- *Corresponding Author:
- D. V. Kohli
Department of Pharmaceutical Sciences, Dr. H. S. Gour Vishwavidyalaya, Sagar-470 003, India.
E-mail: amitnayak2001@yahoo.com
| Date of Submission | 07 May 2004 |
| Date of Revision | 25 February 2006 |
| Date of Acceptance | 26 June 2006 |
| Indian J Pharm Sci, 2006, 68 (4): 415-419 |
Abstract
Spliceosome-mediated RNA trans-splicing is a new technology for gene therapy that exploits the expressed genetic differences between normal and diseased cells. This technology may be applied to a wide range of diseases that involve the expression of unique or mutated genes. Spliceosome-mediated RNA trans-splicing technology can be employed to control the expression of any delivered gene by the presence of the chosen target pre-mRNA.
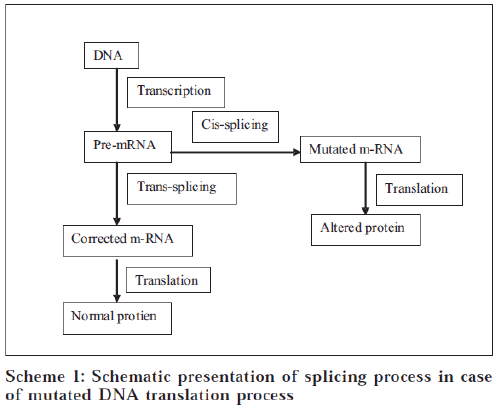
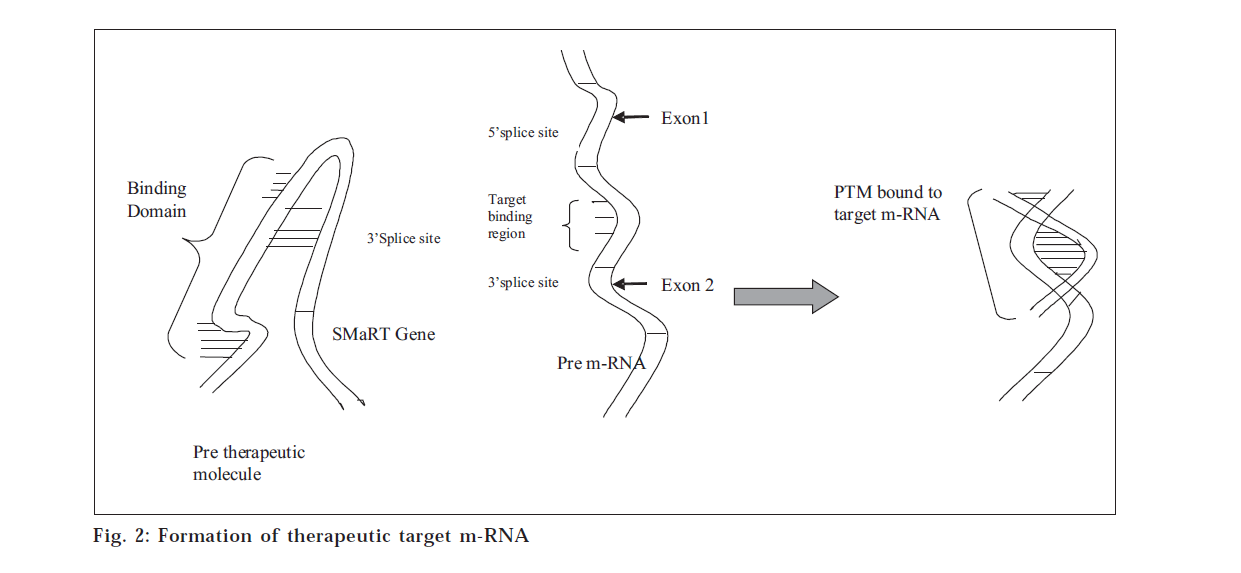
The term SMaRT stands for Spliceosome-mediated RNA trans-splicing. This technique targets and repairs the messenger RNA (mRNA) transcripts copied from the trans-splicing between the pre-therapeutic molecule (PTM) and its pre-mRNA target mutated gene [1]. Instead of attempting to replace the entire gene, this technique repairs only that section of the mRNA transcript which contains the mutation.
