- *Corresponding Author:
- S. R. Sahoo
Acharya & BM Reddy College of Pharmacy, Soldevanahalli, Hesaraghatta Bengaluru-560 107, India
E-mail: sahu.samir070@gmail.com
| Date of Submission | 23 October 2018 |
| Date of Revision | 16 May 2019 |
| Date of Acceptance | 08 August 2019 |
| Indian J Pharm Sci 2019;81(5):892-899 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Thirty halophilic bacteria were isolated from the soil of Karwar mangrove regions, Karnataka. These were examined for the production of pigments, antibiotic and proteolytic activities. KA16SPiv produced fluorescent green pigment and KA16SK2HS produced pink pigment. The optimal pigment production was at pH 8, 37°. Pigments are extractable in chloroform and methanol. Absorption spectra suggested phenazine skeleton for green and prodigiosin skeleton for pink pigment. A polyphasic taxonomic study showed KA16SPiv to be Pseudomonas aeruginosa whereas KA16SK2HS to be Salinicoccus roseus with accession no: MF377544 and MF377542, respectively.
Keywords
Halophilic bacteria, Karwar mangrove, Pigments
Pigments are the inseparable components of human life, which are associated with almost all articles being used today. It is a vital constituent, as a color is probably one of the first characteristics to be perceived by senses. Synthetic pigments are often carcinogenic and non-biodegradable. These limitations of synthetic pigments have caused the need for a fresh, enthusiastic look out for natural pigments, which are found in plants, insects, and microbes. Microbes after plants are the second largest[1,2] source for obtaining natural color.
Bacteria among all the microbes have immense potential to produce diverse pigments. The microbial pigments are of industrial interest as they are more stable and with a high solubility factor than those of plant and animal origin[3]. Besides having controllable and predictable yield these microbial pigments are season-independent, often of food grade may also exhibit medicinal properties. Various pigments like carotenes, melanins, flavones, quinines and violacein[4-6] are produced by different bacterial genera like Serratia, Streptomyces, Pseudomonas, Bacillus, Vibrio and Cytophyga that play an important role in their molecular and physiological processes and help taxonomist to identify and classify the bacteria, as they differ in chemical structure with specific chromophores. Carotenoids such as bacterioruberin, which plays a role as a biological pigment and in bacterial metabolite have been isolated from Halorubrum sp., Haloarcula sp. from the western coast of India by Raghavan and Furtado; Kibilan et al.[7,8]. In humans, these pigments act as protective agents against oxidative damage[9] and demonstrate pro-vitamin A function that protects from lifestyle-related disorders such as cardiovascular disease and age-related muscular degeneration[10].
The present study was taken up with an objective to isolate industrially useful pigments from mangrove microbes of Karwar mangrove region. Terrestrial freshwater and marine halophilic environments existing together in the mangrove forests create an extremophilic environment and make the microbes different from those of either environment. Two pigment producing isolates identified as P. aeruginosa and Salinicoccus roseus with accession numbers MF377544 and MF377542 were taken up for the study of various parameters to optimize pigment production.
Materials and Methods
UV spectrophotometer (Carry 60, Agilent Technology) was used for spectrophotometric studies. PCR (T100, Bio-Rad), horizontal electrophoresis- gel unit (Bio- Rad), universal hood II- Gel-Doc (Bio-Rad) were used. All media were of HiMedia and all chemicals used were of analytical grade. Molecular analysis was carried out in Trans-Disciplinary University of Health Science and Technology and Eurofins Genomics India, Bangalore. All solvents used were of analytical grade and HPLC grade methanol (HiMedia, purity 99.7 %), chloroform of Fisher Scientific (purity 99.7 %).
Isolation of bacteria:
The bacterial strains were isolated from the sediments collected during post-monsoon (October-November) season from the coastal Mangrove region of Karwar, Karnataka (Latitude- 14.84587° and Longitude- 74.11872°). Sediment samples were collected at a depth of 5 and 40 cm from the root region of various species of trees like Sonneratia alba, Kandelia candel. The sediment samples were incubated in HiMedia halophilic broth (M591-500G) for 12 d for enriching and isolation of extreme halophiles. Following tenfold serial dilution technique in halophilic agar plates, halophilic bacterial isolation was carried out[11] by incubating aerobically at 37° for 24-48 h[12]. A greenish color was observed in KA16SPiv and a pink color in KA16SK2HS cultures. Pure cultures were obtained by repeated streaking over the nutrient agar plates. Then the isolates were preserved in glycerol at –20°[13] and on NA slants at 4° for further use.
Biochemical characterization of bacterial isolates:
Morphological and physiological properties of the isolates were investigated on the basis of colony characteristics on the nutrient agar medium and Gram’s reaction. The isolates were subjected to various biochemical tests followed by enzymatic and sugar utilization test as per Bergy’s manual of determinative bacteriology[14]. The online ABIS tool-based on these results helped in identifying the close prokaryotic relative of the isolates[15].
