriptional regulator. However, these compounds are not very active against P. aeruginosa QS system which can be inhibited by the synthetic furanones[52].
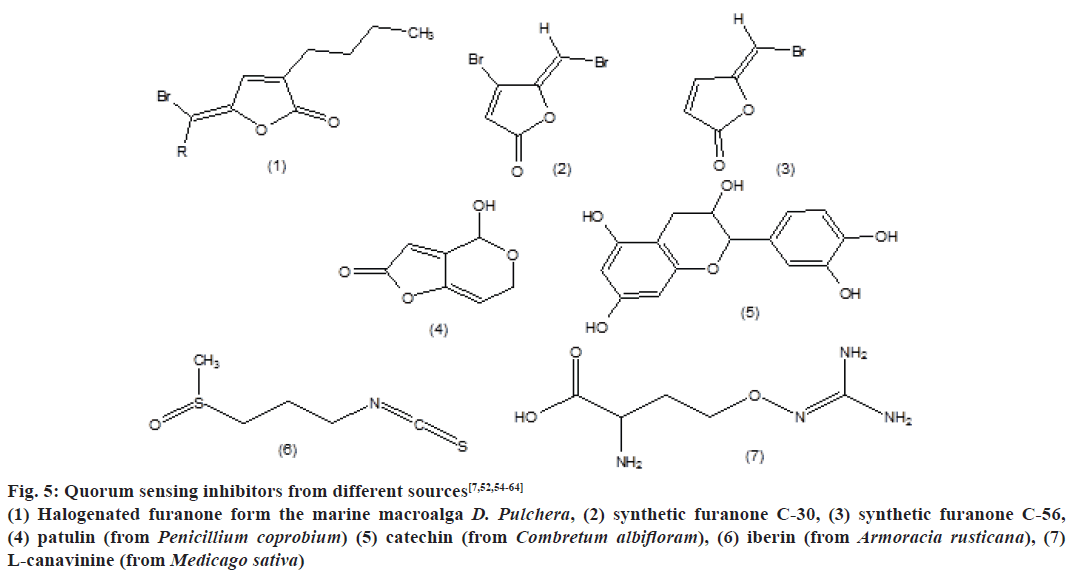
QSI’s from different sources are mentioned in Fig. 5, which include halogenated furanone form the marine macroalga D. Pulchera, synthetic furanone C-30, synthetic furanone C-56, patulin (from Penicillium coprobium), catechin (from Combretum albifloram), iberin (from Armoracia rusticana) and L-canavinine (from Medicago sativa).
Figure 5: Quorum sensing inhibitors from different sources[7,52,54-64]
(1) Halogenated furanone form the marine macroalga D. Pulchera, (2) synthetic furanone C-30, (3) synthetic furanone C-56,
(4) patulin (from Penicillium coprobium) (5) catechin (from Combretum albifloram), (6) iberin (from Armoracia rusticana), (7)
L-canavinine (from Medicago sativa)
Synthetic QSIs:
A team of researchers were involved in synthesizing a series of novel QSIs through the laboratory exercise to get better result on inhibition of biofilm formation by infectious microorganisms[10]. The synthetic halogenated furanone was reported for its effect on the architecture of the biofilm and enhancement property in the process of bacterial detachment, leading to a loss of bacterial biomass from targeted layer. This furanone compound is capable of interfering with AHLmediated QS in P. aeruginosa, which is a derivative of secondary metabolites produced by the Australian macroalga Delisea pulchra[54]. In the literature, C30 and C56 synthetic furanone compounds were reported for their interference with N-acyl homoserine lactone and suppression of bacterial QS in lungs. Moreover, the synthesized compound N-decanoyl cyclopentyl amide was also reported for its potent inhibitory activity against QS in P. aeruginosa[7].
Natural QSIs:
The fact that plants and microorganisms have coexisted for millions of years and accordingly plants may have devised some mechanisms to combat microbial virulence led to the search of QSIs from plants and other natural sources. Following is a brief account of some such QSIs from natural sources[55].
Penicillium species:
Two active secondary metabolites penicillic acid (from P. radicicola) and patulin (from P. coprobium) were identified as QSIs. It is reported that patulin targets 45 % and penicillic acid targets 60 % of the QS genes in P. aeruginosa indicating that these compounds target the LasR and RhlR QS regulators[56].
Flavonoids from Combretum albifloram bark:
C. albifloram belongs to the Combretaceae family, the members of which are used in traditional medicine for bacterial infections. Various species of the genus Combretum are known to produce a wide variety of secondary metabolites including tannins, flavonoids, terpenoids, and stilbenoids. Screening of extracts from C. albifloram is reported to reduce the production of virulence factors which are controlled by QS. During research flavonoid, flavan-3-ol-catechinwas identified for their QSI activities. Catechin is reported to down regulate several QS regulated genes like LasI, LasR, RhlI, RhlR[57].