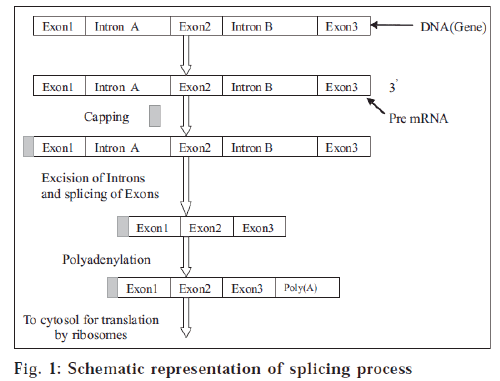
DNA sequences contain the genetic information that encodes proteins. The decoding process of DNA to protein occurs through a nucleic acid intermediate called messenger RNA (mRNA) [2]. The blocks of genetic sequence that encode amino acids (protein-building blocks) are known as exons. Exons are separated from each other by blocks of sequence called introns. Cells use a large molecular complex called a spliceosome to clip out the introns and splice together the exons. The resulting messenger RNA molecule is the final template that cells use to make a protein [3]. SMaRT involves three steps (fig. 1): 1. Delivery of an RNA strand that pairs specifically with the intron next to the mutated segment of mRNA. Once bound, this RNA strand prevents spliceosomes from including the mutated segment in the final spliced RNA product. 2. Simultaneous delivery of a correct version of the segment to replace the mutated piece in the final mRNA product. 3. Translation of the repaired mRNA to produce the normal functional protein.
In human cells, the majority of the primary transcripts synthesized by RNA polymerase II contain interruptions in the coding sequences, which are known as introns. Most messenger RNA precursors (pre-mRNA) undergo cis-splicing in which introns are excised and the adjoining exons from a single pre-mRNA are ligated together to form mature mRNA. This reaction is driven by a complex known as the spliceosome (a macromolecular enzyme). Spliceosomes can also combine sequences from two independently transcribed pre-mRNAs in a process known as trans-splicing. SMaRT is an emerging technology in which RNA pre-therapeutic molecules (PTMs) are designed to recode a specific pre-mRNA by suppressing cis-splicing while enhancing trans-splicing between the PTM and its pre-mRNA target.
Most protein-encoding RNAs have to be processed by RNA splicing to generate fully functional messenger RNAs [4]. This discovery engendered the concept of RNA repair for therapeutic applications. The basic idea is that since RNAs are continuously being revised in human cells, perhaps the RNA revisionist machinery could be redirected to repair mutant RNAs associated with disease [5].
Splicing of m-RNA
Introns are removed from nuclear mRNA precursors via a two-step trans-esterification pathway in complex ribonucleoprotein particles called spliceosomes. In the first step, the 2'-hydroxyl group of an adenosine which is sited near the 3' end of the intron attacks the 5' splice site, producing 5'-exon and lariat intron-3’exon intermediates. In the second step, the 3'-hydroxyl of the 5'-exon intermediate attacks the 3' splice site to give the spliced mRNA and lariat intron products of splicing. The spliceosomal small nuclear ribonucleoprotein particles (snRNPs) are major structural and functional components of the splicing machinery. The small nuclear RNA components (U1, U2, U4, U5 and U6 snRNAs) of the snRNPs play diverse roles in intron recognition and splice site definition and may be intimately involved in spliceosomal catalysis.
Splicing between two independently transcribed premRNAs is termed trans-splicing. This has been described in trypanosomes, nematodes, flatworms and plant mitochondria. In vitro trans-splicing has been used as a model system to examine the mechanism of splicing. Trans-splicing of pre-mRNAs in human cells has been postulated to account for some rare events. Until now, the practical application of targeted trans-splicing to modify specific mRNAs within human cells has been limited to those based upon tetrahymena group I ribozyme-based mechanisms.
The process of pre-mRNA splicing can be divided into three stages [1]
In the first stage, formation of the commitment complex takes place, which is the precise recognition of intron-exon junctions (splice sites) and the correct pairing of the 5’ splice site with its cognate 3’ splice site is critical for splice site selection. It is during the formation of the commitment complex that splice sites are first recognized by spliceosomal components with the aid of certain nonspliceosomal proteins.