16S rRNA sequence analysis:
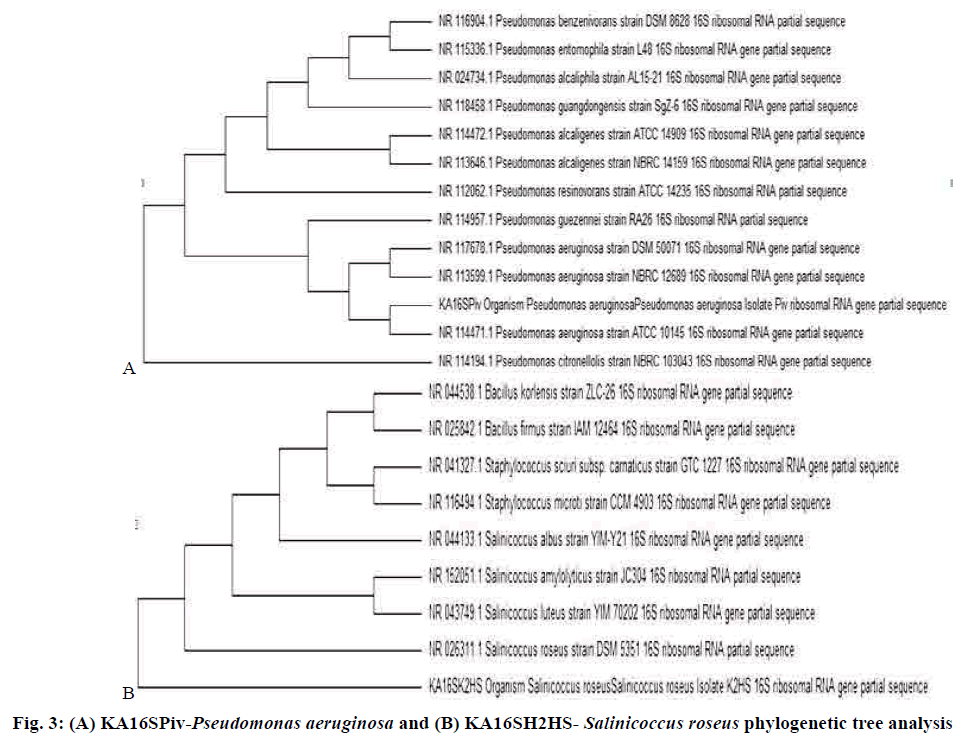
Further identification was carried out using 16S rRNA gene sequencing. Bacterial genomic DNA was extracted[16]. The DNA was used as a template for PCR using universal primers. These purified products are a template in cycle sequencing[17]. The amplified 16S rRNA gene was purified with Qiagen Inc. kit and electrophoreses on 1 % agarose gel sequencing was carried out in Eurofins[18-20]. BLAST program was used to access the DNA similarities and multiple sequence alignment and molecular phylogeny were performed using BioEdit software.
Growth parameters:
Overnight grown bacterial cultures were inoculated in different Erlenmeyer flasks; each containing nutrient broth with different salt concentration (0, 5, 10, 15, 20, 25, 30 and 35 %) for a period of 24 h and streaked on to nutrient agar plates with the same concentration of salts as above. After a period of 24 h, the tubes without visible growth were kept incubated for a period of another 72 h for confirmation. The organisms were set incubated with varying pH environments (i.e. 2, 4, 6, 8 and 10) for a period of 24 h. pH was adjusted with the help of 1 N HCl, 1 N NaOH. 5 ml of bacterial suspensions from each of the flasks was pipetted out and analyzed at 620 nm.
