- *Corresponding Author:
- W. Zhang
International Peace Maternity and Child Health Hospital
School of Medicine, Shanghai Jiao Tong University, China
E-mail: weizhang611@163.com
| This article was originally published in a special issue, “Therapeutic Perspectives in Biomedical Research and Pharmaceutical Sciences and their Nursing Methods” |
| Indian J Pharm Sci 2021:83(4)Spl issue “115-121” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
To determine the correlation between deoxyribonucleic acid methylation and gene expression of cytochrome P450 family 19 subfamily A polypeptide 1 cytochrome P450 family 19 subfamily a member 1, anti-Mullerian hormone and anti-Mullerian hormone receptor type 2, which regulate follicle development and androgen metabolism in polycystic ovary syndrome pathogenesis. The levels of cytochrome P450 family 19 subfamily a member 1, anti-Mullerian hormone and anti-Mullerian hormone receptor type 2 messenger ribonucleic acid and protein expression were evaluated by various techniques in polycystic ovary syndrome patients. Estradiol, androstenedione and anti-Mullerian hormone levels were also tested in follicular fluid. In order to quantify the connection between deoxyribonucleic acid methylation and gene expression, we conducted Pearson product-moment correlation coefficient analyses. The estradiol level did not alter, however the androstenedione level was increased. Polycystic ovary syndrome granulosa cells were hypermethylated cytochrome P450 family 19 subfamily a member 1, but hypomethylated anti- Mullerian hormone and anti-Mullerian hormone receptor type 2. In general, the greater the amount of deoxyribonucleic acid methylation, the lower the level of downstream messenger ribonucleic acid and protein expression. Aromatase enzyme activity is regulated by deoxyribonucleic acid methylation and a hormone called follicular estradiol/androstenedione ratio is linked with it. A prior study reported that anti- Mullerian hormone inhibits aromatase in granulosa cells and therefore the inverse connection between cytochrome P450 family 19 subfamily a member 1 and anti-Mullerian hormone confirms that finding. Our study suggests alterations in deoxyribonucleic acid methylation as the underlying mechanism of aberrant gene expression of cytochrome P450 family 19 subfamily a member 1, anti-Mullerian hormone and anti- Mullerian hormone receptor type 2 and polycystic ovary syndrome pathogenesis.
Keywords
Deoxyribonucleic acid methylation, androgen, methylation specific polymerase chain reaction, granulosa cells, in vitro fertilization, aromatase
Polycystic ovary syndrome (PCOS) is a common endocrine disorder among adolescents and women of reproductive age. PCOS is characterized by excessive ovarian and/or androgen secretion, chronic anovulation and abnormal follicle development leading to menstrual irregularity, masculinization and sub-infertility[1-3]. PCOS patients exhibit high luteinizing hormone (LH) and low follicle-stimulating hormone (FSH) levels that promote the accumulation of antral follicles with arrested growth and development[4]. Metabolic issues such as hypertension, obesity, glucose intolerance and increased risk of developing endometrial cancer and diabetes mellitus type 2 in later life are often associated with PCOS[5,6]. Although family history is thought to contribute to PCOS development[5], inheritance patterns have been inconclusive and no genes have been confirmed to modulate PCOS development[7]. Environmental factors as well as gene-environment interactions have also been purported to influence predisposition to PCOS pathogenesis[6,7]. Given that PCOS is an incurable disorder and has higher prevalence especially among South Asian women, as high as about 50 %[7]; thus, it is imperative to clarify and elucidate its disease etiology.
As genetic and environmental factors have been implicated in PCOS development, epigenetic dysregulation has been touted as a probable mechanism underlying abnormal changes in the expression of androgen metabolism-related genes and androgen levels[7]. This is supported by a deoxyribonucleic acid (DNA) methylome profiling study by next-generation sequencing, which revealed genes with altered DNA methylation patterns in PCOS[8]. Moreover, in an earlier DNA methylome profiling study of PCOS, we identified several genes including cytochrome P450 family 19 subfamily A polypeptide 1 (CYP19A1), anti- Mullerian hormone (AMH) and AMH receptor type 2 (AMHR2) with altered DNA methylation profiles[9]. All three genes have been implicated in hyperandrogenism and follicular dysplasia[10].
