- *Corresponding Author:
- K. Lu
School of Material and Chemical Engineering, Henan University of Engineering, Zhengzhou, Henan, China
E-mail: lukui126@126.com
| Date of Submission | 01 November 2016 |
| Date of Revision | 09 March 2017 |
| Date of Acceptance | 08 September 2017 |
| Indian J Pharm Sci 2017;79(6): 893-899 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The interaction of cell-penetrating peptide human immunodeficiency virus transacting activator of transcription, peptide Tat (49–57), which is the minimal transduction domain of human immunodeficiency virus Tat protein, with calf thymus DNA was investigated with UV/Vis spectroscopy, fluorescence spectroscopy, circular dichroism, and viscometry. Peptide Tat (49–57) could interact with DNA via the groove binding mode, which is accompanied with electrostatic interaction. The fluorescence experiments revealed that the binding constant was 2.63×105 l mol−1. The UV/Vis spectroscopy and circular dichroism results revealed that the interaction of Tat (49–57) binding with calf thymus DNA disturbed the acting force of accumulation between DNA base pairs. The antimicrobial study using Tat (49–57) against bacteria proved that Tat (49–57) possessed antimicrobial activity against both Gram-positive and Gram-negative bacteria with low hemolysis.
Keywords
Human immunodeficiency virus Tat (49-57), synthesis, calf thymus DNA, binding mode, antibacterial activity
Cell-penetrating peptides, also called protein transduction domains (PTDs), are short cation peptides that can traverse the cell membranes [1-3]. During their cellular uptake, they can deliver various molecules into cells; these molecules include peptides, proteins, oligonucleotides, and nanoparticles [4-8], most of which otherwise cannot cross the cell membranes by their own. Transacting activator of transcription protein transduction domain (Tat-PTD) is derived from the Tat protein of human immunodeficiency virus (HIV) and is widely used to deliver different molecules into cells with causing non cellular injury and no toxicity [3, 9,10]. The cell-penetrating property of Tat-PTD results from its basic epitope (amino acids 49-57), which is the minimal transduction domain of Tat protein, includes two lysine’s and six arginine’s within a linear sequence of nine amino acid residues (RKKRRQRRR). Tat (49-57) is a short cationic peptide, possessing a high net positive charge at physiological pH. Recent studies proved that TAT can form transient pores and translocate across the membranes by diffusing on these pores, on account of the strong electrostatic attraction between positively-charged Tat (49-57) and negativelycharged phospholipid [3,11].
After penetrating the cell membranes, Tat-PTD locates in the cell nucleus [12,13], and the uptake into the nucleus precedes its digestion [14], thereby suggesting that the intracellular Tat-PTD can bind to DNA. DNA possesses a crucial role in the whole biological process; consequently, DNA becomes a pharmacological target for various kinds of antimicrobial drugs [15]. Thus, illuminating the binding mode of Tat-PTD with DNA can help understand the molecular mechanisms of drug action and can provide modification guidelines to enhance the clinical efficacy and bioavailability of Tat- PTD and its derivatives.
High affinity of the peptide Tat for DNA has been investigated [14]. However, the binding mode and main type of non-covalent interactions in the binding process between Tat (49-57) and DNA have not yet been illustrated in detail. Therefore, we systematically investigated the binding mode and stability of Tat (49-57) to double-stranded DNA by using various techniques. Furthermore, peptide Tat (49-57) is an Arg-rich cationic peptide that is capable of penetrating the membrane to target nucleus, which indicates the potential of Tat (49-57) as a novel antimicrobial peptide [16]; hence, the antimicrobial activities of Tat (49-57) were also tested.
Materials and Methods
Fmoc-Arg (Pbf)-Wang resin, Fmoc-Arg (Pbf)-OH, Fmoc-lys (Boc)-OH, Fmoc-Gln (Trt)-OH, 2-(7-Aza-1Hbenzotriazole- 1-ly)-1,1,3,3-tetramethyluronium hexa fluorophosphate (HATU), N,N-diisopropylethylamine (DIEA), and trifluoroacetic acid (TFA) were purchased from GL Biochem, Shanghai, China. Calf thymus DNA (ct-DNA) was obtained from Sigma Ltd., Shanghai, China, and ethidium bromide (EB) was purchased from Fluka Chemie.