Pigment production and extraction:
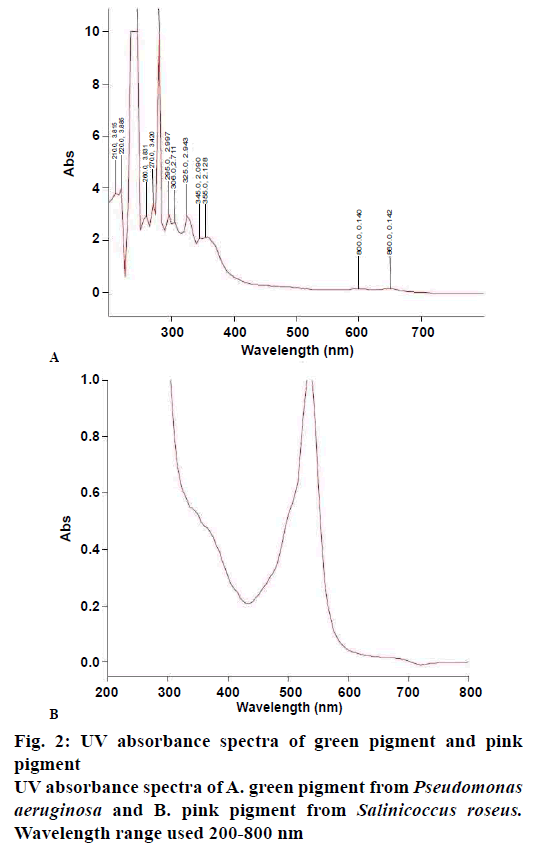
The pigment was extracted following the method described by Frank and DeMoss[21] for blue-green pigment and by Maekel and Kester[22] for pink pigment. The selected bacterial isolates P. aeruginosa and S. roseus were cultured in nutrient broth at pH 7±0.1 and temperature 37°. The colour of nutrient broth has changed to dark green colour, which was inoculated with KA16SPiv at the end of the incubation period and the extraction of pigment was carried out using chloroform by centrifuging it at 8000×g for 10 min at 4° (the process was repeated three times). The pink pigment from KA16SK2HS was found in the cell biomass, which was separated from the medium by centrifugation at 6000×g for 10 min at 4°. Methanol was added to the pellet and centrifuged at 6000× g for 7 min at 4°. Hexane, ethyl acetate and n-butanol, proved ineffective in extracting these pigments. Salting out of the pigment was also found to be ineffective. The crude pigments extracted were primarily identified by measuring their OD in a range of 200-800 nm.
Antibacterial assay of the pigment:
The assay was carried out by well diffusion technique for both green and pink pigments against clinical pathogenic strains such as Staphylococcus aureus, Bacillus subtilis (Gram-positive bacteria), Proteus mirabilis, Escherichia coli, Salmonella typhimurium (Gramnegative bacteria), Candida glabrata, C. albicans, C. parasilosis and Fibulobasidium inconspicuum (fungal strains), chloroform and methanol were kept as control. The isolates were swabbed on to the surface of the Muller-Hinton agar plates with wells dug into it. 10 μl each of the pigments were added to the wells and incubated for 24 h at 37°. Standard antimicrobial were taken as comparison by implementing as disc diffusion method[23]. The antibacterial activity of the pigments was determined by measuring the zones of growth inhibition around the well.
Screening for protease activity:
The proteolytic activity of the bacterial isolates was tested by using skimmed milk medium supplemented with 5 % NaCl and 1 % casein. The plates were incubated overnight at 37°. P. aeruginosa was found to produce protease as identified based on clear zone formation on the plates. S. roseus proved negative for proteolytic activity.
Assay of proteolytic activity:
Fresh inocula of protease producing strain were added to the nutrient broth containing 5 % NaCl and 1 % casein at pH 7 and incubated in a shaker incubator in 150×g at 37° for 24 h. After incubation, the cultures were centrifuged at 10 000×g for 15 min at 4°. The cell-free supernatant was used for protease assay by Lowry method used by Kembhavi et al.[24]. The reaction mixture containing 1 ml of the cell-free supernatant was added to 1 ml of casein (1 % w/v in 50 mM potassium phosphate buffer, pH 7.5) and incubated for 10 min at 37°. 2 ml of 10 % trichloroacetic acid reagent was added to terminate the reaction, kept for 30 min at room temperature and centrifuged at 10 000×g for 15 min at 4°. Five millilitres of 500 mM sodium carbonate solution was added to the 2 ml of filtrate and absorbance was taken at 570 nm. One unit of enzyme activity was defined as the amount of enzyme required to liberate 1 μmol of tyrosine per min under the defined assay conditions.
Pigments as food and textile colorant:
As per the spectrophotometer analysis, a single peak was seen in the case of pink pigment while multiple peaks were recorded in the case of the green pigment. The pink pigment was further analysed for colour retaining ability test for food and textiles. Test for textile grade colour was carried out by taking several 5 cm of white cotton cloth strips and treating each of them with 2 ml of pink pigment and drying them under laminar airflow for 90 min. Once dried, the cloth was boiled in 20 ml of water. Food colour grade test was carried out by taking 2 ml of the pink colour crude extract and diluting it with 10 ml of distilled water mixed with 1.5 % agar and heated till boiled then cooled for solidification. These tests were made to ensure the stability of the pigment and its colouring ability[25].
