- *Corresponding Author:
- S. B. Prasad
Cell and Tumor Biology Laboratory, Department of Zoology, North-Eastern Hill University, Shillong-793 022, India
E-mail: sbnehu@gmail.com
| Date of Submission | 20 December 2018 |
| Date of Revision | 26 March 2019 |
| Date of Acceptance | 19 June 2019 |
| Indian J Pharm Sci 2019;81(4):720-728 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Rutin-induced changes in apoptosis and reduced glutathione levels was studied in murine ascites Dalton’s lymphoma cells under different treatment conditions followed by the binding affinity of rutin and cis-diamminedichloroplatinum (II) with two antiapoptotic proteins and two glutathione-related enzymes was examined through molecular docking analysis. Rutin, as well as cis-diamminedichloroplatinum (II) induced apoptosis and a decrease in reduced glutathione level in Dalton’s lymphoma cells. The molecular interactions of rutin, cis-diamminedichloroplatinum (II) and a known inhibitor of the protein/enzyme showed that the binding free energies (ΔG) with antiapoptotic proteins, Bcl-xL and c-FLIP, and glutathione-related enzymes such as glutathione S-transferase and glutathione reductase were closer and comparable, which might suggest inhibitory potential of rutin on these proteins/enzymes and could play a role in its anticancer activity.
Keywords
Rutin, apoptosis, glutathione, anticancer, molecular docking
Rutin or rutoside or sophorin is quercetin-3-rutinoside, a citrus flavonoid glycoside found in a wide variety of plants and has been reported to exhibit anticancer effect against many cancers[1-3]. Molecular docking is a frequently used tool in computer-aided structure-based rational drug design, which may help in evaluating how a small molecule called ligand and the target macromolecule (proteins/enzymes) fit together[4,5]. The anticancer activity of rutin involves inhibition of cell proliferation, a decrease in reduced glutathione (GSH) and induction of apoptosis in cancer cells[2,6]. Cis-diamminedichloroplatinum (II), (CDDP) is a well-known platinum-based cancer chemotherapeutic drug and its anticancer properties have been credited to its DNA binding ability, induction of apoptosis and decrease in glutathione level in cancer cells[7-9]. Therefore, to have a comparative study, CDDP was used as a reference anticancer drug in the present study.
Apoptosis is a genetically regulated programmed cell death and the efficacy of anticancer drugs is often measured by their ability to selectively promote apoptosis in cancer cells[10]. B-cell lymphoma-extralarge (Bcl-xL), a member of the Bcl-2 family of proteins, is an antiapoptotic protein, which leads to caspase activation and ultimately, programmed cell death[11]. c-FLIP (cellular FLICE, a FADD-like IL-1β-converting enzyme-inhibitory protein) is a major antiapoptotic protein and an important factor that suppresses cytokine- and chemotherapy-induced apoptosis[12]. GSH, a tri-peptide of L-cysteine, L-glutamic acid and glycine, plays important roles in antioxidant defense, nutrient metabolism, and regulation of cellular processes, including cell differentiation, proliferation and apoptosis[13]. Glutathione S-transferases (GST) catalyzes the conjugation of GSH to xenobiotic substrates for the purpose of detoxification. Glutathione reductase (GR) catalyzes the reduction of glutathione disulphide to GSH[14].
Known inhibitors such as ABT-737, plumbagin, ethacrynic acid, and ajoene for Bcl-xL, c-FLIP, GST and GR, respectively were also used for comparative in silico study. ABT-737 is a small molecule drug and a known inhibitor of Bcl-2 and Bcl-xL protein[15]. Plumbagin inhibits and down-regulates c-FLIP expression[16]. Ethacrynic acid is a potent inhibitor of GST family members, which are enzymes involved in xenobiotic metabolism[17]. Ajoene is a covalent inhibitor as well as a substrate of human GR[18].
On the basis of the significance of changes in apoptosis and glutathione in cancer chemotherapy, the present study was aimed to determine apoptosis and GSH level in murine ascites Dalton’s lymphoma (DL) cells under different treatment conditions and subsequently to perform in silico binding analysis using rutin, CDDP and a known inhibitor as a ligand with two antiapoptotic proteins, Bcl-xL and c-FLIP and two GSH-related enzymes, GST and GR.
Materials and Methods
Rutin (CAS No. 153-18-4, rutin hydrate) with ≥94 % purity was purchased from Sigma (St. Louis, MO, USA). CDDP solution (1 mg/ml of 0.9 %, NaCl) was obtained from Biochem Pharmaceutical Industries, Mumbai, India.