Bacterial strains, including Gram-positive Staphylococcus aureus (ATCC 29213), S. epidermidis (KCTC 1917) and Bacillus subtilis (KCTC 3068) and Gram-negative bacteria Escherichia coli (ATCC 25922) and Salmonella typhimurium (CMCC 50013) were provided by the School of Biological Engineering, Henan University of Technology.
Peptide synthesis
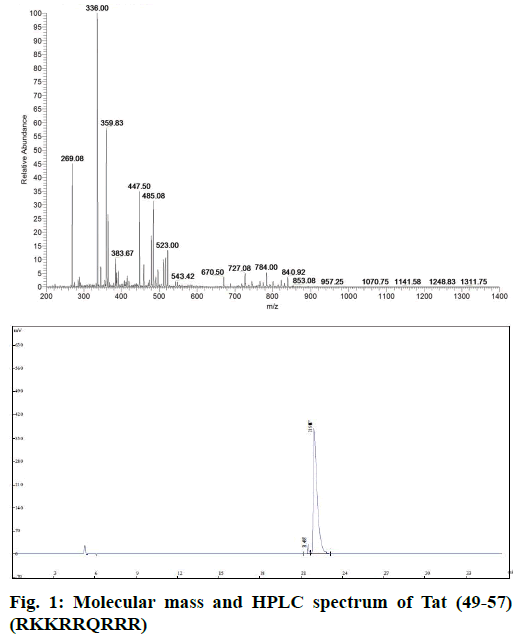
Peptide Tat (49-57) was synthesized in accordance with standard Fmoc solid-phase peptide synthesis procedures [17]. The crude peptide was purified by reversed phase high-performance liquid chromatography (HPLC) with a semi-preparative Agilent C18 column (pore size: 300 Å, partial size: 5 μm, 250 mm×9.4 mm). The buffer system consisted of buffer A: 0.1% (v) TFA in MeCN/H2O 10:90 (v/v) and buffer B: 0.1% (v) TFA in MeCN/H2O 50:50 (v/v). Gradient elution with buffer A to buffer B in 20 min was used at a rate of 2 ml×min−1. The molecular mass of peptide Tat (49-57) was confirmed using LCQ Fleet MS (Thermo Scientific, USA).
UV spectroscopic method
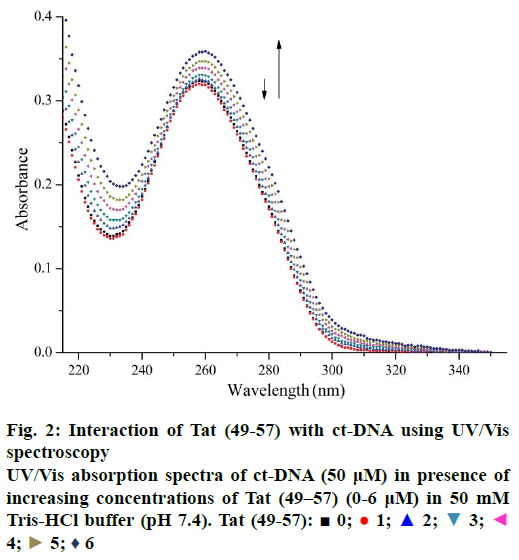
UV/Vis spectra were recorded on UV-3600 spectrometer (Shimadzu Corporation, Japan) using quartz cuvettes (10 mm path length). Absorption titration experiments were conducted by maintaining the concentration of ct-DNA (50 μM) and changing the Tat (49-57) concentration. The UV/Vis measurements were carried out after the solutions were equilibrated for 10 min. During absorption measurements, equal concentration of Tat (49-57) was subtracted to eliminate the background absorption in the range of 190-350 nm.
Fluorescence measurements
Fluorescence spectra were recorded in 10 mm quartz cells on a Cary Eclipse spectrofluorometer (Agilent, Australia). EB was used in fluorescence measurements as fluorescence probe. About 3 ml of ct-DNA-EB complex solution containing ct-DNA (50 μM) and EB (5 μM) was constant and titrated with peptide Tat (49- 57). The mixtures in all experiments were allowed to stand for 10 min to equilibrate before measurement. Emission spectra were measured in the range of 500- 750 nm at the excitation wavelength of 262 nm.
DNA melting studies were carried out via monitoring the fluorescence intensities of DNA-EB and DNAEB- Tat (49-57) complex at 20-100°. The temperature was monitored by a thermostat attached to the sample holder. The melting temperature (Tm) of DNA was determined as the transition midpoint.