Results and Discussion
Thirty halophiles were isolated from the mangrove sediments, 2 of them showed pigmented characters over NA plates, which were selected for further study. After studying the morphological characters followed by staining procedure and observation under the microscope, the 2 bacterial strains were found to be different, were subjected to biochemical tests as shown in Table 1 in order to determine their physiology. Below 5 % NaCl no growth could be found; luxuriant growth was observed within a range of 5-15 % of NaCl concentration, whereas in 20-30 % of NaCl mild growth was seen and above 35 % NaCl no growth was observed. Optimum pH is shown in fig. 1, which was observed at 620 nm.
| Test conducted | Isolate designation | |
|---|---|---|
| KA16SPiv | KA16SK2HS | |
| Shape | Rod | Coccus |
| Gram Stain | Negative | Positive |
| Oxidase | Positive | Positive |
| Indole | Negative | Negative |
| Methyl Red | Positive | Negative |
| VP Test | Negative | Negative |
| Citrate | Positive | Positive |
| Hydrogen Sulfide Production | Negative | Negative |
| Urease | Positive | Positive |
| Motility | Positive | Negative |
| Gelatine liquefaction | Positive | Negative |
| Nitrate reduction | Negative | Positive |
| Catalase | Positive | Positive |
| Amylase | Negative | Negative |
Table 1: Biochemical test of the isolates
P. aeruginosa was found positive towards proteolytic activity via clear zone formation. S. roseus did not display protease activity. P. aeruginosa was further subjected to the protease assay. The observation was made based on absorbance value at 570 nm quantifying the amount of protease to be 5.266 U/mg.
Extraction of green pigment was carried out by using chloroform and methanol was used for pink. In our study crude extract containing pink pigment from the bacterial isolate gave maximum absorption spectra at 535 nm, which is comparable to prodigiosin skeleton whereas green gave maximum absorption at 280, 270, 260, 365 and 310 nm as shown in fig. 2A and B, which are comparable to phenazine skeleton present in 1-hydroxyphenazine, phycocyanin, and oxychlororaphine.
The antimicrobial activity exhibited by the pigments was compared with that of the standard clinicallyused antimicrobial drugs against bacterial and fungal pathogens. The selected bacterial strains, KA16SPiv, and KA16SK2HS belonged to the genus Pseudomonas and Salinicoccus as revealed from ABIS analysis. These purified products are a template in cycle sequencing. 16S rRNA gene sequences from the isolates were used to determine the closest prokaryotic relative using NCBI BLASTN, which was found to be P. aeruginosa and S. roseus. Sequences deposited in GenBank under accession number MF377544 and MF377542, respectively. Sequence alignments were carried out using Bioedit (version 7.2.6). The evolutionary history was inferred using the UPGMA method with a tree evaluation by bootstrap analysis for 3000 replication in MEGA 6.0 (Molecular Evolutionary Genetics Analysis)[26,27], which provides confidence estimation through phylogenetic tree topologies that the isolates are P. aeruginosa and S. roseus. 16S rRNA sequences from major bacterial clusters obtained from GenBank were included in the phylogenetic analysis, which is represented in fig. 3A and B. The P. aeruginosa and S. roseus isolate have shown similarities with ATCC 10145 and DSM 5351 respectively[25,28] (Tables 2-4).
| Bacterial strains | Zone of inhibition (mm) | ||||
|---|---|---|---|---|---|
| Ampicillin (30 mg) | Tetracycline (10 mg) | Streptomycin (10 mg) | Green pigment (1 mg/ml) | Pink pigment (1 mg/ml) | |
| S. aureus | 25 | 28 | 23 | 18 | 14 |
| B. subtilis | 26 | 24 | 21 | 16 | 17 |
| P. mirabilis | 21 | 10 | 18 | 10 | 10 |
| E. coli | 17 | 15 | 20 | 19 | 15 |
| S. typhimurium | 24 | 15 | 21 | 11 | 10 |
Table 2: Antibacterial assay
| Fungal strains | Zone of inhibition (mm) | ||||
|---|---|---|---|---|---|
| Ciprofloxacin (10 μg) | Ketoconazole (10 μg) | Fluconazole (10 mg) | Green pigment (1 mg/ml) | Pink pigment (1 mg/ml) | |
| Candida glabrata | 28 | 10 | 10 | 18 | 12 |
| Candida albicans | 10 | 29 | 30 | 12 | 17 |
| Fibulobasidium inconspicuum | 31 | 10 | 10 | 16 | 12 |
Table 3: Antifungal assay
| Test Isolates | NaCl Concentration (%) | |||||||
|---|---|---|---|---|---|---|---|---|
| 0 | 5 | 10 | 15 | 20 | 25 | 30 | 35 | |
| P. aeruginosa | Nil | +++ | +++ | +++ | +++ | ++ | + | Nil |
| S. roseus | Nil | +++ | +++ | +++ | +++ | ++ | + | Nil |
Table 4: Salt tolerance test
Microbial fibrinolytic protease is used by the detergent and pharmaceuticals industry and in medical diagnosis for resolving serious medical threats like disorders of blood clotting and fibrinolysis[16,17]. Skimmed milk agar[29] and other enzyme-substrate been incorporated in solid media[27-30] are widely used as a screening agent. Proteolytic isolate screening was carried out with skimmed milk media. Tri-chloroacetic acid[30] and tannic acid[31] applied by several authors to determine the enzyme activity. It was observed that the P. aeruginosa has a protease, an activity of 5.266 U/mg. These results showed similarity with those reported by Morihara et al., who have isolated a protease from Pseudomonas sp., which inactivated S human plasma alpha1-proteinase[32,33]. The intracellular fibrinolytic protease (2.5 U/g) was found in case of G. lucidum isolated by Kumaran et al.[34]. B. subtilis isolated from the marine environment by Rym et al. produced 20.72-292.36 U/mg protease activity when tested under various conditions and changing the media[26]. The production of microbial pigments is affected by varying pH[35,36]. The optimum pH of the bacterial isolate was found to be at an alkaline pH i.e. 8.0 and growth was reduced at pH 10. An acidic pH has an adverse effect on both of the isolates. Results of the present investigation showed similarities to the results reported by Tibor[37] and Aberoumand[38], who observed optimum temperature and pH for growth of pigmented bacterial isolates to be 37° and pH 7.