Iberin from Horseradish (Armoracia rusticana):
Iberin is anisothiocyanate, produced by A. rusticana, a member of the Brassicaceae family. Chemically, iberin is 1-isothiocyanato-3-(methyl-sulfinyl) propane. It was revealed by DNA microarray analysis that iberin specifically and effectively targets the LasIR and RhlIR QS networks and down regulate the QS controlled production of rhamnolipid, a virulence factor expressed by P. aeruginosa which destroys the polymorphonuclear leucocytes, the first line of defence in the host[58,59].
South Florida medicinal plants:
In a study, extracts from six South Frorida plants namely Conocarpus erectus (family-Combretaceae), Chamaesyce hypericifolia (family- Euphorbiaceae), Callistemon viminalis (family- Myrtaceae), Bucida buceras (family- Combretaceae), Tetrazygia bicolour (family- Melastomataceae), Quercus virginiana (family- Fagaceae) were found to inhibit QS controlled factors like LasA protease, LasB elastase, and pyoverdin. The extracts also caused reduction in the expression of LasI, LasR, RhlI, and RhlR genes in P. aeruginosa and reduction in the production of signal molecules. However, the extracts had marginal effects on bacterial growth, indicating that the quorum quenching activity is not related to bacteriostatic or bacteriocidal effect[60,61].
Herbs from traditional Chinese medicine (TCM):
The growth of TCM herbs along with QS bacteria led to the postulation that these plants may have evolved to produce QSIs to reduce the pathogenicity of bacteria. Six compounds namely rhein (from Rheum palmatum L.), chrysophanol (from Rheum officinale Baill.), nodakenetin (from Peucedanum decursivum Maxim), shikonin (from Lithospermum erythrorhizon Sieb.), emodin (from Rheum palmatum L.), fraxin (from Fraxinus chinensis Roxb.) found in TCM were identified as potential QSIs and studied for activity in P. aeruginosa, Stenotrophomonas maltophila and Escherichia coli. It was found that among these compounds emodin significantly inhibited biofilm formation in P. aeruginosa and caused proteolysis of an AHL binding protein. Emodin also caused increased activity of ampicillin against P. aeruginosa[12,62,63].
L-Canavinine from alfalfa (Medicagosativa):
It was reported that L-Canavinine from the seeds of alfalfa (Medicago sativa) inhibited the production of violacein in Chromobacterium violaceum, which is a QS regulated process with AHL as the QS signal. It was also reported to inhibit the QS regulated production of exopolysaccaride EPS II in the symbiotic bacteria Sinorhizobium meliloti[19,64].
Increasing resistance of bacteria towards antibiotics is a major challenge today before the health care professionals and medicinal chemists. Biofilm mode of growth of bacteria provides them extra safety and tolerance as compared to planktonic lifestyle, which is a severe problem in medicine and industry. It is very difficult to eradicate the bacteria in biofilm mode of growth even with the most advanced antibiotics. Under such conditions, inhibition of bacterial QS system offers an attractive way to check bacteria in fields like medicine, industry and agriculture. Inhibition of bacterial QS system can be achieved by targeting the QS signal generator, the signal, or the signal receptor. The coexistence of bacteria with plants and other microbes provides a clue to search for QSIs among them. Traditional herbal medicine is a vast field, which can be explored for the QSIs and till date has provided a number of QSIs. Another advantage of bacterial QSI is that it renders the bacteria more prone to antibiotics. So, treatment of bacterial infection by antibiotics along with QSIs remains a vast field of research.
Conflicts of interest:
There are no conflicts of interest.
References
- Bjarnsholt T, Jensen PO, Rasmussen TB, Christophersen L, Calum H, Hentzer M, et al. Garlic blocks quorum sensing and promotes rapid clearing of pulmonary Pseudomonas aeruginosa infections. Microbiology 2005;151(Pt 12):3873-80.
- Junker LM, Clardy J. High-throughput screens for small-molecule inhibitors of Pseudomonas aeroginosa biofilm development. Antimicrob Agents Chemother 2007;51(10):3582-90.
- Yang L, Rybtke MT, Jakobsen TH, Hentzer M, Bjarnsholt T, Givskov M et al. Computer-aided identification of recognized drugs as Pseudomonas aeruginosa quorum-sensing inhibitors. Antimicrob Agents Chemother 2009;53(6):2432-43.