Circular dichroism (CD) studies
CD measurements were conducted on a Mos-500 spectropolarimeter (Bio-Logic, France). The ct-DNA solution (50 μM) in Tris-HCl buffer (pH 7.4) was titrated with Tat (49-57). Equal concentrations of Tat (49-57) were subtracted from the corresponding spectra of samples. The CD spectra were recorded in the range of 220-320 nm using 10 mm path length cell at 37°.
Viscosity measurements
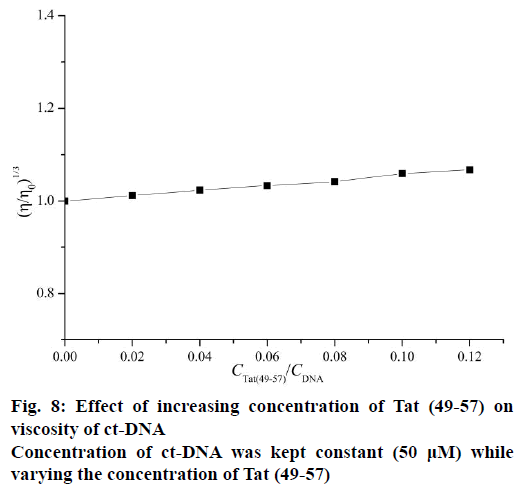
Viscosity measurements were conducted via Ubbelodhe viscometer maintained at 25±0.1° in a thermostat water bath. Different amounts of Tat (49-57) were added to ct-DNA with constant concentration (50 μM). After equilibrium for 15 min, the flow time of samples was measured in triplicate to obtain the concurrent values using a digital stopwatch. The data were presented as (η/η0)1/3 versus CTat (49–57)/CDNA, where η0 and η are the specific viscosity of DNA in the absence and presence of Tat (49-57), respectively.
Antimicrobial activity test
Broth microdilution method [3] was used to examine the antibacterial activities of Tat (49-57). About 100 μl of Tat (49-57) solution (serial two-fold dilutions in sterile water) was added to 100 μl of bacterial suspension (2×104 CFU/ml) in 1% peptone. The medium without bacteria was used as negative control. After incubation at 37° for 20 h, the minimal inhibitory concentration (MIC) was determined.
Haemolysis activity assay
Haemolysis activity of Tat (49-57) was tested against human erythrocytes. Human blood was collected in a tube containing heparin sulphate from a healthy volunteer after written informed consent under a protocol approved by Zhengzhou University Life Science Ethics Committee. The fresh blood was centrifuged (2000×g, 3 min, 4°) to collect erythrocytes and subsequently washed with PBS (10 mM phosphate buffer, pH 7.4) for 5 times [18]. Serial two-fold dilutions of Tat (49-57) were performed in PBS buffer and added to 4% (v/v) erythrocytes dispensed in 96-well plates. The mixture was incubated at 37° for 30 min. Erythrocytes without Tat (49-57) and 0.1% (v/v) Triton X-100 were employed as negative and positive controls, respectively. The supernatants were obtained, and the absorbance was measured at 570 nm. The hemolysis was calculated using the following Eqn., haemolysis (%)=[(ATat (49–57)–APBS)/(ATriton–APBS)]×100.
Results and Discussion
Tat (49-57) was synthesized via solid-phase peptide synthesis procedures on Wang resin and purified through RP-HPLC. The final product was white powder of 97.2% purity and characterized by electrospray ionization mass spectrometry (ESI-MS, Figure 1). The sequence of Tat (49-57) was RKKRRQRRR, and its theoretical molecular weight was 1339.62 g×mol-1. MS (m/e): 670.50 [M+2H]2+, 447.50 [M+3H]3+, 336.00 [M+4H]4+, 269.08 [M+5H]5+, 485.08 [M+3K]3+ was consistent with the molecular peptide Tat (49-57).
