- *Corresponding Author:
- H. M. Niu
Department of Orthopedics, Guanyun County People's Hospital Affiliated to Kangda College of Nanjing Medical University, Lianyungang, Jiangsu 222200, China
E-mail: nhm197809@163.com
| This article was originally published in a special issue, “Transformative Discoveries in Biomedical and Pharmaceutical Research” |
| Indian J Pharm Sci 2023:85(4) Spl Issue “87-92” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The aim of this research was to discover microRNAs and circular RNAs with varying expression in osteoporosis patients who have been given denosumab and to explore the correlation between microRNAs, messenger RNAs and circular RNAs and the drugs. As a first finding, we found differential microRNA and circular RNA expression in patients treated with denosumab for osteoporosis. Our next step was to identify microRNAregulated genes and genes that correspond to circular RNAs and then to perform enrichment analysis on those genes. We explored the correlation between circular RNA, microRNA and messenger RNA for the drugs as a concluding step. Our findings revealed 18 microRNA and 239 circular RNAs that were increased, and 15 microRNAs and 316 circular RNAs that were reduced in osteoporosis patients treated with denosumab in comparison to those who had not been treated. Analysis of Kyoto encyclopedia of genes and genomes pathway revealed that pathways targeted by differentially expressed microRNAs were notably enriched and they are axon guidance, proteoglycans in cancer, sphingolipid signalling and erythroblastic oncogene B signalling. The gene ontology enrichment analysis, meanwhile, revealed that pathways targeted by differentially expressed microRNAs were notably enriched and they are wingless-related integration site-induced cell-cell signalling, glutamatergic synapse and deoxyribonucleic acid-binding transcription activator activity. Analysis of Kyoto encyclopedia of genes and genomes pathway revealed that pathways significantly enriched by differentially expressed circular RNA were Salmonella infection, endocytosis, Fc gamma R-mediated phagocytosis and spinocerebellar ataxia; conversely, gene ontology enrichment analysis indicated that pathways significantly enriched by differentially expressed circular RNA were positive regulation of cellular catabolic process and cadherin binding. Considered to be the most important microRNAs involved the hsa-miR-424-5p, hsa-miR-1299, hsa-miR-195-5, hsa-miR-320d and hsa-miR-548am-3p are in the functions of denosumab in osteoporosis. A number of critical circular RNAs have been identified, such as hsa_circ_0005044, hsa_ circ_0005027, hsa_circ_0005657, hsa_circ_0005988 and hsa_circ_0013386.
Keywords
Osteoporosis, denosumab, micro ribonucleic acid, circular ribonucleic acids, messenger ribonucleic acid
Osteoporosis, a metabolic bone disorder, is a condition that occurs when bone density diminishes and fragility increases. In the world, more than 75 million people suffer from osteoporosis, a disease caused by both genetics and environment[1]. Osteoporosis, occurs among 70 % in those aged 80 y and above, and 15 % in those aged 50 y and above, which is a common occurrence[2]. When calcium and vitamin D are lacking in the diet, as well as inactivity, osteoporosis is more likely to occur. Denosumab (marketed as Prolia® or Pralia®), a recently approved new medicine, is a key mediator of bone resorption and Receptor Activator of Nuclear Factor Kappa-Β Ligand (RANKL) is its key. It is also a potentially effective molecular targeted agent to treatosteoporosis. Clinical guidelines dictate that denosumab should be administered subcutaneously every 6 mo for a variety of reasons, such as the treatment of postmenopausal women with osteoporosis who are at a high risk of fracture or who are unable to tolerate or fail other osteoporosis treatments. Despite the importance of these drugs in treating osteoporosis, their exact targets remain largely unknown and no studies have directly compared the downstream effects of circular Ribonucleic Acid (circRNA)-microRNA (miRNA)-messenger RNA (mRNA) expression profiling for the drug. Endogenous genes encode miRNAs, which are small, single-stranded and noncoding RNAs, and these are responsible for controlling cells. It is possible for miRNAs to bind to complementary sequences in the 5' and 3' ends of target mRNA, thereby repressing its expression of the target mRNAs through post-transcriptional regulation, forming a complex regulatory network[3,4]. miRNAs, with a major part in many essential biological activities[5], control a third of human genes. Moreover, circRNAs were first discovered in mammalian cells and have a loop structure that makes them more durable than linear RNAs that prevents exonuclease-mediated degradation[6]. As miRNA sponges, circRNAs bind abundantly to miRNAs. Interactions between disease-related miRNAs and circRNAs, thus have a role in the development of illness[7]. Currently, we don’t know the extent to which miRNA-circRNA-mRNA networks are regulated in osteoporosis, particularly during treatment with denosumab. By using bioinformatics methods, this article will describe the role that miRNAs, circRNAs and mRNAs play in denosumab treatment for osteoporosis patients.