CYP19A1 encodes the cytochrome P450 aromatase enzyme that regulates androgen metabolism[2]. Specifically, aromatase catalyzes the conversion of androgen (androstenedione (A) and testosterone(T)) to estrogen (estrone and estradiol (E2)) in ovarian granulosa cells upon stimulation by FSH[6]. The increase in E2 level enhances follicle sensitivity to FSH, thereby promoting the growth and development of follicles, enabling oocyte maturation. Suppression of aromatase activity causes androgen accumulation in the follicular fluid (FF) and serum and gives rise to PCOS[2] and other ovulation disorders[4]. In an earlier study on CYP19A1, we showed that the CYP19A1 promoter DNA hypermethylation is associated with downregulation of its gene expression[11]. AMH plays important functions in modulating antral follicle maturation and dominant follicle selection[12] and oocyte quality[13]. Abnormally high AMH and AMHR2 expression in granulosa cells and increased AMH FF and serum levels were found in PCOS patients[14]. Elevated AMH secretion in PCOS reportedly counteracts insulin-induced stimulation of aromatase activity in granulosa cells[14].
Hence in this study, we determined the correlation of aberrant DNA methylation with the downstream expression of CYP19A1, AMH and AMHR2 genes in PCOS. In ovarian granulosa cells of patients stratified into PCOS and control groups were collected during in vitro fertilization (IVF) treatment at the International Peace Maternity and Child Health Hospital, China. We first determined the changes in the DNA methylation patterns of these three genes in PCOS using methylation-specific polymerase chain reaction (MSP). Quantitative polymerase chain reaction (qPCR) and western blotting were used to monitor changes in gene expression levels in PCOS. Finally, the association between the DNA methylation status of these three genes and their corresponding changes in gene expression were determined using Pearson correlation analysis. Basal hormone levels (serum LH/FSH and E2/T, FF, E2/A and AMH) were also compared between PCOS and control groups and their association with DNA methylation status was investigated. Our results suggest aberrant DNA methylation as the mechanism underlying changes in downstream gene expression and consequent PCOS pathogenesis. It may be worth further exploring these three genes as therapeutic targets in PCOS therapy.
Materials and Methods
Patient selection and grouping:
Patients who sought fertility treatment at the International Peace Maternity and Child Health Hospital (Shanghai, China) between May 2017 and August 2018 were recruited for the study. Ovarian granulosa cells were collected from 30 PCOS patients and 30 normal patients with tubal or male factor infertility (control) during IVF-embryo transfer (IVF-ET) and were flash frozen in liquid nitrogen for storage for subsequent use. The study protocol was approved by the Ethics Committee of the Obstetrics and Gynecology Hospital of Fudan University (Shanghai, China) and was initiated after obtaining signed informed consent from all participants. Patients in the PCOS and control groups were selected according to the inclusion and exclusion criteria. PCOS diagnosis was based on the revised standards of the American Society of Reproductive Medicine and the Rotterdam criteria[15]. Satisfaction of any two of the three criteria constituted a positive PCOS diagnosis and excluded other hyperandrogenic disorders: no ovulation or rare ovulation, clinical and biochemical changes in androgen levels and polycystic ovarian changes.
Participants were excluded based on the following criteria: complications including endometriosis, tumor, diabetes, cardiovascular disease, hypertension and endocrine diseases like hyperthyroidism; abnormal prolactin level, hematuria and liver and kidney function; no obstetrics and gynecology complications; no medical and surgical complications; and no medical history of hormone drugs in the past 3 mo. Given the known influence of age and body mass index (BMI) on DNA methylation level[3,6,16], 1:1 matching of the PCOS group to control group was performed according to age (±3 y) and BMI (±3 kg/m2).
Patient information collected included medical history, weight, and height. Ovarian volume and number of follicles were determined from ultrasound examination. Basal hormone (FSH, LH, E2, testosterone, prolactin (PRL), androstenedione and AMH) levels in patient sera were measured using the enzyme-linked immunosorbent assay (ELISA) and E2/T and LH/FSH ratios were calculated. Menstrual blood was collected during the follicular phase on the day of human chorionic gonadotropin (hCG) injection before in IVF-ET, and basal hormone levels (A, AMH, E2, E2/A) in the FF were measured by ELISA.
Ovulation induction and oocyte collection:
Ovulation was stimulated via gonadotropin-releasing hormone agonist (GnRHa)/Gn/hCG injection and ovarian granulosa cells, and FF was collected as previously described[7]. Intradermal injection of GnRHa (dafilin; 0.1 mg/d) was given for pituitary desensitization during the mid-luteal phase (~20th d of menstruation) of the menstrual cycle before the ovulation cycle. In the next menstrual cycle, gonadotropin (Gn) was given from 2-3 d to stimulate ovulation. Transvaginal ultrasound scan of the ovaries was used to monitor antral follicular development and Gn was discontinued when the average diameter of 2-3 follicles reached 18±2 mm indicating mature follicles. Intramuscular injection of 10 000 IU hCG was immediately performed when follicles were mature during IVF-ET and oocyte retrieval (collection of ovarian granulosa cells and FF) was performed on the same day under the guidance of vaginal ultrasound 34-36 h later.