UV/Vis spectroscopy is an effective technique to detect interactions between DNA and small molecules. The mode of the interactions is reflected from the changes in the position of the peak and absorptive intensity [19]. The UV spectra of ct-DNA with different concentrations of Tat (49-57) are shown in Figure 2. A slight reduction in the absorption band at 260 nm occurred upon the initial addition of Tat (49-57). Hyperchromism was observed upon the subsequent addition of Tat (49-57) to ct-DNA solution. Hypochromism was due to the electrostatic interaction of DNA phosphate groups with the side chains of Tat (49-57). This interaction tightened the double strands of DNA to prevent aromatic bases from becoming increasingly exposed and resulted in the reduced absorption intensity [20,21]. With the increase in the concentration of Tat (49-57), when the groove of ct-DNA was padded by Tat (49-57), the interaction of side chains containing guanidine and amino groups of Tat (49-57) destabilized the base pairs of DNA, caused the aromatic bases to become exposed, and resulted in hyperchromism of DNA [22]. The structure of Tat (49-57) very closely resembles DNA minor groove binders netropsin [23,35], possessing positive charges and linearity. Therefore, according to the experimental data, minor groove binding interactions might have occurred between ct-DNA and Tat (49-57), and the interactions of Tat (49-57) with ct-DNA disrupted the accumulative force between DNA base pairs.
Fluorescence probe should be utilized because of the non-fluorescence of Tat (49-57) and the minimal practical usefulness of intrinsic fluorescence from DNA [24]. In this work, EB was used as the fluorescence probe. The fluorescence intensity of DNA can be enhanced after interaction with EB because EB can intercalate into the DNA base pairs [25-27].
Figure 3 shows the fluorescence spectra of ct-DNAEB with different concentrations of Tat (49-57). The fluorescence intensity at 600 nm weakened remarkably upon the addition of Tat (49-57) to ct-DNA-EB complex. This result suggests that EB in the DNA-EB complex might have been substituted by Tat (49-57) and led to the decrease in fluorescence intensity or a new DNA-EB-Tat (49-57) complex that did not have any fluorescence was formed by Tat (49-57) binding with DNA-EB. The high affinity of EB with DNA was hardly be disrupted by Tat (49-57) because the binding constant (2.63×105 l×mol-1) between Tat (49-57) and DNA was lower than that of the classical intercalation binding of DNA-EB complex (2×106 l×mol-1) [22]. Therefore, the formation of non-fluorescence complex DNA-EB-Tat (49-57) was the probable cause of the decrease in the fluorescence intensity of DNA-EB complex. We therefore deduce that the interaction between Tat (49-57) and DNA is not intercalation binding. The quenching mechanism of DNA-EB by Tat (49-57) was analysed with the Stern-Volmer Eqn., F0/F=1+Kqτ0[Q], where, F and F0 are the fluorescence intensities of DNA-EB system in the presence and absence of Tat (49-57), respectively. Kq is the quenching rate constant, and Q is the concentration of Tat (49-57). τ0 is the average lifetime of fluorophore without Tat (49-57), and its value is approximately 10-8 s [28].
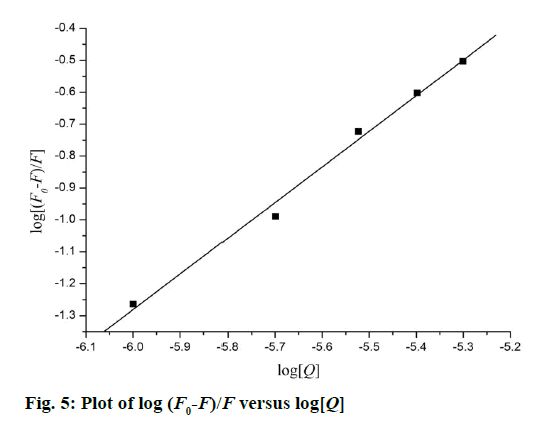
The quenching type, which can be classified as dynamic quenching and static quenching, was analysed with the Stern-Volmer Eqn. Kq was obtained from the intercept of the linear of F0/F versus Q (Figure 4). For Tat (49-57), the value of Kq was 6.41×1012 l×mol-1×s-1, which was much higher than the value of the maximum scatter collision quenching constant of various quenchers with biomolecules (2.0×1010 l×mol-1×s-1) [29]. This finding suggests that the quenching type of Tat (49-57) binding with DNA is static quenching. The binding constant (K) and binding site (n) of Tat (49-57) with the DNA EB complex was evaluated with the following Eqn., log[(F0-F)/F] =logK+n log [Q]. Log [(F0-F)/F] versus log [Q] was plotted, and a line with a good linear correlation was obtained (Figure 5). According to this line, binding constant K was determined to be 2.63×105 l×mol-1, and n was 1.12. K was similar to the value that had been well-established among groove binding agents [30].