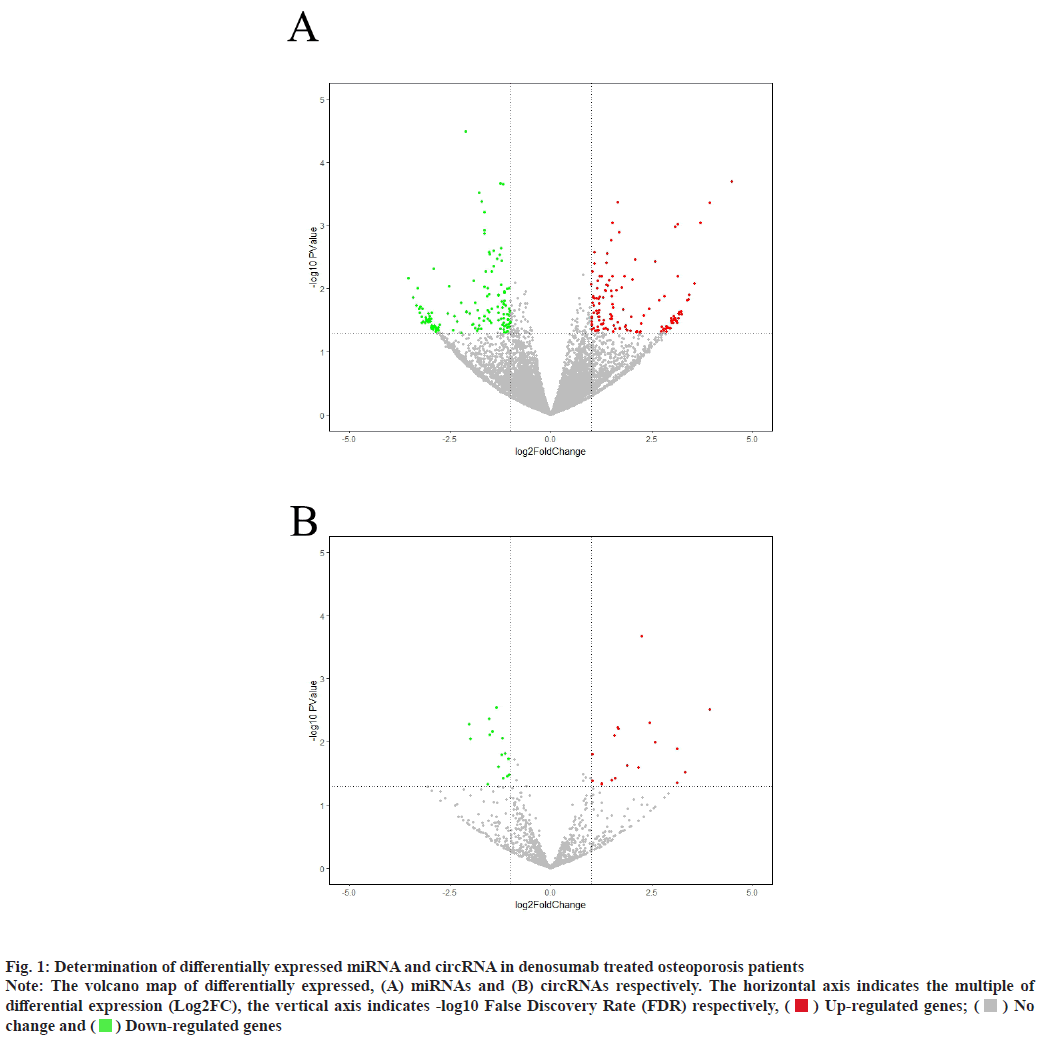
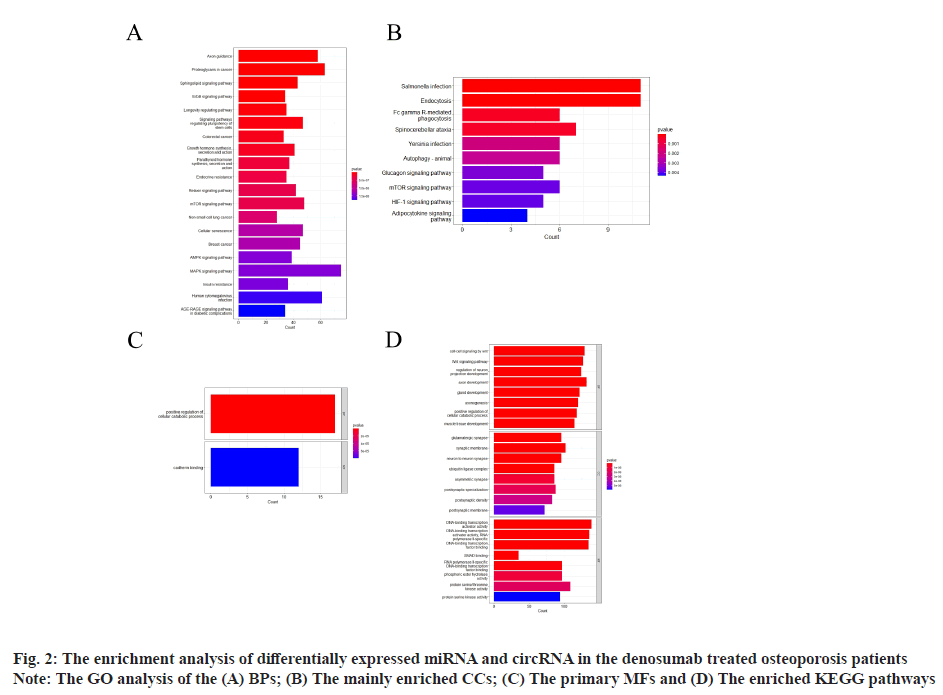
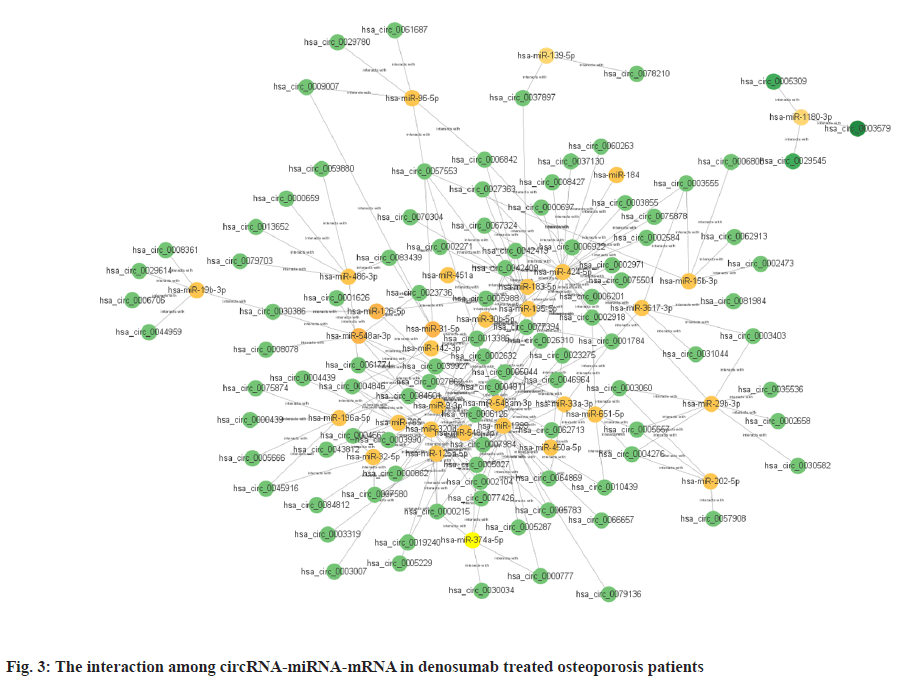
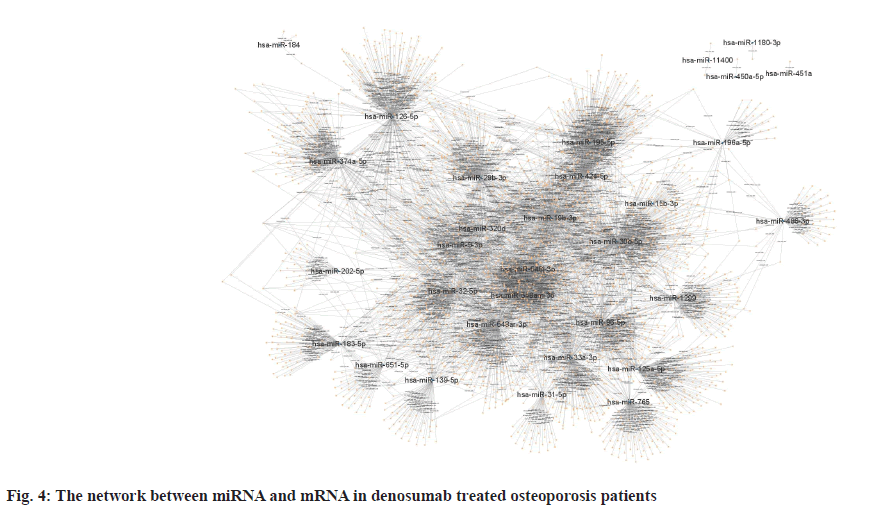
Sources of data were explained here. The osteoporosis dataset GSE159121 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE159121) and GSE141614 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE141614) were obtained from the Gene Expression Omnibus (GEO) database. Their circRNA and miRNA expression was obtained. Differential gene analysis of the study was explained here. EdgeR from the R language was employed to analyze the dissimilar expression of circRNAs and miRNAs between osteoporosis patients and healthy individuals. To screen for miRNA and circRNA, an absolute Log-transformed differential expression multiple (Log2 Fold Change (FC)) greater than 1 and p-value of 0.05 was employed as a threshold[8]. Functional enrichment analysis of the study was reported here. Using the clusterProfiler package in R language, a Gene Ontology (GO) analysis was conducted and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was also conducted, with a p-value of less than 0.05 being deemed statistically significant[9]. Prediction of miRNA target genes and circRNA target miRNAs was done based on the microRNA Target Prediction Database (miRDB) (http://mirdb.org/index). Predictions of target genes for miRNAs were made in HTML version 6.0. By using the circBank (http://www.circbank.cn/) database, target miRNAs of circRNAs were predicted. In the meantime, a regulatory network of miRNA-mRNAs and circRNA-miRNAs was visualized with Cytoscape (https://cytoscape.org/, version 3.9.1). Determination of differentially expressed miRNA and circRNA in denosumab treated osteoporosis patients were shown in fig. 1. Firstly, we downloaded a data GSE159121 from GEO database, including 3 osteoporosis patients and 3 normal people. The miRNA and circRNA expression profile were provided in the dataset. Analysis of the difference in miRNA and circRNA expression