Isolation of granulosa cells:
FF was first separated from egg cells by centrifugation at 2000 rpm for 10 min at room temperature. EDTA and phosphate-buffered saline (PBS) were added to obtain cell suspensions (1×108 ml). Ficoll solution (GE Healthcare, Chicago, IL, USA) was slowly added to cell suspensions at a 1:1 volume ratio for lymphocyte separation before centrifugation at 1500 rpm for 15 min. The white granulosa cell layer was aspirated, rinsed with PBS and centrifuged to get rid of any remaining Ficoll solution. Red blood cell lysate was added to the cell pellet at a 1:3 volume ratio and the mixture was gently inverted and mixed for 1 min before letting it stand at room temperature for 10 min. Then the mixture was centrifuged at 5000 rpm for 1 min and the granulosa cells obtained were used for subsequent experiments.
Measurement of hormone levels by ELISA:
E2, A and AMH hormone levels in FF were measured using E2 and A ELISA kits (Cusabio, Hubei, China) and the AMH ELISA Kit (Yanyu Chemical Reagent Co. Ltd., Shanghai, China) according to the manufacturer’s instructions. The optical density of each well was measured sequentially with a microplate reader at a wavelength of 450 nm.
Methylation-specific polymerase chain reaction:
Genomic DNA of granulosa cells was extracted using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Bisulfite modification of genomic DNA was performed using the EZ DNA Methylation-GoldTM Kit according to the manufacturer’s instructions (Zymo Research, Irvine, CA, USA). Next, bisulfite-treated genomic DNA was amplified using MSP primers designed using Methyl Primer Express v1.0 and synthesized by Shanghai Shengong Technology Service Co., Ltd. (Shanghai, China). The primer sequences are shown in Table S1. MSP was performed using the following conditions: denaturation at 95° for 5 min, denaturation at 94° for 45 s, annealing at a gradient temperature of 65° to 50° for 45 s (2° temperature decrease every 3 cycles), extension at 72° for 45 s, cycle reaction for 35 cycles, extension at 72° for 7 min. PCR products were analyzed on a 2 % agarose gel. Lymphocyte genomic DNA from peripheral blood that had been treated with SssI-CpG-methylase (NEB, Cambridge, UK) before MSP was used as the positive control, whereas sulfitetreated genomic DNA for the negative control was not treated with SssI-CpG-methylase. Semi-quantitative assessment of the extent of methylation was scored using this formula: relative methylation level=gray value of methylated band (M)/(gray value of methylated band (M)+gray value of unmethylated band (U)).
Total ribonucleic acid extraction and quantitative polymerase chain reaction:
Total RNA was extracted from granulosa cells using Trizol reagent (Invitrogen, Carlsbad, CA, USA) and the RNA concentration was measured. Reverse transcription to cDNA was performed using the RT-PCR kit (Invitrogen) according to the manufacturer’s instructions. PCR primers were designed by Primer Express 2.0 design and synthesized by Shanghai Shenggong Technical Service Company (Shanghai, China). The primer sequences used were as follows: CYP19A1: for, 5'- GCTGGAAATGATCTTTACCCCA -3', rev, 5'- TGTAGCCTGGTTCTCTGGTGTG -3'; AMH: for, 5'-GACAGCAATGGCAGCAGCTC -3', rev, 5'-CTCCTGGAACCTCAGCGAGG -3'; AMHR2: for, 5'-TCCGATTTATCACTGCCAGCC -3', rev, 5'-TCCATCTTCCCGAATGAGCAC -3'. The qPCR reaction was performed using the SYBR Green Real- Time PCR kit (Qiagen) according to the manufacturer’s instructions. The conditions used for qPCR were as follows: 94° pre-denaturation for 5 min; 35 cycles (94°, denaturation for 30 s, 60°, annealing for 30 s, 72°, extension for 30 s); 72°, extension for 10 min and 4° incubation. GAPDH was used as an internal reference control. Relative quantitative analysis was performed using the 2-△△Ct method.