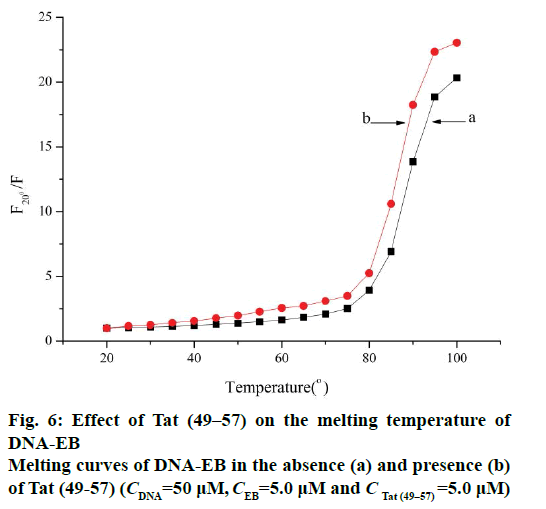
In this experiment, the melting temperature (Tm) was measured with the fluorescence technique. The intercalative binding mode can stabilize the helix structure of DNA and causes the melting temperature to increase by about 5-8°, but the electrostatic and groove binding interactions cause no obvious or minimal change in Tm [15,24]. The melting curves (Figure 6) showed that the Tm of ct-DNA-EB in the absence (Figure 6a) of Tat (49-57) was 87.3±1°, and the Tm of ct- DNA-EB-Tat (49-57) complex (Figure 6b) was 85.6±1° under the experimental conditions. The interaction of Tat (49-57) with ct-DNA caused Tm to decrease. The interaction of Tat (49-57) decreased the stability of ct- DNA double helix structure and affected the affinity of EB with DNA. The change in the Tm of ct-DNA-EB complex after the addition of Tat (49-57) was less than 5°, revealing that the interaction of Tat (49-57) with DNA is electrostatic or groove binding.
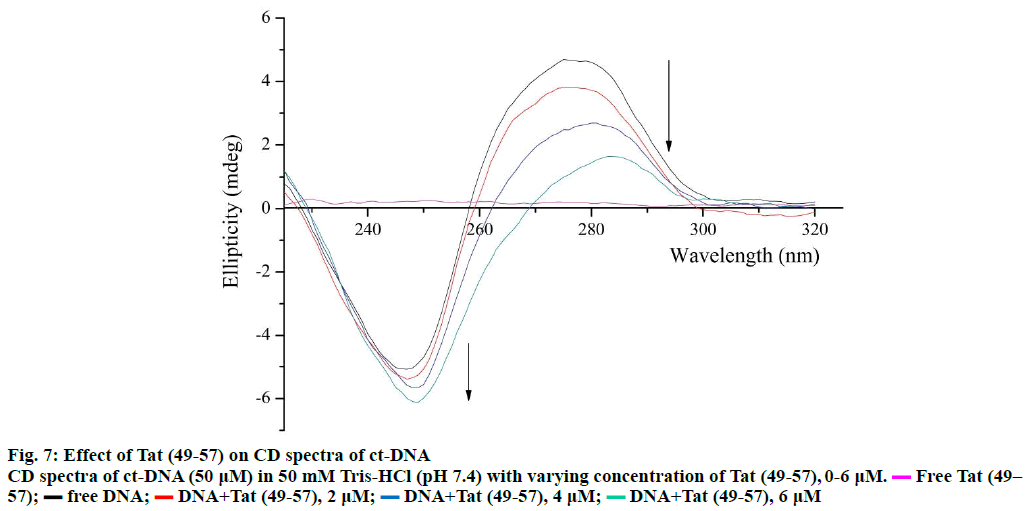
The changes in the secondary structure of DNA during molecule-DNA interactions can be determined through CD spectroscopy [31]. The specific CD spectrum of B-form DNA consists of a negative band at 247 nm caused by the right-handed helicity of DNA and a positive band at 275 nm caused by the accumulation of base pairs [15]. The CD spectra of ct-DNA with Tat (49- 57) are shown in Figure 7. The positive band at 275 nm decreased remarkably with an apparent red shift, and the band at 245 nm gained slight negativity without a major red shift. The decrease in the positive band at 275 nm indicated that the binding of Tat (49-57) weakened the stacking interactions between base pairs of DNA [32], and the slight increase in the negative band at 245 nm revealed that the interaction of Tat (49-57) with DNA tightened the double helix of DNA. These results are consistent with the UV/Vis spectroscopy results.