was done to choose those that were significantly differentially expressed. Next, the specific expressed miRNAs and circRNAs were further compared with denosumab treated patients in dataset of GSE141614. Both |LogFC|>1 and p<0.05 was set as a selecting threshold. As a result, 18 miRNAs and 239 circRNAs are upregulated in osteoporosis patients. Meanwhile, 15 miRNAs and 316 circRNAs are downregulated in osteoporosis patients. To visualize miRNA and circRNA expression difference analysis results, a volcano plot was drawn as shown in fig. 1A and fig. 1B. Enrichment analysis of differentially expressed miRNA and circRNA were shown in fig. 2. Then we used the genes regulated by differentially expressed 33 miRNAs and 415 circRNAs in denosumab treated osteoporosis patients to conduct enrichment analysis (fig. 2). As for miRNA-mRNA analysis, we used microRNA Sequence Database (miRBase). Through the mirBase database, miRNA regulated genes were obtained and genes with TargetScore>90 were selected. Meanwhile, for circRNA-mRNA analysis, circBank database was used. Among 415 circRNAs, 211 circRNAs regulate corresponding mRNA. Subsequently, genes regulated by those miRNA and circRNA were used for GO and KEGG enrichment analysis. In the osteoporosis patients group, Biological Processes (BPs) of differentially expressed miRNAs determined by GO analysis were mainly cell-cell signaling by Wingless-related integration site (Wnt), Wnt signaling pathway, regulation of neuron projection development and axon development (fig. 2A) and the mainly enriched Cell Components (CCs) include glutamatergic synapse, synaptic membrane, neuron to neuron synapse and ubiquitin ligase complex (fig. 2B). The primarily enriched Molecular Functions (MFs) were Deoxyribonucleic Acid (DNA)-binding transcription activator activity, DNA-binding transcription activator activity, RNA polymerase II-specific, DNA-binding transcription factor binding and Suppressor of Mothers against Decapentaplegic (SMAD) binding (fig. 2C). Analysis of KEGG