Tumor maintenance and drugs treatment schedule:
Originally, inbred Swiss albino mice were procured from Pasteur Institute, Shillong, India and inbred mice colony was maintained at 24±2° in a standard laboratory condition by providing food pellets (Amrut Laboratory, New Delhi) and drinking water to the animals ad libitum. Ascites DL tumor was maintained in vivo in 10-12 w old mice of both sexes weighing 25-30 g by serial intraperitoneal (ip) transplantation of viable tumor cells (1×107 cells in 0.25 ml phosphatebuffered saline (PBS, pH 7.4) to the animals as per the established procedure in the lab. Following tumor transplantation, an increase in abdomen size and body weight associated with sluggish movement of animals was noted from 3-4 d onwards, which was an early sign of tumor progression and malignancy[19,20]. Tumor-transplanted hosts usually survive for 19-21 d. The maintenance, use of the mice and the experimental protocols has been approved by the Institutional Animal Ethics Committee (No. NEC/IEC/2018/005, dated October 01, 2018) of the North-Eastern Hill University, Shillong, India.
Rutin was dissolved in dimethyl sulfoxide (DMSO, 1 mg/20 ml), then diluted in PBS to get the desired concentrations in which the final concentration of DMSO did not exceed 0.5 %. The CDDP solution (1 mg/ml of 0.9 %, NaCl) was obtained from Biochem Pharmaceutical Industries, Mumbai, India. The doses, the day of treatment of the drugs were based on earlier reports from the laboratory[3], and accordingly, rutin (30 mg/kg, ip) or CDDP (8 mg/kg, ip) were administered to tumor-transplanted mice. The experiments were carried out in 3 groups, group I- mice serving as tumorbearing control received vehicle only (PBS). Group II- mice were treated with rutin (30 mg/kg, ip) on d 8 and 10 post-tumor transplantation. Group III- mice were treated with a single therapeutic dose of CDDP (8 mg/kg, ip) on d 10 post-tumor transplantation.
Fluorescence-based apoptosis determination:
Apoptosis was determined in DL cells collected from mice under different treatment conditions using acridine orange and ethidium bromide (AO/EB) staining method as described previously[21]. The DL cells were collected at different time intervals from the intraperitoneal cavity of tumor-bearing hosts in different groups using an insulin syringe. Tumor cells were washed twice with PBS and treated with AO/EB (100 μg/ml PBS of each dye) for 5 min. The cells in different treatment groups were thoroughly examined under a fluorescent microscope (Leitz) and photographed using a digital camera (A1000IS-Canon). One thousand cells were analyzed and percent apoptotic cells was calculated from 15 selected view fields under a microscope and compared with that of control.
Estimation of GSH:
Total GSH in DL cells was determined using the method of Sedlak and Lindsay[22]. Briefly, 5 % homogenates of DL cells were prepared in 0.02 M EDTA (pH 4.7). One hundred microlitres of the homogenate or pure glutathione was added to 1.0 ml of 0.2 mol/l Tris-EDTA buffer, pH 8.2. To this, 0.9 ml of EDTA (0.02 mol/l, pH 4.7) with 20 μl of Ellman’s reagent (10 mM DTNB in methanol) were added. After 30 min of incubation at room temperature, the reaction mixture was centrifuged at 3000 g for 15 min and the absorbance of the clear supernatant was read against a reagent blank at 412 nm in a Varian Carey-50 spectrophotometer. The results were read from a standard curve prepared from 1 mM solution of GSH.
In silico study:
The 3-D structures of the ligands, rutin (CID 5280805), CDDP (CID 2767) and inhibitors ABT-737 (CID 11228183), plumbagin (CID 10205), ethacrynic acid (CID 3278) and ajoene (CID 5386591) were obtained from PubChem Database[23]. The 3-D structures of the proteins Bcl-xL (2YXJ), c-FLIP (3H13), glutathione transferase (2A2R) and GR (3DK8) were obtained from Protein Database Bank (PDB)[24].
These retrieved structures were modified and used for carrying out molecular docking studies using Autodock 4.2 software[25]. The proteins were individually loaded in Autodock 4.2 software. Any bound ligands and heteroatoms like water and ions were removed. Polar hydrogen bonds and Kollman charges were added to these proteins. The coordinates of the grid box were defined for individual proteins so that it covered the entire protein. Lamarckian Genetic Algorithm[25], was implemented in the docking study where the ligand, rutin, and CDDP were docked against these proteins individually. The binding energies of the protein-ligand complexes were evaluated/calculated in Autodock 4.2 based on the Eqn., ΔG = (VL-L[bound]–VL-L[unbound])+(VPP[ bound]–VP-P[unbound])+(VP-L[bound]–VP-L[unbound]+ΔSconf), where, P refers to the protein; L refers to the ligand; V represents the pair-wise evaluations and S denotes the loss of conformational entropy upon binding. The binding free energies obtained for all ligands and receptor interactions are shown in Table 1. Further, the ligplot analysis was carried out[26], for rutin-receptor complexes to analyse the protein residues involved in the interactions, as rutin was the main drug to explore the binding interactions.