The intercalative binding mode generally causes DNA viscosity to increase significantly because the intercalative mode requires the DNA base pairs to separate sufficiently at intercalative sites to accommodate the binding molecule; this separation increases the effective length of DNA. However, the non-intercalative binding mode causes less change in the viscosity of the DNA solution [26,30]. As shown in Figure 8, the viscosities of ct-DNA increased slightly upon continued addition of Tat (49-57); this increase is not as pronounced as that observed in the classical intercalator [33] but very similar to distamycin [34] a well-known classical DNA minor groove binder. This result confirmed that Tat (49-57) preferred to interact with DNA through groove binding mode rather than classical intercalative interaction.
The broth microdilution method was utilized to measure the antibacterial activities of Tat (49-57). The MIC values of Tat (49-57) against bacterial strains, including two Gram-negative and three Gram-positive species, are summarized in Table 1. Compared with melittin, Tat (49-57) displayed potent antibacterial activity, with the MIC value within the 8-16 μM range. We also examined the haemolytic activity of Tat (49-57) against human erythrocytes. It did not show haemolysis of human erythrocytes in all the tested concentrations (Table 2). The side chains of Tat (49-57) have two cationic tail groups (guanidine). The structural features of Tat (49-57) are similar to head-to-head pyrrole tetraamides [34,35], that can display antibacterial activity and also bind to the minor groove of DNA. Several other studies also revealed that agents that can bind to DNA.
| Peptide | MIC (µM) | ||||
|---|---|---|---|---|---|
| E. coli | S. typhimurium | B. subtilis | S. epidermidis | S. aureus | |
| Tat (49–57) | 8.04±0.08 | 16.12±0.08 | 15.93±0.10 | 8.07±0.17 | 16.25±0.12 |
| Melittin | 2.13±0.13 | 4.15±0.18 | 2.02±0.12 | 1.97±0.13 | 1.96±0.25 |
Table 1: Antimicrobial Activity of Peptides
| Peptides (µM) | 256 | 128 | 64 | 32 | 16 | 8 | 4 | 2 | |
| % Haemolysis | Tat (49–57) | 4.16 | 3.29 | 1.88 | 1.41 | 0.47 | 0 | 0 | 0 |
| Melittin | 100 | 100 | 100 | 100 | 98 | 91 | 92 | 68 | |
Table 2: Hemolytic Activity of Peptides
In the proposed study, the interaction between Tat (49-57) and ct-DNA was studied through UV/ Vis spectroscopy, fluorescence spectroscopy, CD spectroscopy and viscosity measurements. The results indicated that Tat (49-57) can bind to DNA through minor groove binding. The results were supported by competitive binding assay with EB, DNA melting, and viscosity measurement studies. Tat (49-57, RKKRRQRRR) is an Arg-rich peptide, similar to Q/ RGR-motif peptides [36]. Its side chains have the first and last residues (R). Tat (49-57) may directly contact the minor groove of AT-rich sequences via inserting guanidine and amino groups from Tat (49-57) side chains into the minor groove. Tat (49-57) also displayed antibacterial activity, with an MIC value within the 8-16 μM range; it showed no haemolysis of human erythrocytes in all the tested concentrations. This study on the antibacterial activity and DNA binding of Tat (49-57) is expected to provide valuable information on the mechanism of the antimicrobial activity of Tat (49- 57) and could be useful for peptide drug design with highly effective bioactivity.
Financial assistance
The paper was supported by the National Natural Science Foundation of China (Nos. 21572046, 21402040, 21301050).
Conflicts of interest
There are no conflicts of interest.
References
- Tiriveedhi V, Butko P. A fluorescence spectroscopy study on the interactions of the TAT-PTD peptide with model lipid membranes. Biochemistry 2007;46:3888-95.
- Said Hassane F, Saleh AF, Abes R, Gait MJ, Lebleu B. Cell penetrating peptides: overview and applications to the delivery of oligonucleotides. Cell Mol Life Sci 2010;67:715-26.
- Song J, Zhang Y, Zhang W, Chen J, Yang X, Ma P, et al. Cell penetrating peptide TAT can kill cancer cells via membrane disruption after attachment of camptothecin. Peptides 2015;63:143-9.
- Nakase I, Tanaka G, Futaki S. Cell-penetrating peptides (CPPs) as a vector for the delivery of siRNAs into cells. MolBiosyst 2013;9:855-61.
- Presente A, Dowdy SF. PTD/CPP peptide-mediated delivery of siRNAs. Curr Pharm Design 2013;19:2943-7.