pathways uncovered that the miRNAs expressed differently were mainly concentrated in pathways such as axon guidance, proteoglycans in cancer, sphingolipid signaling and Erythroblastic Oncogene B (ErbB) signaling. In osteoporosis patients group, the functions enriched by all differentially expressed circRNA regulated genes using GO analysis are positive regulation of cellular catabolic process and cadherin binding. KEGG analysis show that the functions enriched by all differentally expressed circRNA regulated genes are Salmonella infection, Endocytosis, Fc gamma R-mediated phagocytosis and spinocerebellar ataxia (fig. 2D). Interactions among circRNA-miRNA-mRNA for denosumab treated osteoporosis patients were shown in fig. 3. Then we find interaction between circRNA, miRNA and mRNA in osteoporosis patients. We found correlation between circRNA and miRNA using circBank database. We discovered 211 circRNAs after discovering their corresponding circRNA in 33 distinctially expressed miRNAs and interacting with them in osteoporosis patients. Subsequently, a miRNA-mRNA network was constructed by mirBase, with specific miRNAs and genes with TargetScore>90 selected to construct the network as shown in fig. 4. We found that 32 differentially expressed miRNAs (except hsa- miR-11400) interacted with corresponding differentially expressed circRNAs in osteoporosis patients. The greatest amount of target circRNAs is regulated by Homo sapiens (human) hsa-miR-424- 5p, while hsa-miR-1299 has 20. The target circRNAs regulated by hsa-miR-195-5p, hsa-miR-320d and hsa-miR-548am-3p, meanwhile, are 14, 13 and 11 respectively (fig. 4)-a noteworthy distinction. Base on these facts, the five miRNAs (hsa-miR-424-5p, hsa-miR-1299, hsa-miR-195-5p, hsa-miR-320d and hsa-miR-548am-3p) are suggested to be the more critical miRNAs in denosumab treated osteoporosis patients. Meanwhile, as for differently expressed circRNA and miRNA in osteoporosis patients, hsa_ circ_0005044 can interact with the the most miRNAs, the number of