The second stage involves creation of catalytic sites. Here a number of dynamic interactions, including snRNApre-mRNA interactions as well as pre-mRNA-protein and protein-protein interactions, bring the reactive sites on the pre-mRNA together and create the catalytic sites for the trans-esterification reactions.
Finally in the third stage, which involves trans-esterification reactions, the cleavage and ligation reaction required for intron removal and exon ligation proceeds via two trans-esterification reactions. In the first reaction, the 5’-exon is cleaved and the 5’ end of the intron is joined to the branch point creating the intron lariat structure. The second reaction occurs when the free 3’ end of the 5’ exon is joined to the downstream exon, resulting in exon ligation and release of the intron sequence. This mechanism is similar to that used by a class of self-splicing introns called Group II introns. The Group II introns are found in certain ribosomal RNAs (rRNA) and transfer RNAs (tRNA) and can undergo self-splicing in the absence of any proteins. The similarity in the mechanisms of spliceosome-mediated splicing and Group II intron self-splicing has led to the hypothesis that the catalytic core of the spliceosome functions are similar to an RNA enzyme (ribozyme).
Cis-splicing
Components of the spliceosome recognize the exon-intron boundaries at the 5' and 3' splice sites, excise the intron and ligate the adjoining exons. In most cases, splicing joins a 5' splice site and a 3' splice site within the same pre-mRNA molecule, termed cis-splicing [6].
Two trans-esterification reactions remove each intron and ligate the adjacent exons. Spliceosome catalyzes the selection of the splice sites and the subsequent trans-esterification reaction. The spliceosome is composed of four ribonucleoprotein (RNP) particles and scores of protein. Each small RNP particle contains one or two U-rich small nuclear RNAs (snRNAs). These are named as U1 snRNP, U2 snRNP, U5 snRNP, U4/6 snRNP. U1 snRNP recognizes 5’ splice site’s consensus sequences in part by an RNA-RNA interaction between the premRNAs. Similarly, the U2 snRNP recognizes the branch point sequences of the major class introns; that is why these introns are known as U2 introns [7].
Cis-splicing versus trans-splicing
The splicing reactions described above most commonly involve splice sites located within one RNA molecule and are thus considered to occur in cis manner. In trypanosomes, flatworms and nematodes, however, specialized spliceosomes can mediate the trans-splicing between highly structured short-leader RNAs and many different pre-mRNAs. These specialized trans-splicing reactions are very efficient. In mammals, however, trans-splicing of conventional pre-mRNAs appears to be exceedingly rare, suggesting a mechanistic barrier to trans-splicing [8-11]. In principle, the low level of naturally occurring trans-splicing could be due to the presence of trans-acting inhibitors or to the lack of specific trans-activators in mammalian cells. These hypotheses were ruled out when it was shown that specialized Short-Leader RNAs could trans-splice to pre-mRNAs in mammalian cells [12]. A block of efficient trans-splicing would also be expected if spliceosomes had to scan conventional introns in order to achieve splice site juxtaposition and subsequent splicing. Scanning was essentially ruled out when pre-mRNAs that included obstacles to scanning were shown to splice in vitro with unaltered efficiency [13]. A variation of the scanning objection remained plausible, however, since in vivo splicing occurs cotranscriptionally and the elongating RNA polymerase II is likely tethered to spliceosome components that recognize the splice sites [14,15]. This tethering of the splice sites can significantly reduce the likelihood that splice sites in two nascent transcripts will meet. This potential barrier for naturally occurring trans-splicing has to be carefully considered when designing a targeted SMaRT effector.
Applications Of Smart Gene Therapy
Gene therapies are constructs derived from nature. The manipulations needed to create genetic therapy add enormous complexity to considerations of safety and preclinical toxicity testing; and for every intended consequence of a complex biologic product, there are unintended consequences. Biologic products, like all products, carry risks along with benefits [1].
SMaRT is an emerging technology in which RNA pre-therapeutic molecules (PTMs) are designed to recode a specific pre-mRNA by suppressing cis-splicing (fig. 2) while enhancing trans-splicing between the PTM and its pre-mRNA target [16]. SMaRT may represent a general approach for reprogramming the sequence of targeted transcripts, providing a novel approach to gene therapy [6].