- Nasrollahi SA, Fouladdel S, Taghibiglou C, Azizi E, Farboud ES. A peptide carrier for the delivery of elastin into fibroblast cells. Int J Dermatol 2012;51:923-9.
- Mo RH, Zaro JL, Shen WC. Comparison of cationic and amphipathic cell penetrating peptides for siRNA delivery and efficacy. Mol Pharm 2011;9:299-309.
- Xia H, Gao X, Gu G, Liu Z, Hu Q, Tu Y, et al. Penetratin-functionalized PEG–PLA nanoparticles for brain drug delivery. Int J Pharm 2012;436:840-50.
- Rizzuti M, Nizzardo M, Zanetta C, Ramirez A, Corti S. Therapeutic applications of the cell-penetrating HIV-1 Tat peptide. Drug Discov Today 2015;20:76-85.
- Brooks H, Lebleu B, Vivès E. Tat peptide-mediated cellular delivery: back to basics. Adv Drug Deliver Rev 2005;57:559-77.
- Chen X, Sa'adedin F, Deme B, Rao P, Bradshaw J. Insertion of TAT peptide and perturbation of negatively charged model phospholipid bilayer revealed by neutron diffraction. BiochimBiophysActa 2013;1828:1982-8.
- Vives E, Brodin P, Lebleu B. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J BiolChem 1997;72:16010-7.
- Suzuki T, Futaki S, Niwa M, Tanaka S, Ueda K, Sugiura Y. Possible existence of common internalization mechanisms among arginine-rich peptides. J BiolChem 2002;277:2437-43.
- Ziegler A, Seelig J. High affinity of the cell-penetrating peptide HIV-1 Tat-PTD for DNA. Biochemistry 2007;6:8138-45.
- Hegde A H, Prashanth S N, Seetharamappa J. Interaction of antioxidant flavonoids with calf thymus DNA analyzed by spectroscopic and electrochemical methods. J Pharm Biomed Anal 2012;63:40-6.
- Jung HJ, Park Y, Hahm KS. Biological activity of Tat (47–58) peptide on human pathogenic fungi. BiochemBiophys Res Commun 2006;345:222-8.
- Zhao D, Ma L, Lu K. Syntheses of valpromide dipeptide derivatives and interactions of derivatives with ctDNA. Res ChemIntermed 2015;41:8591-601.
- Verdon J, Berjeaud JM, Lacombe C. Characterization of anti-Legionellaactivity of warnericin RK and delta-lysin I from Staphylococcus warneri. Peptides 2008;29:978-84.
- Jaumot J, Gargallo R. Experimental methods for studying the interactions between G-quadruplex structures and ligands.Curr Pharm Design 2012;18:1900-16.
- Sirajuddin M, Ali S, Badshah A. Drug-DNA interactions and their study by UV-Visible, fluorescence spectroscopies and cyclic voltametry. J PhotochemPhotobiol B 2013;124:1-19.
- Shi JH, Chen J, Wang J, Zhu YY. Binding interaction between sorafenib and calf thymus DNA: spectroscopic methodology, viscosity measurement and molecular docking. SpectrochimActa A MolBiomolSpectrosc 2015;136:443-50.
- Charak S, Jangir DK, Tyagi G. Interaction studies of epirubicin with DNA using spectroscopic techniques. J MolStruct 2011;1000:150-4.
- Fuchs JE, Spitzer GM, Javed A, Biela A, Kreutz C, Wellenzohn B, et al. Minor groove binders and drugs targeting proteins cover complementary regions in chemical shape space. J ChemInf Model 2011;51(9):2223-32.
- Bi S, Zhang H, Qiao C, Sun Y, Liu C. Studies of interaction of emodin and DNA in the presence of ethidium bromide by spectroscopic method. SpectrochimActa A MolBiomolSpectrosc 2008;69:123-9.
- Sahoo BK, Ghosh KS, Bera R. Studies on the interaction of diacetylcurcumin with calf thymus-DNA. ChemPhys 2008;351:163-9.
- Rehman SU, Yaseen Z, Husain MA, Sarwar T, Ishqi HM, Tabish M. Interaction of 6 mercaptopurine with calf thymus DNA-deciphering the binding mode and photo-induced DNA damage. PLoS One 2014;9(4):e93913.
- Sarwar T, Rehman SU, Husain MA, Ishqi HM, Tabish M. Interaction of coumarin with calf thymus DNA: deciphering the mode of binding by in vitrostudies. Int J BiolMacromol 2015;73:9-16.