Western blot analysis:
Total protein was extracted from granulosa cell lysates as previously described. Briefly, total protein was isolated using RIPA lysis buffer (Shenneng Bocai Biotechnology) and protein concentration was determined using the BCA assay. Next, proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto a polyvinylidene fluoride membrane before being probed with primary antibodies against CYPR19A1 (1:200, sc374176; Santa Cruz Biotechnology, Dallas, TX, USA), AMH (1:200, cat. no. MAB1737; R&D Systems, Minneapolis, MN, USA) or AMHR2 (1:200, cat. no. 4749-MR; R&D Systems) overnight at 4°. Subsequently, the membrane was incubated with horseradish peroxidase-labeled secondary antibody (1:2000; Santa Cruz) at room temperature for 1 h. β-actin (Sigma, St. Louis, MO, USA) was used as the internal reference control. A two-color infrared laser scanning imaging system was used for band visualization, and band intensities were determined using Image Pro-Plus 3.0.1 software (Media Cybernetics, Silver Spring, MD, USA).
Statistical analysis:
Statistical analysis was performed using statistical package for the social sciences (SPSS) v16.0 software (SPSS Inc., Chicago, IL, USA). The mRNA level of the target gene relative to the internal reference gene GAPDH is expressed as the median (minimum-maximum) and the relative methylation level of the gene promoter region and relative protein level are expressed as the mean±standard deviation (SD). Student’s paired t-test or t-test was applied for data analysis assuming a normal distribution for comparing between the PCOS and control groups. Pearson correlation analysis was performed to determine the correlation between examined parameters. p<0.05 was considered statistically significant.
Results and Discussion
PCOS patients had a significantly higher LH/FSH ratio (p<0.01; Table S2) and ovarian volume but significantly lower E2/T level (p<0.01). No significant difference was observed for age, BMI, FSH, E2 and PRL indicators between the PCOS and control groups (Table S2). Elevated A and AMH levels (p<0.05; Table S3) and lower E2/A level (p<0.05) in the FF were observed for the PCOS group. No significant difference in FF E2 level between the two groups was observed (Table S3).
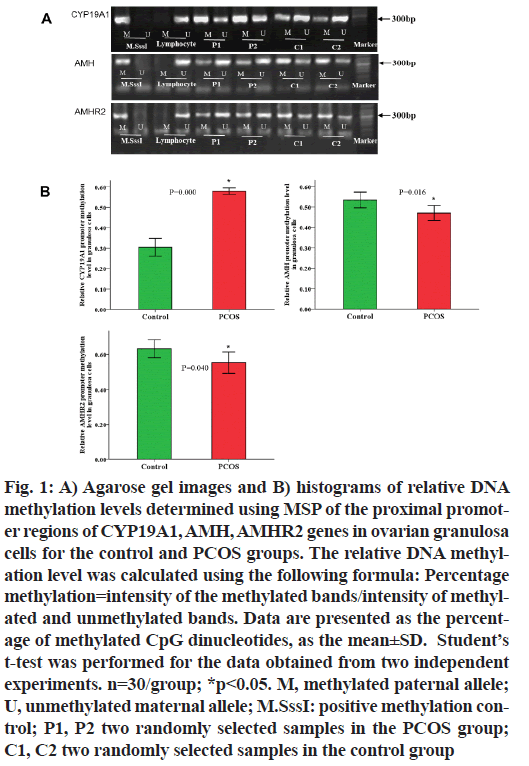
We performed MSP to determine the DNA methylation profiles of CYP19A1, AMH, and AMHR2 genes for both PCOS and control groups. Hypermethylation of CYP19A1 gene promoter (PCOS vs. control: 0.58±0.04 % vs. 0.30±0.12 %; n=30; p<0.001) and hypomethylation of AMH and AMHR2 gene promoters were observed in PCOS granulosa cells compared with the control group (PCOS vs. control: AMH: 0.47±0.10 % vs. 0.53±0.09 %, AMHR2: 0.55±0.16 % vs. 0.63±0.14; n=30; p<0.05; Table 1 and fig. 1). As DNA methylation suppresses gene transcriptional activity, these observations indicate that the hypermethylated CYP19A1 gene lead to reduced gene transcription, whereas the hypomethylated AMH and AMHR2 genes enable increased transcriptional activity.