Results and Discussion
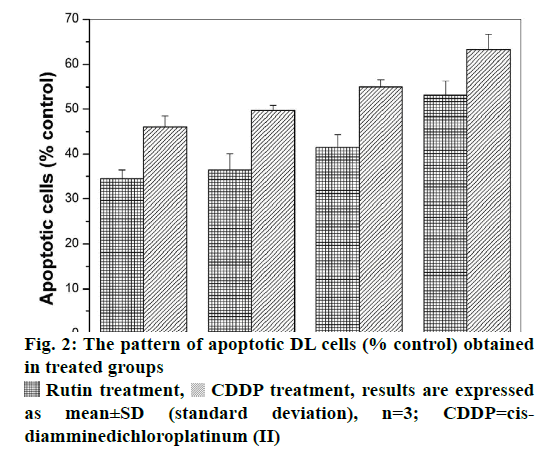
Viable DL cells nuclei stain green due to the permeability of only AO whereas apoptotic cells appear red/orange due to co-staining of AO and EB dyes. Rutin treatment induced various apoptotic features in DL cells. The prominent apoptotic features such as membrane blebbing (MB), apoptotic bodies (AB) and chromatin condensation (CC) were observed in rutin as well as CDDP-treated groups in a time-dependent manner (fig. 1). The apoptotic cells (% control) obtained in the rutin-treated group was ~53 % whereas in the CDDPtreated group was ~64 % (fig. 2). Rutin, as well as CDDP treatment, caused a substantial decrease in GSH level in DL cells (fig. 3).
Figure 1: Morphological features of apoptotic and viable DL cells stained with acridine orange and ethidium bromide
(A) Dalton’s lymphoma (DL) cells from tumor-bearing mice treated with rutin showing 24-96 h duration. (B) DL cells from tumorbearing mice treated with CDDP for the 24-96 h duration. Prominent apoptotic features such as membrane blebbing (MB), apoptotic bodies (AB), chromatin condensation (CC), early apoptosis (EA) and late apoptosis (LA) were noted. Green nuclei are viable cells (VC) and red/orange nuclei indicate apoptotic cells (EA/LA). Control=DL cells from tumor-bearing mice without any treatment; CDDP=cis-diamminedichloroplatinum (II). Scale bar of each picture is 50 μm (magnification: x100)
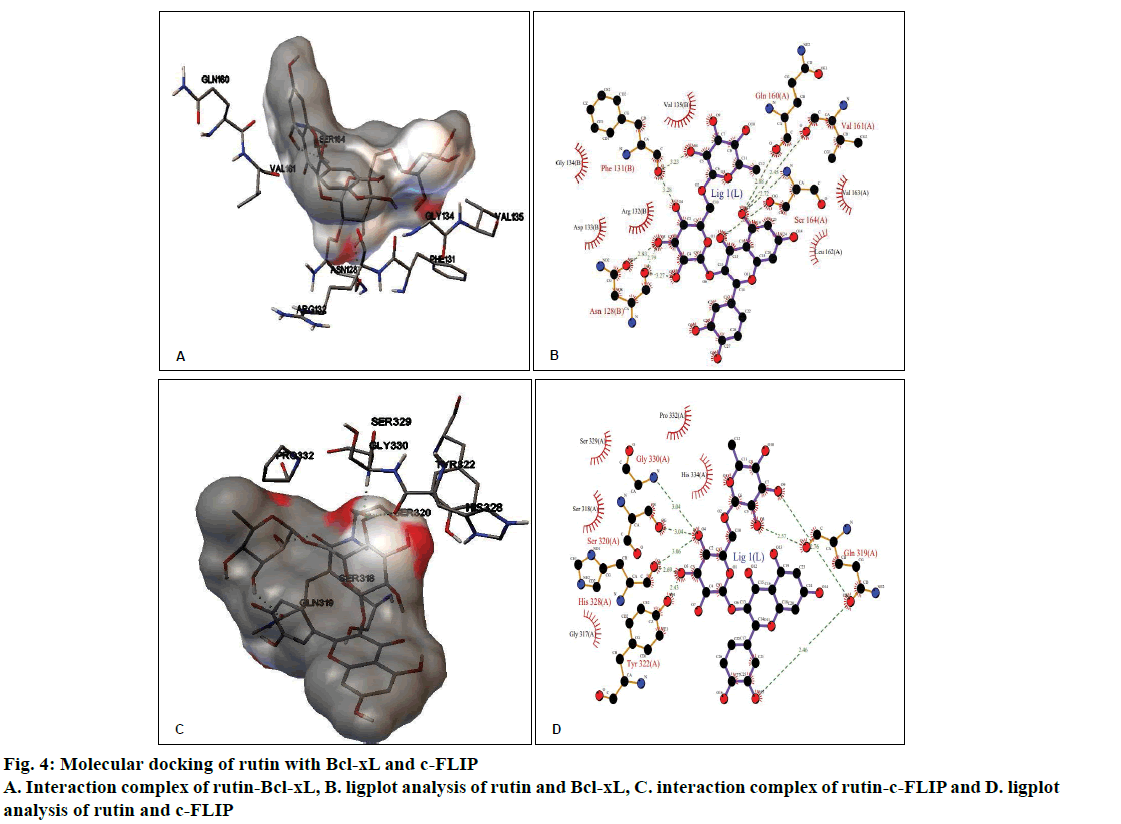
The molecular docking of different ligands and the proteins depicting their binding energy are shown in Table 1. The lowest binding energy between the protein and the ligand represents the best binding conformation and the present findings showed that the binding energy for rutin with GR was lowest (–5.36 kcal/mol). The ligplot analysis showed the interacting residues between the ligand and the protein. Rutin binds to Bcl-xL with ΔG of –3.27 kcal/mol, which was comparable with CDDP (–3.10 kcal/mol) and ΔG obtained with known inhibitor ABT-737 was –10.88 kcal/mol. The binding modes analysis of rutin-Bcl-xL revealed that it formed 9 hydrogen bonds and the number of nonbonded contacts was 96. The residues involved in the interaction mediated by hydrogen bonds are Asn128, Phe131, Gln160, Val161, and Ser164; while the residues mediated by hydrophobic contacts or nonbonded contacts are Arg132, Asp133, Gly134, Val135, Leu162 and Val163 (fig. 4A and B, Table 2).