This might be a result of a molecular adaptation by bacterial isolates as well as increases the activity of an enzyme for optimal growth. At 37°, the isolates were incubated for a period of 7 d to check optimum time to produce the pigment. It was observed that the production starts after 24 h. 72 h of incubation was the optimal time period for pigment production in case of P. aeruginosa. In the case of S. roseus pigment production started from 48 h optimal period required is 96 h. The isolates are strictly aerobe since both pigment production and growth increased with agitation.
Maximum growth and pigment production occurred in 100 ml medium per 250 ml Erlenmeyer flask. Increasing the volume of the medium keeping the flask size same decreased the growth and pigment production by the cells, which might be a result of decreased dissolved oxygen volume. Asker and Ohta[39] have reported similar observations. Yokoyama et al.[40] reported that varying the volume of the medium controlled the growth of and carotenoid production by Agrobacterium auranticum. Various solvents were used to extract the colours, except for chloroform in case of green colour and methanol for pink, those solvents were found ineffective. Both pigment solutions were scanned from 200-800 nm. Spectral data of all microbial pigments were collected. Pigment structures for the two relevant genera were selected. The spectra of the pigments isolated were compared with literature report to arrive at their structural features.
The pink pigment from S. roseus showed a single peak with the maximum absorbance at 535 nm, which was clear in speculating the compound to be similar to that of prodigiosin skeleton, whereas the green compound showed various peaks similar to different compounds like 1-hydroxyphenazine, phycocyanin, and oxychlororaphine, which are similar to phenazine skeletons. This result was corroborated by many authors[41,42], who observed red colored pigment production using Serratia sp., Streptomyces sp. and Salinicoccus sp. and blue-green pigment from Pseudomonas sp corroborated with many reported studies[43-48]. Both the pigments showed a distinct antimicrobial effect against the test pathogens and are effective as antibacterial and antifungal towards a few clinical pathogenic species. Generally, the pigments are ineffective against most fungal pathogens. Results obtained in the present study also corroborated with studies that reported isolation of antifungal compound 1,6-dihydroxy-2- chlorophenazine from Streptosporangium sp. and P. aeruginosa[49,50]. Similar observation was made from P. chlororaphis showing an inhibition zone of 21 mm against Pythium asphanidermatum and when the concentration was increased complete inhibition of Macrophomina phaseolina, R. solani and S. rolfsi was observed[51]. Phenazine from Pseudomonas sp. has been reported to be an antibacterial agent[52,53]. Pink pigment isolated by Darah et al., from Serratia marcescens has completely inhibited P. aeruginosa and 13 more bacterial strains[51]. Results of the present study agreed with those reported by Pierson and co-workers[54-56]. The pink pigment produced a good coloration when mixed with agar; it gets deteriorated to light yellowish pink on boiling but had created no negative effect on agar binding property[57]. The cotton cloth treated with pink pigment has shown a lighter shade of pink after 20 min of boiling but has retained its color which can be said effective for textile coloration. Even on exposure to sunlight for 60 min no visible change in colour degradation was found, which was similar to those reported by Shirata and co-workers[58].
The idea behind testing the antimicrobial activity of the pigments against food and waterborne pathogens is to check whether an additional protective coating can be laid to the food and textiles when these are used as a colorant. Not many details of Salinicoocus sp. could be found but the isolated pigment has shown many spectrophotometrical similarities with Serratia sp., which helped to carry out studies on purification of the pigments through TLC, HPLC method and identifying the compound through NMR, LCMS, and MS-MS for future and also test the antimicrobial effect of the pure compounds. As P. aeruginosa is a proteolytic organism, further studies would be based on extraction and purification of protease and test it in vitro.