target miRNAs regulated by hsa_ circ_0005044 is 7. The target miRNAs regulated by hsa_circ_0005027, hsa_circ_0005657, hsa_ circ_0005988 and hsa_circ_0013386 are 6. Those 5 circRNAs are the most important circRNAs in denosumab treated osteoporosis patients. Osteoporosis, a common malady of the human bone, is marked by a decrease in bone mass, microarchitectural deterioration and fractures of fragility. The lack of calcium and vitamin D in food, as well as a lack of exercise, can heighten the likelihood of this condition. In industrialized countries, osteoporosis has become a major public health issue[10]. The aging population is predicted to cause a rise of 25 % in fractures over the next 10 y[11], a condition that is prevalent among post-menopausal women, with nearly 1 in 3 women over which 50 affected. The formation of bone by osteoblast, as well as osteolysis mediated by osteoclast, is a multifaceted process of bone metabolism. Osteoblasts, mainly in the inner and outer periosteum and bone marrow, are distinguished from mesenchymal progenitor cells. A variety of biologically active substances are specifically secreted by these cells. The Wnt signaling pathway, one of several pathways in bone tissue, is responsible for the regulation of bone homeostasis and its promotion[12]. Osteoblasts, the most essential cells for bone formation and remodeling, are also responsible for the synthesis, secretion and mineralization of the bone matrix. Therefore, the differentiation and activity of osteoblasts are closely linked to the onset and development of osteoporosis[13]. Derived from blood mononuclear macrophages, osteoclasts are terminally differentiated cells whose primary purpose is to facilitate bone resorption. These cells are essential for bone development, growth, repair and remodeling. Abnormal bone resorption can result from abnormal osteoclast function. Osteoclast differentiation plays an important role in osteoporosis, as hyperfunction can lead to degenerative bone diseases such as