| Relative DNA methylation level | PCOS group (n=30) | Control group (n=30) | p |
|---|---|---|---|
| (mean ±SD) | (mean ±SD) | ||
| CYP19A1 | 0.58±0.04 | 0.30±0.12 | 0.000** |
| AMH | 0.47±0.10 | 0.53±0.09 | 0.016* |
| AMHR2 | 0.55±0.16 | 0.63±0.14 | 0.040* |
*p<0.05, **p<0.001
Table 1: Relative DNA Methylation Level of CYP19A1, AMH, AMHR2 Genes in Ovarian Granulosa Cells by MSP
Fig. 1: A) Agarose gel images and B) histograms of relative DNA methylation levels determined using MSP of the proximal promoter regions of CYP19A1, AMH, AMHR2 genes in ovarian granulosa cells for the control and PCOS groups. The relative DNA methylation level was calculated using the following formula: Percentage methylation=intensity of the methylated bands/intensity of methylated and unmethylated bands. Data are presented as the percentage of methylated CpG dinucleotides, as the mean±SD. Student?s t-test was performed for the data obtained from two independent experiments. n=30/group; *p<0.05. M, methylated paternal allele; U, unmethylated maternal allele; M.SssI: positive methylation control; P1, P2 two randomly selected samples in the PCOS group; C1, C2 two randomly selected samples in the control group
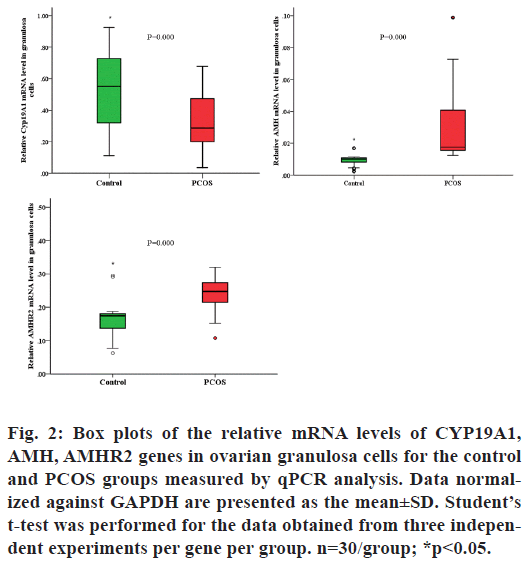
To confirm the expression of the three genes from the MSP analyses, we examined the expression of these three genes at the mRNA and protein levels using qPCR and western blot analyses, respectively. Upregulated AMH and AMHR2 mRNA levels (p<0.001) and downregulated CYP19A1 mRNA level (p<0.001; Table 2 and fig. 2) were observed in the granulosa cells using qPCR.
| Relative mRNA level | PCOS group (n=30) | Control group (n=30 | p |
|---|---|---|---|
| Median(min-max) | Median(min-max) | ||
| CYP19A1 | 0.32 (0.13-0.51) | 0.54 (0.29-0.78) | 0.000** |
| AMH | 0.02 (0.03-0.04) | 0.01 (0.002-0.02) | 0.000** |
| AMHR2 | 0.25 (0.19-0.31) | 0.16 (0.11-0.2) | 0.000** |
*p<0.05, **p<0.00
Table 2: Relative MRNA Levels of CYP19A1, AMH, AMHR2 Genes in Ovarian Granulosa Cells
Fig. 2: Box plots of the relative mRNA levels of CYP19A1, AMH, AMHR2 genes in ovarian granulosa cells for the control and PCOS groups measured by qPCR analysis. Data normalized against GAPDH are presented as the mean±SD. Student?s t-test was performed for the data obtained from three independent experiments per gene per group. n=30/group; *p<0.05.
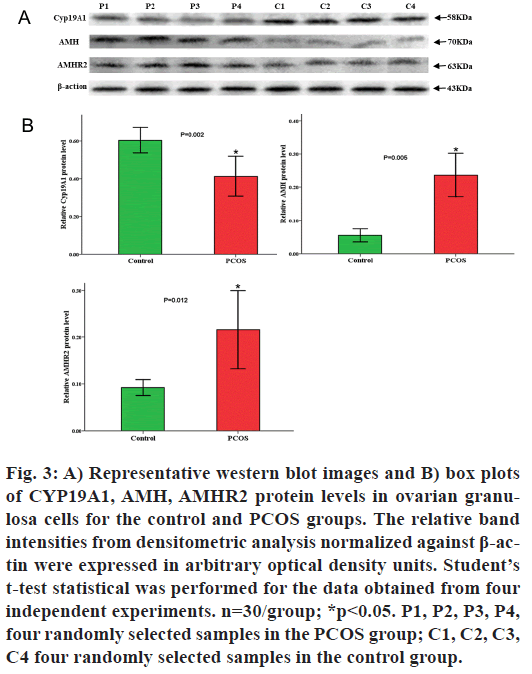
Indeed, the qPCR results were consistent with our observations from MSP analysis and supported our conclusions. Similarly, western blot analyses revealed elevated AMH and AMHR2 protein levels (p<0.05; Table 3 and fig. 3) and downregulated CYP19A1 protein expression (p<0.05).