The present study was an attempt to demonstrate the multi-functionality found within the estuarine environment strains isolated from the Mangrove region. The isolate Pseudomonas sp. and Salinicoocus sp. are potential producers of blue-green and pink colour pigment, which are comparable with 1-hydroxyphenazine, phycocyanin and oxychlororaphine and prodigiosin. This report highlights the key role of bacterial pigmentation, growth in varying pH of such hypersaline region, with promising food colorant potential[38]. This was also observed in the present study, that both strains have different performing property and required cheap raw material having the ability of rapid growth rate leading to short fermentation cycle times which is an advantage for industrial production. Thus the mangrove environment seems to be a fruitful environment to explore.
Acknowledgments:
The work was funded by RGUHS, grant no. RGU;R&D; Res.Wing 2014-15, dated 13/03/15. Authors wish to thank the management of Acharya Institutes for providing the facilities, Dr. P. Mesta, Marine Biologist of IISc field station at Kumta and his team for their support during the fieldwork. Authors also thank Dr. S. P. Balasubramani, Molecular biologist of Trans- Disciplinary University, Bangalore, for his help in the identification of isolates and the author is greatly indebted to his Mother who was always with him in his efforts.
Conflicts of interest:
The authors have no conflict of interest to declare.
References
- Ryu BH, Park BG, Chi YE, Lee JH. Production of purplish-red pigment in mixed culture of Streptomyces propurpuratus and Bacillus sp. Korean J Appl Microbiol Bioeng 1989;17:327-3.
- Mizukami H, Konoshima M, Tabata M. Variation in pigment production in Lithospermum erythrorhizon cultures. Phytochemistry 1978;17:95-7.
- Gunasekaran S, Poorniammal R. Optimization of fermentation conditions for red pigment production from Penicillium sp. under submerged cultivation. Afr J Biotechnol 2008;7(12):1894- 8.
- Dufosse L. Microbial production of food grade pigments. J Food Technol Biotechnol 2006;44(3):313-21.
- Ahmad WA, Wan Ahmad WY, Zakaria, ZA, Yusof NZ. Isolation of Pigment-Producing Bacteria and Characterization of the Extracted Pigments. In: Ahmad WA, Wan Ahmad WY, Zakaria, ZA, Yusof NZ, editors. Application of Bacterial Pigments as Colorant. 8th ed. Germany: Springer; 2012. p. 25-44.
- Harley JP, Prescott LM, Klein DK. Microbiology. 7th ed. NewYork: McGraw-Hill Education; 2007.
- Raghavan TM, Furtado I. Expression of carotenoid pigments of Haloarcheal cultures exposedto aniline. Environ Toxicol 2005;20;165-9.
- Kabilan M, Bhakti BS, Judith MB. Culturable halophilic archea at the initial and crystallization stages ofsalt production in natural solar salts Goa, India. Aquat Biosyst 2012;8;1-8.
- Scolnik PA, Bartley GE. Nucleotide sequence of a putative geranyl pyrophosphate synthase from Arabidopsis. Plant Physiol 1995;104:1469-70.
- Young AJ, Lowe GM. Antioxidant and prooxidant properties of carotenoids. Arch Biochem Biophys 2001;385:20-7.
- Rath CC, Subramanyam VR. Thermo tolerant enzyme activities of Bacillus species isolated from the hot springs of Orissa. Microbios 1996;86(348):157-61.
- Das S, De. M, Ray R, Chowdhury C, Jana TK , De TK. Microbial Ecosystem in Sunderban Mangrove Forest Sediment, North-East Coast of Bay of Bengal, India. Geomicrobiol J 2012;27(7):656-66.
- Dubey RC, Maheshwari DK. Practical Microbiology. 2nd ed. Hyderabad, India: S. Chand Publishing; 2007.
- Holt JG, Krieg NR, Sneath PHA, Staley JT , Williams ST. Bergey’s Manual of Determinative Bacteriology. 9th ed. Balitomore: Williamsons and Wilkins; 1994.
- Bryant TN. PIBWin software for probabilistic identification. J Appl Microbiol 2004;97:1326-7.
- Gupta R, Beg QK, Lorenz P. Bacterial alkaline proteases: molecular approaches and industrial applications. Appl Microbiol Biotechnol 2002b;59:15–32.
- Peng Y, Yang X, Zhang Y. Microbial fibrinolytic enzymes; an overview of source, production, properties and thrombolytic activity in vivo. Appl Microbiol Biotechnol 2005;69:126-32.
- Suganthi C, Mageswari A, Karthikeyan S, Anbalagan M, Sivakumar A, Gothandam KM. Screening and optimization of protease production from a halotolerant Bacillus licheniformis isolated from saltern sediments. J Genet Eng Biotechnol 2013;11(1):47-52.
- Zhou J, Bruns MA, Tiedje JM. DNA recovery from soils of diverse composition. Appl Environ Microbiol 1996;62(2):316-322
- Achenbach L, Woese C. 16S and 23S rRNA-like primers. In: Sowers KR, Schreier HJ, editors. Archaea: A Laboratory Manual, Methanogens. 3rd ed. New York: Cold Spring Harbor Laboratory Press, Cold Spring Harbor; 1995. p. 521-2.