- Stapper AP, Narasimhan G, Ohman DE, Barakat J, Hentzer M, Molin S, et al. Alginate production affects Pseudomonas aeruginosa biofilm development and architecture, but is not essential for biofilm formation. J Med Microbiol 2004;53(Pt 7):679-90.
- Dekimpe V, Deziel E. Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: the transcriptional regulator RhlR regulates LasR-specific factors. Microbiology 2009;155(Pt 3):712-23.
- Skindersoe ME, Alhede M, Phipps R, Yang L, Jensen PO, Rasmussen TB, et al. Effects of Antibiotics on Quorum Sensing in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2008;52(10):3648-63.
- Ishida T, Ikeda T, Takiguchi N, Kuroda A, Ohtake H, Kato J. Inhibition of Quorum Sensing in Pseudomonas aeruginosa by N-Acyl cyclopentylamides. Appl Environ Microbiol 2007;73(10):3183-88.
- Schuster M, Urbanowski ML, Greenberg EP. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. Proc Natl Acad Sci USA 2004;101(45):15833-39.
- Patriquin GM, Banin E, Gilmour C, Tuchman R, Greenberg EP, Poole K. Influence of quorum sensing and iron on twitching motility and biofilm formation in Pseudomonas aeruginosa. J Bacteriol 2008;190(2):662-67.
- Wu H, Song Z, Hentzer M, Andersen JB, Molin S, Givskov M, et al. Synthetic furanones inhibit quorum-sensing and enhance bacterial clearance in Pseudomonas aeruginosa lung infection in mice. J Antimicrob Chemother 2004;53(6):1054-61.
- Louise D, Christensen LD, Gennip MV, Jakobsen TH, Alhede M, Hougen HP, et al. Synergistic antibacterial efficacy of early combination treatment with tobramycin and quorum-sensing inhibitors against Pseudomonas aeruginosa in an intraperitoneal foreign-body infection mouse model. J Antimicrob Chemother 2012;67(5):1198-206.
- Ding X, Yin B, Qian L, Zeng Z, Yang Z, Li H, et al. Screening for novel quorum-sensing inhibitors to interfere with the formation of Pseudomonas aeruginosa biofilm. J Med Microbiol 2011;60(Pt 12):1827-34.
- Imperi F, Massai F, Pillai CR, Longo F, Zennaro E, Rampioni G, et al. New life for an old drug: the anthelmintic drug niclosamide inhibits Pseudomonas aeruginosa quorum sensing. Antimicrob Agents Chemother 2013;57(2):996-1005.
- Moreau-Marquis S, Bomberger JM, Anderson GG, Swiatecka-Urban A, Ye S, O’Toole GA, et al. The F508-CFTR mutation results in increased biofilm formation by Pseudomonas aeruginosa by increasing iron availability. Am J Physiol Lung Cell Mol Physiol 2008;295(1):L25-37.
- More MI, Finger LD, Stryker JL, Fuqua C, Eberhard A, Winans SC. Enzymatic synthesis of a quorum-sensing autoinducer through use of defined substrates. Science 1996;272:1655-58.
- Christensen LD, Moser C, Jensen PO, Rasmussen TB, Christophersen L, Kjelleberg S, et al. Impact of Pseudomonas aeruginosa quorum sensing on biofilm persistence in an in vivo intraperitoneal foreign-body infection model. Microbiology 2007;153(Pt 7):2312-20.
- Lu Y, Zeng J, Wu B, Wang L, Cai R, Zhang N, et al. Quorum Sensing N -acyl Homoserine Lactone s -SdiA Suppresses Escherichia 310 coli -Pseudomonas aeruginosa conjugation through inhibiting traI Expression. Microbiology 2017;7:1-8.
- Diggle SP, Gardner A, West SA, Griffin AS. Evolutionary theory of bacterial quorum sensing: when is a signal not a signal? Philos Trans R Soc Lond B Biol Sci 2007;362(1483):1241-49.
- Rutherford ST, Bassler BL. Bacterial Quorum Sensing: It’s Role in virulence and possibilities for its control. Cold Spring Harb Perspect Med 2012;2(11): a012427.
- Popat R, Crusz SA, Diggle SP. The social behaviours of bacterial pathogens. Br Med Bull 2008;87:63-75.
- Gonzalez JE, Keshavan ND. Messing with Bacterial Quorum Sensing. Microbiol Mol Biol Rev 2006;70(4):859-75.
- Scheie AA, Petersen FC. The Biofilm Concept: Consequences for future prophylaxis of oral diseases. Crit Rev Oral Biol Med 2004;15(1):4-12.