Rutin binds to c-FLIP with ΔG of –3.17 kcal/mol, which was closer with CDDP interaction (–3.07 kcal/ mol) and comparable with known inhibitor plumbagin (–6.00 kcal/mol). The binding modes analysis of rutin with c-FLIP revealed that it formed 8 hydrogen bonds and a number of non-bonded contacts were 54. The residues involved in the rutin-c-FLIP interaction mediated by hydrogen bonds are Gln319, Ser320, Tyr322, His328, and Gly330; while the residues mediated by hydrophobic contacts or non-bonded contacts are Gly317, Ser318, Ser329, Pro332 and His334 (fig. 4C and D, Table 2).
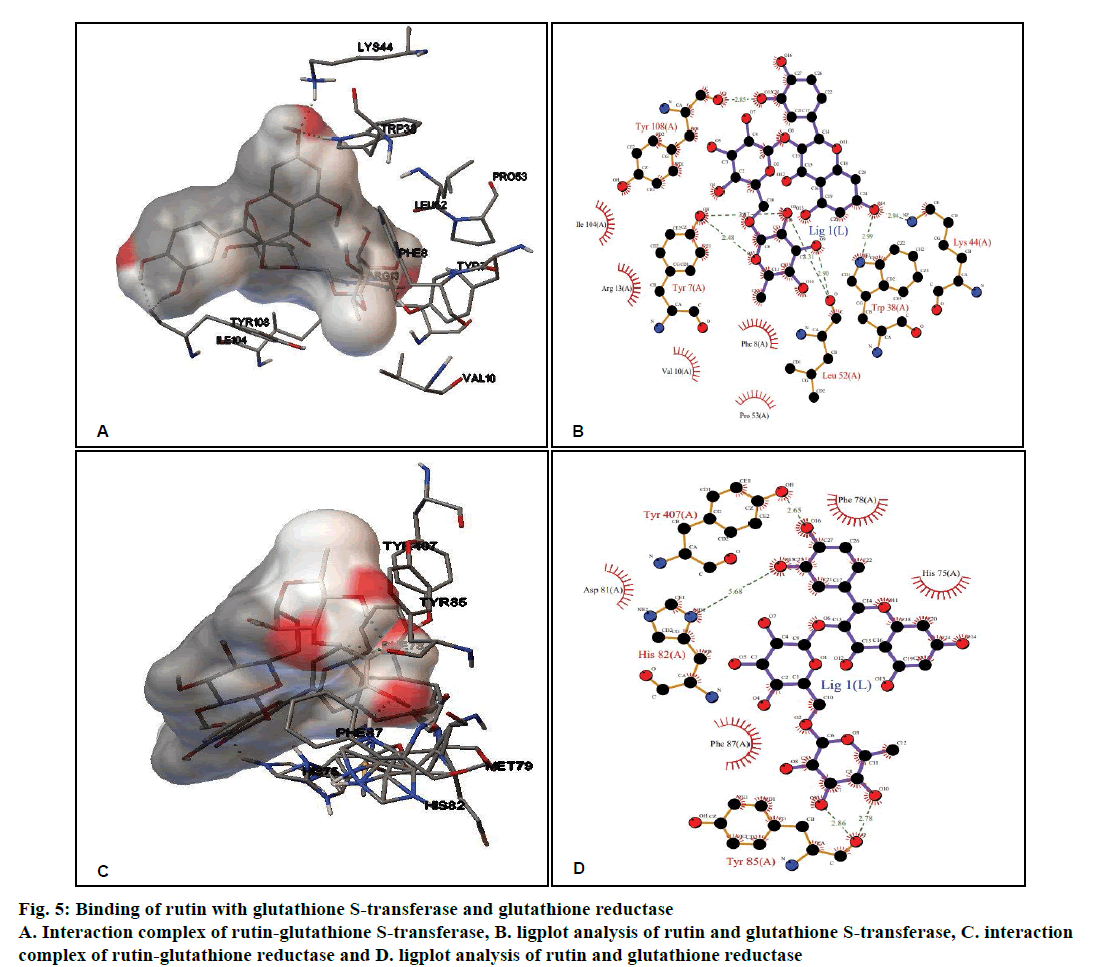
Rutin, CDDP or known inhibitor ethacrynic acid binds to GST with ΔG of –4.58 kcal/ mol, –3.29 kcal/mol and –6.65 kcal/mol, respectively. The binding modes analysis of rutin with GST revealed that it formed 8 hydrogen bonds and a number of non-bonded contacts were 56. The residues involved in the interaction mediated by hydrogen bonds are Tyr7, Trp38, Lys44, Leu52, and Tyr108; while the interactions mediated by hydrophobic contacts or non-bonded contacts are Phe8, Val10, Arg13, Pro53 and Ile104 (figs. 5A and B, Table 2).
Figure 5: Binding of rutin with glutathione S-transferase and glutathione reductase
A. Interaction complex of rutin-glutathione S-transferase, B. ligplot analysis of rutin and glutathione S-transferase, C. interaction complex of rutin-glutathione reductase and D. ligplot analysis of rutin and glutathione reductase
Rutin showed better binding energy with GR (–5.36 kcal/mol) as compared to CDDP (–2.83 kcal/ mol) and known inhibitor ajoene (–4.92 kcal/mol). This might suggest that rutin has inhibitory potential comparable to that of the known inhibitor. Rutin and GR interactions formed 5 hydrogen bonds and a number of non-bonded contacts were 59. The residues involved in the interaction mediated by hydrogen bonds are His82, Tyr85 and Tyr407, while the residues mediated by hydrophobic contacts or nonbonded contacts are: His75, Phe78, Asp81, and Phe87 (fig. 5C and D, Table 2).