- Ling X, Zhong W, Huang Q, Ni K. Spectroscopic studies on the interaction of pazufloxacin with calf thymus DNA. J PhotochemPhotobiol B 2008;3:172-6.
- Labieniec M, Gabryelak T. Interactions of tannic acid and its derivatives (ellagic and gallic acid) with calf thymus DNA and bovine serum albumin using spectroscopic method. J PhotochemPhotobiol B 2006;2:72-8.
- Strekowski L, Harden DB, Wydra RL, Stewart KD, Wilson WD. Molecular basis for potentiation of bleomycin‐mediated degradation of DNA by polyamines. Experimental and molecular mechanical studies.J MolRecognit 1989;2:158-66.
- Deiana M, Matczyszyn K, Massin J, Olesiak-Banska J, Andraud C, Samoc M. Interactions of isophorone derivatives with DNA: spectroscopic studies. PloS One 2015;10.
- Li H, Bu X, Lu J, Xu C, Wang X, Yang X. Interaction study of ciprofloxacin with human telomeric DNA by spectroscopy and molecular docking.SpectrochimActa A MolBiomolSpectrosc 2013;107:227-34.
- Vaidyanathan VG, Nair BU. Photooxidation of DNA by a cobalt (II) tridentate complex. J InorgBiochem 2003;94:121-6.
- Barrett MP, Gemmell CG, Suckling CJ. Minor groove binders as anti-infective agents. PharmacolTher 2013;139(1):12-23.
- Dyatkina NB, Roberts CD, Keicher JD, Dai Y, Nadherny JP, Zhang W, Lou L. Minor groove DNA binders as antimicrobial agents. 1. Pyrroletetraamides are potent antibacterials against vancomycin resistant Enteroccoci and methicillin resistant Staphylococcus aureus. J Med Chem 2002;45(4):805-17.
- Gordon BRG, Li Y, Cote A, Weirauch MT, Ding P, Hughes TR, Liu J. Structural basis for recognition of AT-rich DNA by unrelated xenogeneic silencing proteins. ProcNatlAcadSci USA 2011;108(26):10690-5.
 0;
0;  1;
1;  2;
2;  3;
3; 4;
4;  5;
5;  6
6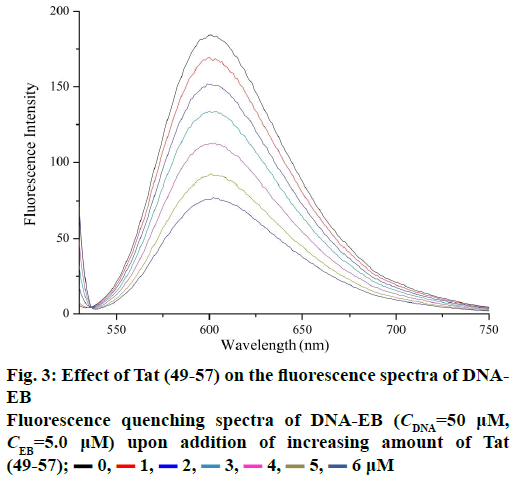
 0,
0,  1,
1,  2,
2,  3,
3,  4,
4,  5,
5,  6 μM
6 μM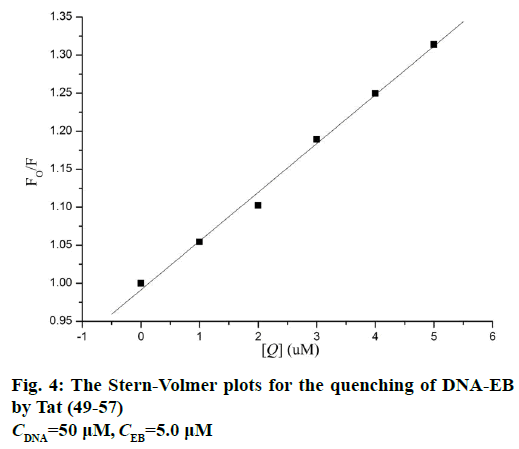
 Free Tat (49–57);
Free Tat (49–57);  free DNA;
free DNA;  DNA+Tat (49-57), 2 μM;
DNA+Tat (49-57), 2 μM;  DNA+Tat (49-57), 4 μM;
DNA+Tat (49-57), 4 μM;  DNA+Tat (49-57), 6 μM
DNA+Tat (49-57), 6 μM