- Shiner EK, Rumbaugh KP, Williams SC. Inter-kingdom signalling: deciphering the language of acyl homoserine lactones. FEMS Microbiol Rev 2005;29(5);935-47.
- Von Bodman SB, Willey JM, Diggle SP. Cell-cell communication in bacteria: united we stand. J Bacteriol 2008;190(13):4377-91.
- Kline T, Bowman J, Iglewski BH, de Kievit TR, Kakai Y, Passador L. Novel synthetic analogs of the Pseudomonas autoinducer. Bioorg Med Chem Lett 1999;9(24):3447-52.
- Mari S, Vrane J. Characteristics and significance of microbial biofilm formation. Period Biolog 2007;109(2):1-7.
- Annous BA, Fratamico PM, Smith JL. Quorum Sensing in Biofilms: Why Bacteria Behave the Way They Do. J Food Sci 2009;74(1): R24-R37.
- Kim J, Hahn JS, Franklin MJ, Stewart PS, Yoon J. Tolerance of dormant and active cells in Pseudomonas aeruginosa PA01 biofilm to antimicrobial agents. J Antimicrob Chemother 2009;63(1):129-35.
- Shih PC, Huang CT. Effects of quoram-sensing deficiency on Pseudomonas aeruginosa biofilm formation and antibiotic resistance. J Antimicrob Chemother 2002;49(2):309-14.
- Nidadavolu P, Amor W, Tran PL, Dertien J, Colmer-Hamood JA, Hamood AN. Garlic ointment inhibits biofilm formation by bacterial pathogens from burn wounds. J Med Microbiol 2012;61(Pt 5):662-71.
- Borriello G, Werner E, Roe F, Kim AM, Ehrlich GD, Stewart PS. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob Agents Chemother 2004;48(7):2659-64.
- Bagge N, Hentzer M, Andersen JB, Ciofu O, Givskov M, Hoiby N. Dynamics and spatial distribution of β-lactamase expression in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 2004;48(4):1168-74.
- Hoffmann N, Lee B, Hentzer M, Rasmussen TB, Song Z, Johansen HK, et al. Azithromycin blocks quorum sensing and alginate polymer formation and increases the sensitivity to serum and stationary-growth-phase killing of Pseudomonas aeruginosa and attenuates chronic P. aeruginosa lung infection in Cftr-/-Mice. Antimicrob Agents Chemother 2007;51(10):3677-87.
- Rajput A, Kumar M. Anti-biofilm Peptides: A New Class of Quorum Quenchers and their Prospective Therapeutic Applications. In: Kalia VC. Editor. Biotechnological applications of quorum sensing inhibitors. 1st ed. Singapore: Springer Singapore; 2018. p. 87-110.
- Zhou L, Zheng H, Tang Y, Yu W, Gong Q. Eugenol inhibits quorum sensing at sub-inhibitory concentrations. Biotech Lett 2013;35(4):631-37.
- Dubois-Brissonnet F, Trotier E, Briandet R. The biofilm lifestyle involves an increase in bacterial membrane saturated fatty acids. Front Microbiol 2016;7:1673.
- Brackman G, Coenye T. Quorum Sensing Inhibitors as Anti-Biofilm Agents. Curr Pharm Des 2015;21(1):5-11.
- Whiteley M, Diggle SP, Greenberg EP. Progress in and promise of bacterial quorum sensing research. Nature 2017;551(7680):313-20.
- Papenfort K, Bassler BL. Quorum sensing signal–response systems in Gram negative Bacteria. Nat Rev Microbiol 2016;14(9):576-88.
- Sauer K. The genomics and proteomics of biofilm formation. Genome Biol 2003;4(6):219.
- Lee J, Zhang L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 2015;6(1):26-41.
- Cornforth DM, Popat R, McNally L, Gurney J, Scott-Phillips TC, Ivens A, et al. Combinatorial quorum sensing allows bacteria to resolve their social and physical environment. Proc Natl Acad Sci USA 2014;111:4280-84.
- MacLehose HG, Gilbert P, Allison DG. Biofilms, homoserine lactones and biocide susceptibility. J Antimicrob Chemother 2004;53(2):180-84.
- Bjarnsholt T, Jensen PO, Burmolle M, Hentzer M, Haagensen JA, Hougen HP, et al. Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology 2005;151(Pt 2):373-83.
- Andersen AS, Joergensen B, Bjarnsholt T, Johansen H, Karlsmark T, Givskov M, et al. Quorum-sensing-regulated virulence factors in Pseudomonas aeruginosa are toxic to Lucilia sericata maggots. Microbiology 2010;156(Pt 2):400-07.