Rutin is one of the phenolic compounds found in a wide variety of plants and has been reported to exhibit anticancer properties against many cancers[1]. The anticancer activity of rutin involves inhibition of cell proliferation, a decrease in GSH and induction of apoptosis in cancer cells[2,6]. The fluorescence based method has been used very commonly for the detection of apoptosis in cells[27]. The fluorescencebased apoptosis determination in DL cells using AO/ EB staining showed that rutin treatment significantly increased CC, nuclei fragmentation, the appearance of more AB and MB in DL cells (fig. 1). The number of apoptotic cells increased significantly after rutin treatment and it was comparable with that of CDDP treatment (fig. 2). Rutin has been known to induce apoptosis in cancers as a mode of its anticancer activity[28-30]. The lower intracellular GSH levels decrease cellular antioxidant capacity whereas elevated GSH levels generally increase antioxidant capacity and resistance to oxidative stress[31]. The GSH level decreased in DL cells after rutin treatment as observed for CDDP treatment also (fig. 3). The observed decrease in GSH content in DL cells in control animals at d 11- 14 of tumor growth might be associated with increased tumor burden along with decreased GSH metabolism/ synthesis in the hosts and has also been reported with a similar trend for Ehrlich ascites tumor cells[32]. However, with drug treatment at these time points observed further decrease may be due to inhibition of the enzymes GST, GR associated with GSH metabolism. The possible inhibitory potential of the drugs on these enzymes as noted in in silico analysis along with the inhibition in antiapoptotic proteins Bcl-xL and c-FLIP should be facilitating deaths in DL cells. The decrease in GSH level in DL cells could deteriorate the defensive ability of these cells and contribute to the tumor cells death (fig. 1). The rutin-mediated induction in apoptosis and decrease in GSH level in DL cells may involve changes in antiapoptotic proteins and GSH related enzymes, thus, these were examined through molecular docking analysis using their known inhibitor and other established anticancer drug, CDDP for comparison.