- Frank LH, DeMoss RD. On the biosynthesis of Pyocuanine. J Bacteriol 1959;77:776-82.
- Meckel RA, Kester AS. Extractability of carotenoid pigments from nonphotosynthetic bacteria with solvents and detergents; Implications for the location and binding of the pigments. J Genet Microbiol 1980;120:111-6.
- Umrania VV. Bioremediation of toxic heavy metals using acido-thermophilic autotrophes. Bioresour Technol 2006;97:1237-42.
- Kembhavi AA, Kulkarni A, Pant A. Salt-tolerant and thermostable alkaline, protease from Bacillus subtilis NCIM No. 64. Appl Biochem Biotechnol 1993;38:83-92.
- Spring S, Ludwig W, Marquez MC, Ventosa A, Schleifer KH. The Genus Halobacillus gen. nov., Description of Halobacillus litoralis sp. nov., Halobacillus trueperi sp. nov. and Transfer of Sporosarcina halophila to Halobacillus halophilus comb.nov. Int J Syst Bacteriol 1996;46:492-6.
- Rym A, Anissa H, Mohamed H, Fakher F, Laila M, Kemel J, et al. Fibrinolytic enzymes from a newly isolated marine bacterium Bacillus subtilis A26; characterization and statistical media optimization. Can J Microbiol 2009;55:1048-61.
- Raja A, Khess N, Pujari N, Bhattacharya S, Das A, Subbaramiah SR. Enhancement of protease production by Pseudomonas aeruginosa isolated from dairy effluent sludge and determination of its fibrinolytic potential. Asian Pac J Trop Biomed 2012;19:1845-51.
- Galdzicka M, Plassmeyer ML, Blaine LD, Pienta PA, Gillevet PM. Comparison of Phenetic and Phylogenetic Classification Systems for the Fluorescent Pseudomonads. Virginia (USA): University Blvd Press; 1998. p. 20110-2209.
- Garriga M, Hugas M, Gou P, Aymerich MT, Arnau J, Monfort JM. Technological and sensorial evaluation of Lactobacillus strains as starter cultures in fermented sausages. Int J Food Microbiol 1996;32(1-2);173-83.
- Medina P, Baresi L. Rapid identification of gelatin and casein hydrolysis using TCA. J Microbiol Methods 2007;69(2):391-3.
- Saran S, Isar J, Saxena RK. A modified method for the detection of microbial proteases on agar plates using Tannic acid. J Biochem Biophys Methods 2007;70(4):697-9.
- Morihara K, Tsuzuki H, Oda K. Protease and elastase of Pseudomonas aeruginosa; inactivation of human plasma alpha 1-proteinase inhibitor. Infect Immun 1979;24(1):188-93.
- Bayoudh A, Gharsallah N, Chamkha M, Dhouib A, Ammar S, Nasri M. Purification and characterization of an alkaline protease from Pseudomonas aeruginosa MN1. J Ind Microbiol Biotechnol 2000;24(4):291-5.
- Kumaran S, Palani P, Chellaram C, Premanand T, Kaviyarasan V. Screening of fibrinolytic protease from south indian isolatesof Ganoderma lucidum. Int J Pharma BioSci 2011;2(1):419-31.
- Tinoi J, Rakariyatham N, Deming RL. Simplex optimization of carotenoid production by Rhodotorula glutinis using hydrolyzed mung bean waste flour as substrate. Process Biochem 2005;40:2551-7.
- Sanjay K, Kumaresan N, Manohar B, Kumar US, Vijayalakshmi G. Optimization of carotenoid production by Aspergillus carbonarius in submerged fermentation using a response surface methodology. Int J Food Eng 2007;3(5):292-7.
- Tibor C. Liquid Chromatography of Natural pigments and synthetic dyes. J Chromatogr Libr 2007;71:11.
- Aberoumand A. A Review Article on Edible Pigments Properties and Sources as Natural Biocolorants in Foodstuff and Food Industry. World J Dairy Food Sci 2011;6(1):71-8.
- Asker D, Ohta Y. Production of canthaxanthin by extremely halophilic bacteria. J Biosci Bioeng 1999;88:617-21.
- Yokoyama AH, Izumida I, Miki W. Production of astaxanthin and 4-ketozeaxanthin by the marine bacterium Agrobacterium aurantiacum. Biosci Biotechnol Biochem 1994;58:1842-4.
- Samantaray DP, Sethi S, Dash D, Mishra BB. Bioactive Potential Assessment of Hexavalent Chromium Reducing Bacteria A. calcoaceticus & S.marcescens. Global J Appl Environ Sci 2011;1(1):57-64.