- Singh BN, Singh HB, Singh A, Singh BR, Mishra A, Nautiyal CS. Lagerstroemia speciosa fruit extract modulates quorum sensing-controlled virulence factor production and biofilm formation in Pseudomonas aeruginosa. Microbiology 2012;158(Pt 2):529-38.
- Vandeputte OM, Kiendrebeogo M, Rasamiravaka T, Stevigny C, Duez P, Rajaonson S, et al. The flavanone naringenin reduces the production of quorum sensing-controlled virulence factors in Pseudomonas aeruginosa PAO1. Microbiology 2011;157:2120-32.
- Fuqua C. The QscR quorum-sensing regulon of Pseudomonas aeruginosa: an orphan claims its identity. J Bacteriol 2006;188(9):3169-71.
- Willcox MDP, Zhu H, Conibear TCR, Hume EBH, Givskov M, Kjelleberg S, et al. Role of quorum sensing by Pseudomonas aeruginosa in microbial keratitis and cystic fibrosis. Microbiology 2008;154(Pt 8):2184-94.
- Ding Y, Teo JWP, Drautz-Moses DI, Schuste SC, Givskov M, Yang L. Acquisition of resistance to carbapenem and macrolide-mediated quorum sensing inhibition by Pseudomonas aeruginosa via ICETn43716385. Commiun Biol 2018;1:57
- Rasmussen TB, Givskov M. Quorum sensing inhibitors: a bargain of effects. Microbiology 2006;152:895-904.
- Rasmussen TB, Bjarnsholt T, Skindersoe ME, Hentzer M, Kristoffersen P, Kote M, et al. Screening for Quorum-Sensing Inhibitors (QSI) by Use of a Novel Genetic System, the QSI Selector. J Bacteriol 2005;187(5):1799-814.
- Jimenez PN, Koch G, Thompson JA, Xavier KB, Cool RH, Quax WJ. The multiple signalling systems regulating virulence in Pseudomonas aeruginosa. Microbiol Mol Biol Rev 2012;76(1):46-65.
- Hentzer M, Riedel K, Rasmussen TB, Heydorn A, Andersen JB, Parsek MR, et al. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 2002;148(Pt 1):87-102.
- Delago A, Mandabi A, Meijler MM. Natural Quorum Sensing Inhibitors – Small Molecules, Big Messages. Isr J Chem 2015;56(5):310-20.
- Bjarnsholt T, Givskov M. Quorum-sensing blockade as a strategy for enhancing host defences against bacterial pathogens. Philos Trans R Soc Lond B Biol Sci 2007;362(1483):1213-22.
- Vandeputte OM, Kiendrebeogo M, Rajaonson S, Diallo B, Mol A, Jaziri ME, et al. Identification of catechin as one of the flavonoids From Combretum albiflorum bark extract that reduces the production of quorum-sensing-controlled virulence factors in Pseudomonas aeruginosa PAO1. Appl Environ Microbiol 2010;76(1):243-53.
- Jakobsen TH, Bragason SK, Phipps RK, Christensen LD, Van Gennip M, Alhede M, et al. Food as a Source for Quorum Sensing Inhibitors: Iberin from horseradish revealed as a quorum sensing inhibitor of Pseudomonas aeruginosa. Appl Environ Microbiol 2012;78(7):2410-21.
- Jakobsen TH, Tolker-Nielsen T, Givskov M. Bacterial Biofilm Control by Perturbation of Bacterial Signaling Processes. Int J Mol Sci 2017;18(9):1970.
- Adonizio AL, Downum K, Bennett BC, Mathee K. Anti-quorum sensing activity of medicinal plants in southern Florida. J Ethnpharmacol 2006;105(3):427-35.
- Adonizio A, Kong KF, Mathee K. Inhibition of Quorum Sensing-Controlled Virulence Factor Production in Pseudomonas aeruginosa by South Florida Plant Extracts. Antimicrob Agents Chemother 2008; 52(1):198-203.
- Koh KH, Tham F. Screening of traditional Chinese medicinal plants for quorum-sensing inhibitors activity. J Microbiol Immunol Infect 2011;44(2):144-48.
- Tham F, Yeo SS. Anti-quorum sensing and antimicrobial activities of some traditional Chinese medicinal plants commonly used in South-East Asia. Malay J Micro 2012;8(1):11-20.
- Keshavan ND, Chowdhary PK, Haines DC, González JE. l-Canavanine made by Medicago sativa interferes with Quorum Sensing in Sinorhizobium meliloti. J Bacteriol 2005;187(24):8427-36.