Molecular docking is of ample importance in the study and design of new drugs due to its ability to predict the binding-conformation of small molecule ligands to the appropriate target binding site. The interaction property, in turn, may also be used to predict the binding orientations of drug candidates to protein/ enzyme targets in order to presume the affinity and activity of the drug molecule[33]. The in silico docking analysis indicated that rutin strongly binds to the active site of antiapoptotic proteins i.e., Bcl-xL and c-FLIP, which were comparable with that of CDDP (Table 1). As the binding energy of rutin and CDDP is very close to each other, their binding affinities may also be similar. However, the binding energy of their known inhibitors was much lower than rutin or CDDP, this may indicate stronger inhibitory activity (fig. 4; Table 1). Binding energy is correlated with the probability of affinity and may also predict the bioactivity value for a ligand to the corresponding receptor. The ligplot analysis revealed that rutin interacts with Bcl-xL and c-FLIP involving 9 and 8 hydrogen bonds, respectively (Table 2). During the effect of an apoptotic related drug generally antiapoptotic and proapoptotic proteins activity is affected in the opposite way i.e. during the increase in the activity of a proapoptotic protein there is a simultaneous decrease in the antiapoptotic protein activity. A study has shown that combined administration of CDDP and bortezomib caused up-regulation of caspase‑3, ‑ 8 and ‑ 9 and simultaneously down-regulated Bcl‑2 and Bcl‑xL protein expression levels in T24 cells[34]. The in silico docking study using 5 derivatives of gallic acid against Bcl-xL protein, it was noted that 3,4,5-trimethoxycis- 2-hexenylgallate serves as a potential inhibitor of antiapoptotic Bcl-xL protein and could be a promising candidate to develop as an anticancer agent for treating breast cancer[35]. Gorakh et al.[36] reported the interaction of the c-FLIP with the natural and synthetic inhibitors that stop the activity of c-FLIP with highest binding shown by droxinostat and chyrisin and inhibition of c-FLIP could help in increasing the apoptosis of cancer cells. Thus, the present observation of rutin’s interaction with Bcl-xL and c-FLIP (Table 2) might also involve a similar effect in increasing the apoptosis in DL cells.
| Receptor protein | PDB ID | Binding energy (ΔG) with rutin (kcal/mol) | Binding energy (ΔG) with CDDP(kcal/mol) | Binding energy (ΔG) with a known inhibitor (kcal/mol) |
|---|---|---|---|---|
| Bcl-xL | 2YXJ | –3.27 | –3.10 | –10.88 |
| c-FLIP | 3H13 | –3.17 | –3.07 | –6.00 |
| GST | 2A2R | –4.58 | –3.29 | –6.65 |
| GR | 3DK8 | –5.36 | –2.83 | –4.92 |
CDDP=cis-diamminedichloroplatinum (II); BCL-xL=B-cell lymphoma-extra-large; c-FLIP=cellular FLICE (FADD-like IL-1Β-converting enzyme)-inhibitory protein; GST=glutathione S-transferase and GR=glutathione reductase
Table 1: Binding energies of rutin, CDDP and known inhibitor against docked target proteins
GST and GR are frequently over-expressed in different types of cancers[37,38]. It was noted that rutin binds to GST and GR with a binding energy of –4.58 kcal/mol and –5.36 kcal/mol, respectively (Table 1) which was comparable and closer to the binding energy obtained for CDDP and known inhibitor. Rutin interacted with GST and GR involving 8 and 5 hydrogen bonds, respectively as revealed by the ligplot analysis (Table 2). Methotrexate was reported to inhibit GST activity and a molecular docking study showed that methotrexate interacted with the active site of GST[39]. It has been reported that competitive or irreversible inhibition of GR leads to a decrease in GSH levels[40]. Therefore, the interactions of rutin with these enzymes (fig. 5) and observed binding energies (Tables 1 and 2) may suggest the strong binding affinity which may cause the inhibitory effect of rutin on these enzymes leading to an imbalance/decrease in GSH level and facilitating its anticancer activity in DL cells.
| Receptor protein | PDB ID | Binding energy (ΔG) [kcal/mol] | Residues mediated by hydrogen bonds | Residues mediated by hydrophobic non-bonded contacts | Final inter-molecular energy (kcal/mol) | No. of H-bond |
|---|---|---|---|---|---|---|
| Bcl-xL | 2YXJ | –3.27 | Asn128, Phe131, Gln160, Val161 and Ser164 | Arg132, Asp133, Gly134, Val135, Leu162 and Val163 | –8.04 | 9 |
| c-FLIP | 3H13 | –3.17 | Gln319, Ser320, Tyr322, His328, and Gly330 | Gly317, Ser318, Ser329, Pro332, and His334 | –7.95 | 8 |
| GST | 2A2R | –4.58 | Tyr7, Trp38, Lys44, Leu52,and Tyr108 | Phe8, Val10, Arg13, Pro53,and Ile104 | –9.35 | 8 |
| GR | 3DK8 | –5.36 | His82, Tyr85, and Tyr407 | His75, Phe78, Asp81, and Phe87 | –10.14 | 5 |
Table 2: Molecular interactions of rutin with BCL-xL, c-FLIP, GST and GR
The findings revealed that rutin induced apoptosis and decreased antioxidant ability (GSH level) in DL cells. Molecular interactions of rutin with antiapoptotic proteins, Bcl-xL, c-FLIP, and glutathione-related enzymes, GST and GR showed substantial binding energy comparable to that of CDDP and a specific inhibitor, which might indicate the potency of inhibition of these proteins/enzymes. This inhibitory potential could be involved in increasing apoptosis along with a decrease in GSH in DL cells and in the anticancer activity of rutin.