osteoporosis[1]. In this study, firstly we conducted a differential analysis based on the GSE159121 and GSE141614 dataset, sorted differentally expressed 33 miRNAs and 415 circRNAs. By binding to the 3′ Untranslated Region (UTR) of its mRNA, miR-96- 5p can inhibit the phosphorylation of Epidermal Growth Factor Receptor (EGFR), Extracellular Signal-Regulated Kinase 1 (ERK1) and Akt Serine/ Threonine Kinase (AKT), thereby promoting osteogenic differentiation[14]. This is due to the downregulation of human Heparin-Binding EGF- like Growth Factor (HB-EGF) induced by these miRNA. It was proposed in a different trial that miR- 96-5p could activate the Wnt signaling pathway, thereby stimulating osteogenic differentiation during drug administration[15]. An enrichment analysis of GO and KEGG was conducted using genes regulated by upregulated miRNA and circRNA. The most enriched GO items in miRNA-regulating genes were cell-cell signaling by Wnt, Wnt signaling pathway, glumateric synapse, synapstic membrane, DNA- binding transcription activator activity and RNA polymerase II-specific. Enriched KEGG items of miRNA regulating genes are axon guidance, proteoglycans in cancer, sphingolipid signaling pathway and ErbB signaling pathway. Meanwhile, the main enriched items in circRNA regulating genes are Salmonella infection, endocytosis, Fc gamma R-mediated phagocytosis and spinocerebellar ataxia, etc. Then we try to explore correlation among miRNA, circRNA and mRNA. We found that hsa-miR-424-5p, hsa-miR-1299, hsa-miR-195-5p, hsa- miR-320d and hsa-miR-548am-3p can interact with the most circRNA which are differentially expressed in osteoporosis patients. It is proposed that these miRNAs are the most essential in osteoporosis. Meanwhile, hsa_circ_0005044, hsa_circ_0005027, hsa_circ_0005657, hsa_circ_0005988 and hsa_ circ_0013386 can interact with the most miRNA which are differentially expressed in denosumab treated osteoporosis patients and suggested to be the most critical circRNAs in the drug-regulated mechanism.