Splicing may also be derailed by mutations within introns that create novel splice sites, resulting in the inappropriate inclusion of non-coding sequence. This often occurs close to exons but may also occur deep within introns, creating either a novel donor or acceptor site that, in conjunction with a nearby cryptic splice site of the opposite polarity, defines a novel aberrant exon that the spliceosome recognizes and includes into the mature message. Several examples of this mutational mechanism have been shown to underlie inherited diseases, such as β-thalassemia [17-21] CF [22,23], neurofibromatosis type1 [24], multiple breast tumours [25] dihydropteridine reductase deficiency [26], maple syrup urine disease [27], alport syndrome [28] ornithine δ-aminotransferase deficiency [29], β-glucouronidase deficiency [30], ataxiatelangiectasia [31], congenital lipoid adrenal hyperplasia [32] and obesity in mice [33,34]. The true prevalence of mutations of this kind is probably underestimated since few laboratories analyse intron sequences far from coding regions [35].
Pharmaceutical applications
Cystic fibrosis is the most common genetic inherited disease in the Caucasian population that results from a mutated cystic fibrosis transmembrane regulator (CFTR) protein. This protein normally transports chloride ions across cell membranes, thus maintaining cells electrolyte and fluid balance. In turn, this balance allows bacteria to be cleared from the surface of airway cells and prevents infection. A mutated CFTR protein disrupts normal ion transport and people with CF are prone to chronic and often fatal bacterial lung infections.
In addition to providing a doorway for chloride ions to pass across cell membranes, the CFTR protein also appears to be involved in regulating the action of other ion channels. Proper regulation of the CFTR channel, together with all its neighbouring channels, seems to be necessary for normal lung function and has been a challenge for gene therapists approaching genetic treatment of this disease.
Medical applications describe alternative RNA splicing that is observed in a high proportion of genes; it is not surprising that alterations in RNA splicing can cause or be modified by a disease [36]. Characterization of these splice-specific alterations can provide both new therapeutic targets and diagnostics.
Deregulation of RNA splicing can be induced by mutations or polymorphisms within the gene (e.g., CYP2D6, ß-globin and dystrophin) or changes induced because of alterations or mutations in the splicing machinery (e.g., SMN1) or changes in cell signalling pathways (e.g., FGFR2 and Bcl-X). ss
The cytochrome P450 family (CYP) are enzymes required for drug metabolism. One important member of this family is CYP2D6, which is responsible for the oxidation of at least 40 prescribed drugs [37]. Between 5 and 10% of Caucasians are unable to metabolize drugs such as debrisoquin, the antihypertensive drug, because of polymorphisms within this gene. The most frequent change observed in CYP2D6 is a polymorphism that results in an aberrant splice event, which causes truncation and inactivation of the protein [38].
ß-thalassemia, caused by mutations in the ß-globin gene, is one of the most common human genetic diseases. The most frequent mutations are in introns 1 and 2, which result in aberrant splicing of the mRNA. Correction of this aberrant splicing by blocking the incorrect splice sites with antisense oligonucleotides has been demonstrated39 and is likely to provide a powerful therapeutic approach to correct this disorder.
Duchenne muscular dystrophy (DMD) is caused by a frame shift in the dystrophin gene, which results in the insertion of a premature stop codon and the expression of a truncated inactive protein. Antisense oligonucleotides have been successfully used to induce alternative splicing of dystrophin gene [40]. These alternative splice events remove the exon that encodes the stop codon and return the transcript to the correct reading frame, thereby generating a near full-length transcript that has partial activity and reduce the severity of the condition. Antisense strategies may be applicable not only to ß-thalassemia and DMD mutations but also many other genetic diseases and disease-causing alternative splice events.
Spinal muscular atrophy (SMA) is a disorder characterised by progressive loss of spinal cord motor neurons, resulting in paralysis. This disorder occurs due to the deletion of the survivor of motor neuron gene (SMN1) [41]. SMN1 is involved in the assembly of ribonucleoprotein complexes [42]. Deletion of SMN prevents assembly of the U1 ribonucleoprotein complexes in the cytoplasm and results in global defects in pre-mRNA splicing.