- Aslam Z, Lim JH, Yasir M, Chung YR, Lee ST. Salinicoccus jeotgali sp. nov., isolated from jeotgal, a traditional Korean fermented seafood. Int J Syst Evol Microbiol 2007;57:633-8.
- Kanner D, Gerber NN, Bartha R. Phenazine Pigment Production by a Strain of Pseudomonas aeruginosa. J Bacteriol 1978;134(2):690-2.
- Wilson R, Sykes DA, Watson D, Rutman A, Taylor GW, Cole PJ. Measurement of Pseudomonas aeruginosa Phenazine Pigments in Sputum and Assessment of Their Contribution to Sputum Sol Toxicity for Respiratory Epithelium. Infect Immun 1988;9(56):2515-7.
- Chang PC, Blackwood AC. Simultaneous production of three phenazine pigments by Pseudomonas aeruginosa Mac 436. Can J Microbiol 1969;15:439-4.
- Haynes WC, Stodola FH, Locke JM, Pridham TG, Conway HF, Sohns VE, et al. Pseudomonas aureofaciens Kluyver and phenazine a-carboxylic acid, its characteristic pigment. J Bacteriol 1956;72:412-7.
- Takeda R, Nakanishi I. Pseudomonas pigments (VIII) production of pyoluteorin by fermentation with Pseudononas aeruginosa T359. J Ferm Tech 1960;38:9-19.
- Toohey JI, Nelson CD, Krotkov G. Isolation and identification of two phenazines from a strain of Pseudomonas aureofaciens. Can J Bot 1965;43:1055-62.
- Patel M, Hegde V, Horan AC, Gullo VP, Loebenberg D, Marquez JA, et al. A novel phenazine antifungal antibiotic, 1,6-dihydroxy-2-chlorophenazine. Fermentation, isolation, structure and biological properties. J Antibiot 1984;37(9):943-8.
- Kerr JR, Taylor GW, Rutman A, Høiby N, Cole PJ, Wilson R. Pseudomonas aeruginosa pyocyanin and 1-hydroxyphenazine inhibit fungal growth. J Clin Pathol 1999;52:385-7.
- Darah I, Nazari TF, Kassim J, Lim SH. Prodigiosin -an antibacterial red pigment produced by Serratia marcescens IBRL USM 84 associated with a marine sponge Xestospongia testudinaria. J Appl Pharma Sci 2014;4(10):001-6.
- Kanda S, Wirach W, Chanokporn P, Chalerm R. Isolation and Analysis of Antibacterial Substance Produced from P. aeruginosa TISTR 781. KKU Sci J 2009; 37(2):163-72.
- Arivizhivendhan KV, Mahesh M, Regina MR, Sekaran G. Bioactive Prodigiosin Isolated from Serratia marcescens using Solid State Fermenter and its Bactericidal Activity Compared with Conventional Antibiotics. J Microb Biochem Technol 2015;7(5):305-12.
- Pierson LS, Pierson EA. Phenazine antibiotic production in Pseudomonas aerofaciens. Role in Rhizosphere ecology and pathogen suppression. FEMS Microbiol Lett 1995;136:101-8.
- Hodges GR, Degener CE, Barnes WG. Clinical significances of Citrobacter isolates. Am J Clin Pathol 1978;134:690-2.
- Anjaiah V, Koedam N, Nowak B, Thompson JE, Loper M, Hofte JT, et al. Involvement of phenazines and anthranilate in the antagonism of Pseudomonas aeruginosa PNA1 and Tn5 derivatives towards Fusarium sp. and Pythium sp. Mol Plant-Microbe Interact 1998;11:847-54.
- Saha S, Thavasi R, Jayalakshmi S. Phenazine Pigments from Peudomonas aeruginosa and their applications as antibacterial agent and food colourant. Res J Microbiol 2008;3(3):122-8.
- Shirata A, Tsukamoto T, Yasui H, Hata T, Hayasaka S, Kojima A, et al. Isolation of bacteria producing bluish-purple pigment and use for dyeing. Japan Agric Res Q 2000; 34:131–40.
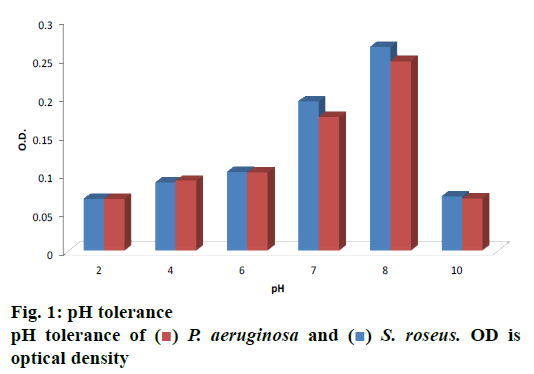
 ) P. aeruginosa and (
) P. aeruginosa and ( ) S. roseus. OD is
optical density
) S. roseus. OD is
optical density