Fig. 1: Determination of differentially expressed miRNA and circRNA in denosumab treated osteoporosis patients
Note: The volcano map of differentially expressed, (A) miRNAs and (B) circRNAs respectively. The horizontal axis indicates the multiple of differential expression (Log2FC), the vertical axis indicates -log10 False Discovery Rate (FDR) respectively,  Up-regulated genes;
Up-regulated genes;  No change and
No change and  Down-regulated genes
Down-regulated genes
Funding:
This study was supported by Lianyungang city Aging Health Research Project (Grant project, No. L202120).
Conflict of interests:
The authors declared no conflict of interest.
References
- Rachner TD, Khosla S, Hofbauer LC. Osteoporosis: Now and the future. Lancet 2011;377(9773):1276-87.
[Crossref] [Google Scholar] [PubMed]
- Schuiling KD, Robinia K, Nye R. Osteoporosis update. J Midwifery Womens Health 2011;56(6):615-27.
[Crossref] [Google Scholar] [PubMed]
- Bellavia D, Salamanna F, Raimondi L, de Luca A, Carina V, Costa V, et al. Deregulated miRNAs in osteoporosis: Effects in bone metastasis. Cell Mol Life Sci 2019;76:3723-44.
[Crossref] [Google Scholar] [PubMed]
- Pu M, Chen J, Tao Z, Miao L, Qi X, Wang Y, et al. Regulatory network of miRNA on its target: coordination between transcriptional and post-transcriptional regulation of gene expression. Cell Mol Life Sci 2019;76(3):441-51.
[Crossref] [Google Scholar] [PubMed]
- Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev 2015;87:3-14.
[Crossref] [Google Scholar] [PubMed]
- Zhao K, Zhao Q, Guo Z, Chen Z, Hu Y, Su J, et al. Hsa_Circ_0001275: A potential novel diagnostic biomarker for postmenopausal osteoporosis. Cell Physiol Biochem 2018;46(6):2508-16.
[Crossref] [Google Scholar] [PubMed]
- Jin D, Wu X, Yu H, Jiang L, Zhou P, Yao X, et al. Systematic analysis of lncRNAs, mRNAs, circRNAs and miRNAs in patients with postmenopausal osteoporosis. Am J Transl Res 2018;10(5):1498-1510.
[Google Scholar] [PubMed]
- Ritchie ME, Phipson B, Wu DI, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43(7):e47.
[Crossref] [Google Scholar] [PubMed]
- Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012;16(5):284-7.
[Crossref] [Google Scholar] [PubMed]
- Lane NE. Epidemiology, etiology and diagnosis of osteoporosis. Am J Obstet Gynecol 2006;194(2):3-11.
[Crossref] [Google Scholar] [PubMed]
- Lane JM, Russell L, Khan SN. Osteoporosis. Clin Orthop Relat Res 2000;(372):139-50.
[Google Scholar] [PubMed]
- Wang F, Wang Y, Zhao Y, Zhan Q, Yu P, Wang J, et al. Sialoglycoprotein isolated from eggs of Carassius auratus ameliorates osteoporosis: An effect associated with regulation of the Wnt/β-catenin pathway in rodents. J Agric Food Chem 2016;64(14):2875-82.
[Crossref] [Google Scholar] [PubMed]
- Yang Y, Yujiao W, Fang W, Linhui Y, Ziqi G, Zhichen W, et al. The roles of miRNA, lncRNA and circRNA in the development of osteoporosis. Biol Res 2020;53(1):40.
[Crossref] [Google Scholar] [PubMed]
- Yang M, Pan Y, Zhou Y. miR-96 promotes osteogenic differentiation by suppressing HBEGF-EGFR signaling in osteoblastic cells. FEBS Lett 2014;588(24):4761-8.
[Crossref] [Google Scholar] [PubMed]
- Ma S, Wang DD, Ma CY, Zhang YD. microRNA‐96 promotes osteoblast differentiation and bone formation in ankylosing spondylitis mice through activating the Wnt signaling pathway by binding to SOST. J Cell Biochem 2019;120(9):15429-42.
[Crossref] [Google Scholar] [PubMed]