The nfibroblast growth factor receptor 2 (FGFR2) represents a clear example of the diagnostic and therapeutic value of monitoring RNA splicing deregulation during disease. The extracellular immunoglobulin-like (Ig) domains of the protein interact with different fibroblast growth factors, setting in motion a cascade of downstream signals, ultimately influencing mitogenesis and differentiation. Two of the alternative RNA isoforms of FGFR2 (IIIb and IIIc) encode altered Ig domains and show differing affinities to the different fibroblast growth factors and therefore differential signalling. Changes in the FGFR2 ratio correlate to prostate cancer progression and are associated with lung, skin and bone defects [43-45]. Control of the alternative splicing of FGFR2 is mediated in part through the action of polypyrimidine tract binding protein (PTB), which acts as an exon-silencing factor that may also have a role in the progression of cancer.
SMaRT may represent a general approach for reprogramming the sequence of targeted transcripts, providing a novel approach to gene therapy. Exon replacement by SMaRT can repair an abnormal premRNA associated with a genetic disease.
References
- Mansfield, S.G., Kole, J., Puttaraju, M., Yang, C.C., Garcia-Blanco,M.A., Cohn, J.A. and Mitchell, L.G., Gene Therapy, 2000, 22, 1885.
- Gorman, L., Suter, D., Emerick, V., Schumperli, D. and Kole, R.,Biochemistry, 1998, 95, 4929.
- Garcia-Blanco, M.A., Puttaraju, M., Mansfield, S.G. and Mitchell, L.G., Gene Therapy Regulation, 2000, 1, 141.
- Burge, C.B., Tuschl, T. and Sharp, P.A., In: The RNA World, ColdSpring Harbor Laboratory Press, New York, 1999, 525.
- Bruce, A.S., J. Clin. Invest.,2003, 112, 310.
- Noguchi, M.D., N. Engl. J. Med. Chem., 2003, 348,193.
- Puttaraju, M., Jamison, S.F., Mansfield, S.G., Garcia-Blanco, M.A. and Mitchell, L.G., Nat. Biotechnol., 1999,17, 246.
- Spritz, R.A., Jagadeeswaran, P., Choudary, P.V., Biro, P.A., Elder,J.T., deRiel, J.K., Manley, J.L., Gefter, M.L., Forget, B.G. andWeissman, S.M., Proc. Natl. Acad. Sci. USA, 1981, 78, 2455.
- Sullivan, P.M., Petrusz, P., Szpirer, C. and Joseph, D. R., J. Biol.Chem., 1991, 266, 143.
- Eul, J., Graessmann, M. and Graessmann, A., EMBO J., 1995, 14,3226, 3235.
- Caudevilla, C., Proc. Natl. Acad. Sci. USA., 1998, 95, 12185.
- Tasic, B., Mol. Cell, 2002, 10, 21.
- Bruzik, J.P. and Maniatis, T., Nature, 1992, 360, 692.
- Pasman, Z. and Garcia-Blanco, M.A., Nucl. Acid. Res., 1996, 24,1638.
- Goldstrohm, A.C., Greenleaf, A.L. and Garcia-Blanco, M.A., Gene,2001, 277, 31.
- Maniatis, T. and Reed, R., Nature, 2002, 416, 499.
- Spritz, R.A., Jagadeeswaran, P., Choudary, P.V., Biro, P.A., Elder,J.T., deRiel, J.K., Manley, J.L., Gefter, M.L., Forget, B.G. andWeissman, S.M., Proc. Natl. Acad. Sci. USA, 1981, 78, 2455.
- Orkin, S.H., Kazazian, H.H., Jr., Antonarakis, S.E., Ostrer, H., Goff, S.C. and Sexton, J.P., Nature, 1981, 300, 768.
- Treisman, R., Orkin, S.H. and Maniatis, T., Nature, 1983, 302, 591.
- Dobkin, C., Pergolizzi, R.G., Bahre, P. and Bank, A., Proc. Natl.Acad. Sci. USA, 1983, 80, 1184.