Acknowledgments:
The authors gratefully acknowledge Department of Zoology and Department of Biotechnology and Bioinformatics, North Eastern Hill University, Shillong for providing the required facilities essential for the completion of the present studies.
Conflict of interest:
The authors declare no conflict of interest.
References
- Sghaier BM, Pagano A, Mousslim M, Ammari Y, Kovacic H, Luis J. Rutin inhibits proliferation, attenuates superoxide production and decreases adhesion and migration of human cancerous cells. Biomed Pharmacother 2016;84:1972-8.
- Elsayed HE, Ebrahim HY, Mohyeldin MM, Siddique AB, Kamal AM, Haggag EG, et al. Rutin as a novel c-Met inhibitory lead for the control of triple negative breast malignancies. Nutr Cancer 2017;69(8):1256-71.
- Prasad R, Prasad SB. Antitumor activity of rutin-cisplatin in combination and its protective effect against hematotoxicity. Res J Life Sci Bioinform Pharm Chem Sci 2018;4(6):42-56.
- Seeliger D, de Groot BL. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput Aided Mol Des 2010;24:417-22.
- Norgan AP, Coffman PK, Kocher JPA, Katzmann DJ, Sosa CP. Multilevel parallelization of AutoDock 4.2. J Cheminform 2011;3:12-15.
- Vadapalli U, Muvvala S, Alluri R, Lakshmi BVS. Anti-proliferative activity of rutin on HeLa cell line induced cervical cancer in rats. Int J Pharm Sci Res 2017;8(11):4803-11.
- Cohen SM, Lippard SJ. Cisplatin: from DNA damage to cancer chemotherapy. Prog Nucleic Acid Res Mol Biol 2001;67:93-130.
- Khynriam D, Prasad SB. Changes in endogenous tissue glutathione level in relation to murine ascites tumor growth and the anticancer activity of cisplatin. Braz J Med Biol Res 2003;36:53-63.
- Amenla, Verma AK, Prasad SB. Dietary ascorbic acid-mediated augmentation of antitumor activity and protection against toxicities induced by cis-diamminedichloroplatinum-(II) in Dalton’s lymphoma-bearing mice. J Cancer Res Update 2013;2:116-30.
- Liu K, Liu P, Liu R, Wu X. Dual AO/EB staining to detect apoptosis in osteosarcoma cells compared with flow cytometry. Med Sci Monit Basic Res 2015;21:15-20.
- Korsmeyer SJ. Regulators of cell death. Trends Genet 1995;11(3):101-5.
- Safa AR. c-FLIP, a master anti-apoptotic regulator. Exp Oncol 2012;34(3):176-84.
- Abdalla MY. Glutathione as a potential target for cancer therapy; More or less is good? Jordan J Biol Sci 2011;4(3):119-24.
- Deponte M. Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes. Biochim Biophys Acta 2012;1830(2013):3217-66.
- Vogler M, Dinsdale D, Dyer MJS, Cohen GM. Bcl-2 inhibitors: small molecules with a big impact on cancer therapy. Cell Death Differ 2008;16:360-7.
- Kong X, Luo J, Xu T, Zhou Y, Pan Z, Xie Y, et al. Plumbagin enhances TRAIL-induced apoptosis of human leukemic Kasumi-1 cells through up-regulation of TRAIL death receptor expression, activation of caspase-8 and inhibition of c-FLIP. Oncology Rep 2017;37:3423-32.
- Ploemen JH, van Ommen B, Bogaards JJ, van Bladeren PJ. Ethacrynic acid and its glutathione conjugate as inhibitors of glutathione S-transferases. Xenobiotica 1993;23(8):913-23.
- Gallwitz H, Bonse S, Martinez-Cruz A, Schlichting I, Schumacher K, Krauth-Siegel RL. Ajoene is an inhibitor and subversive substrate of human Glutathione reductase and Trypanosoma cruzi Trypanothione reductase: Crystallographic, Kinetic, and Spectroscopic Studies. J Med Chem 1999;42:364-72.
- Verma AK, Prasad SB. Changes in glutathione, oxidative stress, and mitochondrial membrane potential in apoptosis involving the anticancer activity of Cantharidin isolated from red-headed Blister Beetles, Epicauta hirticornis. Anticancer Agents Med Chem 2013;13:1096-114.
- Bhaumik A, Prasad SB. Studies on the antitumor potentials of betulinic acid against murine ascites Dalton’s lymphoma. Int J Basic Clin Pharmacol 2016;5(4):1664-71.
- Squier MKT, Cohen JJ. Standard quantitative assays for apoptosis. Mol Biotechnol 2001;19:305-12.
- Sedlak J, Lindsay RH. Estimation of total, protein-bound, and nonprotein sulfhydryl groups in tissue with Ellman’s reagent. Anal Biochem 1968;25:192-205.
- Bolton EE, Wang Y, Thiessen PA, Bryant SH. PubChem: integrated platform of small molecules and biological activities. Annu Rep Comput Chem 2008;4:217-41.
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. Protein data bank. Nucleic Acids Res 2000;28:235-42.
- Rizvi SM, Shakil S, Haneef M. A simple click by click protocol to perform docking: Autodock 4.2 made easy for the non-bioinformatics. EXCLI J 2013;12:831-57.
- Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate a schematic diagram of protein-ligand interactions. Protein Eng 1995;8:127-34.
- Martinez MM, Reif RD, Pappas D. Detection of apoptosis: A review of conventional and novel techniques. Anal Methods 2010;2:996-1004.
- Chen H, Miao Q, Geng M, Liu J, Hu Y, Tian L, et al. Antitumor effect of rutin on human neuroblastoma cell lines through inducing G2/M cell cycle arrest and promoting apoptosis. Sci World J 2013;2013:269165.
- Li Y, Duan S, Jia H, Bai C, Zhang L, Wang Z. Flavonoids from Tartary buckwheat induce G2/M cell cycle arrest and apoptosis in human hepatoma HepG2 cells. Acta Biochim Biophys Sin 2014;46(6):460-70.
- Iriti M, Kubina R, Cochis A, Sorrentino R, Varoni EM, Kabała-Dzik A, et al. Rutin, a quercetin glycoside, restores chemo-sensitivity in human breast cancer cells. Phytother Res 2017;31(10)1529-38.
- Traverso N, Ricciarelli R, Nitti M, Marengo B, Furfaro AL, Pronzato MA. Role of glutathione in cancer progression and chemo-resistance. Oxid Med Cell Longev 2013;2013:1-10.
- Estrela JM, Hernandez R, Terradez P, Asensi M, Puertes IR, Vina J. Regulation of glutathione metabolism in Ehrlich ascites tumour cells. Biochem J 1992;286:257-62.
- Himaja M, Sreekanth K, Ranjitha A, Asif K, Sikarwar MS. Synthesis, docking studies and antioxidant activity of linear tetrapeptide FAYV. Int Res J Pharm 2011;2(7):186-9.
- Konac E, Varol N, Kiliccioglu I, Bilen CY. Synergistic effects of cisplatin and proteasome inhibitor bortezomib on human bladder cancer cells. Oncol Lett 2015;10:560-4.
- Paramita RI, Arsianti A, Radji M. In silico docking studies of alkyl esters derivative of gallic acid on Bcl-xL anti-apoptotic protein of breast cancer. Int J ChemTech Res 2017;10(1):348-55.
- Gorakh WJ, Angshu B, Mithal A. Computational studies on c flip inhibitors as a novel target on breast cancer. World J Pharm Pharm Sci 2016;5(7):1434-44.
- Allocati N, Masulli M, Ilio CD, Federici L. Glutathione transferases: substrates, inhibitors and pro-drugs in cancer and neurodegenerative diseases. Oncogenesis 2018;7:8-22.
- Lorestani S, Hashemy SI, Mojarad M, Shahrestanaki MK, Bahari A, Asadi M, et al. Increased glutathione reductase expression and activity in colorectal cancer tissue samples: An investigational study in Mashhad, Iran. Middle East J Cancer 2018;9(2):99-104.
- Ozgencli I, Kilic D, Guller U, Ciftci M, Kufrevioglu OI, Budak H. A comparison of the inhibitory effects of anti-cancer drugs on thioredoxin reductase and glutathione S-transferase in rat liver. Anticancer Agents Med Chem 2018;18(14):2053-61.
- Rubino FM. Toxicity of Glutathione-binding metals: A review of targets and mechanisms. Toxics 2015;3:20-62.

 Rutin treatment,
Rutin treatment,  CDDP treatment, results are expressed as mean±SD (standard deviation), n=3; CDDP=cisdiamminedichloroplatinum (II)
CDDP treatment, results are expressed as mean±SD (standard deviation), n=3; CDDP=cisdiamminedichloroplatinum (II)
 Control, rutin treatment,
Control, rutin treatment,  CDDP treatment, results are expressed as mean±SD, n=3;
CDDP treatment, results are expressed as mean±SD, n=3; 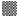 CDDP=cis-diammine dichloroplatinum (II),*p≤0.05 as compared to the corresponding control
CDDP=cis-diammine dichloroplatinum (II),*p≤0.05 as compared to the corresponding control