- Cheng, T.C., Orkin, S.H., Antonarakis, S.E., Potter, M.J., Sexton, J.P.,Markham, A. F., Giardina, P.J., Li, A. and Kazazian, H.H. Jr., Proc.Natl. Acad. Sci. USA, 1984, 81, 2821.
- Highsmith, W.E. Jr., Burch, L.H., Zhou, Z., Olsen, J.C., Boat, T.E.,Spock, A., Gorvoy, J.D., Quittell, L., Friedman, K.J., Silverman, L.M.,Boucher, R.C. and Knowles, M.R., N. Engl. J. Med. Chem.,1994, 331, 974.
- Chillon, M., Dork, T., Casals, T., Gimenez, J., Fonknechten, N., Will,K., Ramos, D., Nunes, V. and Estivill, X., Amer. J. Hum. Genet.,1995, 56, 623.
- Perrin, G., Morris, M.A., Antonarakis, S.E., Boltshauser, E. and Hutter, P., Hum. Mutat.,1996, 7, 172.
- Wang, M., Dotzlaw, H., Fuqua, S.A.W. and Murphy, L.C., BreastCancer Res. Treat., 1997, 44, 145.
- Ikeda, H., Matsubara, Y., Mikami, H., Kure, S., Owada, M., Gough, T.,Smooker, P.M., Dobbs, M., Dahl, H.H.M., Cotton, R.G.H. andNarisawa, K., Hum. Genet.,1997, 100, 637.
- Tsuruta, M., Mitsubuchi, H., Mardy, S., Miura, Y., Hayashida, Y.,Kinugasa, A., Ishitsu, T., Matsuda, I. and Indo, Y., J. Hum. Genet.,1998, 43, 91.
- Knebelmann, B., Forestier, L., Drouot, L., Quinones, S., Chuet, C., Benessy, F., Saus, J. and Antignac, C., Hum. Mol. Gene., 1995, 4, 675.
- Mitchell, G.A., Labuda, D., Fontaine, G., Saudubray, J.M., Bonnefont,J.P., Lyonnet, S., Brody, L.C., Steel, G., Obie, C. and Valle, D., Proc.Natl. Acad. Sci. USA, 1991, 88, 815.
- Vervoort, R., Gitzelmann, R., Lissens, W. and Liebaers, I., Hum.Genet.,1998, 103, 686.
- McConville, C.M., Stankovic, T., Byrd, P.J., McGuire, G.M., Yao,Q.Y., Lennox, G.G. and Taylor, M.R., Amer. J. Hum. Genet.,1996, 59, 320.
- Okuyama, E., Nishi, N., Onishi, S., Itoh, S., Ishii, Y., Miyanaka, H.,Fujita, K. and Ichikawa, Y., J. Clin. Endocrinol. Metab.,1997,82, 2337.
- Chen, H., Charlat, O., Tartaglia, L.A., Woolf, E.A., Weng, X., Ellis, S.J., Lakey, N.D., Culpepper, J., Moore, K. J., Breitbart, R. E., Duyk, G.M., Teppe, R. I. and Morgenstern, J. P., Cell, 1996, 84, 491.
- Lee, G.H., Proenca, R., Montez, J.M., Carroll, K. M., Darvishzadeh,J.G., Lee, J.I. and Friedman, J.M., Nature, 1996, 379, 632.
- Friedman, K.J., Kole J., Cohn J.A., Knowles, M.R., Silverman, andKole, R., J. Biol. Chem., 1999, 274, 36193.
- Faustino, N.A., Genes Dev., 2003, 17, 419.
- Nelson, D.R., Pharmacogenet.,1996, 6, 1.
- Hanioka, N., Amer. J. Hum. Genet.,1990, 47, 994.
- Suwanmanee, T., Mol. Ther., 2002, 6, 718.
- De Angelis, FG.,Proc. Natl. Acad. Sci. USA, 2002, 99, 9456.
- Rodrigues, N.R., Hum. Mol. Genet., 1995, 4, 631.
- Rossoll, W., Hum. Mol. Genet., 2002, 11, 93.
- Naimi, B., Prostate, 2002, 52, 245.
- De Moerlooze, L., Development, 2000, 127, 483.
- Eswarakumar, V.P, Development, 2002 129, 3783.