| Relative protein level | PCOS group (n=16) (mean ±SD) |
Control group (n=16) (mean ±SD) |
p |
|---|---|---|---|
| CYP19A1 | 0.41±0.06 | 0.60±0.04 | 0.002* |
| AMH | 0.24±0.04 | 0.06±0.01 | 0.005* |
| AMHR2 | 0.22±0.05 | 0.09±0.01 | 0.012* |
*p<0.05
Table 3: Relative Protein Levels of CYP19A1, AMH, AMHR2 in Ovarian Granulosa Cells
Fig. 3: A) Representative western blot images and B) box plots of CYP19A1, AMH, AMHR2 protein levels in ovarian granulosa cells for the control and PCOS groups. The relative band intensities from densitometric analysis normalized against ß-actin were expressed in arbitrary optical density units. Student?s t-test statistical was performed for the data obtained from four independent experiments. n=30/group; *p<0.05. P1, P2, P3, P4, four randomly selected samples in the PCOS group; C1, C2, C3, C4 four randomly selected samples in the control group.
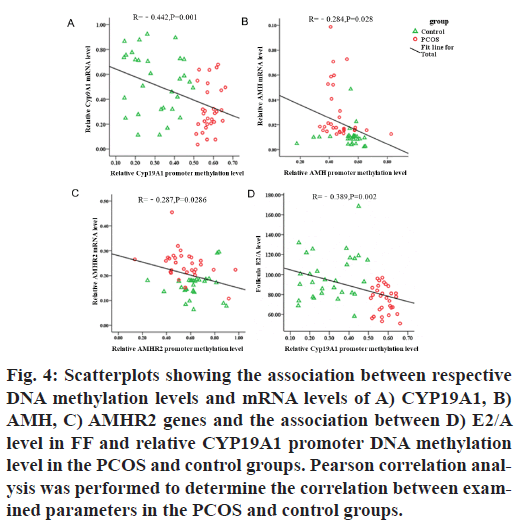
Pearson correlation analysis revealed a negative correlation between CYP19A1 (γ=-0.442; p=0.001), AMH (γ=-0.284; p=0.028), AMHR2 (γ=-0.287; p=0.0286) promoter DNA methylation status and their downstream gene expression in PCOS granulosa cells (p<0.05; fig. 4A-fig. 4C), wherein high DNA methylation level was associated with low enzyme expression level. CYP19A1 DNA methylation status was negatively correlated with the FF E2/A ratio (γ=- 0.389; p=0.002; fig. 4D).
Fig. 4: Scatterplots showing the association between respective DNA methylation levels and mRNA levels of A) CYP19A1, B) AMH, C) AMHR2 genes and the association between D) E2/A level in FF and relative CYP19A1 promoter DNA methylation level in the PCOS and control groups. Pearson correlation analysis was performed to determine the correlation between examined parameters in the PCOS and control groups.
Hyperandrogenism and chronic anovulation are two important pathological changes of PCOS[4]. Given the roles of CYP19A1, AMH, and AMHR2 in follicle development and androgen metabolism, we first clarified their DNA methylation status in PCOS patients who underwent IVF treatment using MSP. We observed hypermethylation of CYP19A1 and hypomethylation of AMH and AMHR2 genes in PCOS ovarian granulosa cells. These results indicate the importance of DNA methylation of genes related to androgen anabolism and follicular development in PCOS development. Given that DNA methylation silences gene transcription, we noted downregulation of CYP19A1 mRNA and protein levels, consistent with the observed CYP19A1 DNA hypermethylation. Pearson correlation analysis confirms the negative correlation between DNA hypermethylation and expression of CYP19A1, confirming our previous results in ovarian tissue[11]. Similarly, hypomethylation of AMH and AMHR2 genes translated to upregulation of their mRNA and protein expression in PCOS. Pearson correlation analyses confirmed the negative correlation between gene expression and DNA methylation status for AMH and AMHR2. Thus, our results suggest that aberrant gene methylation underlies the abnormal expression of CYP19A1, AMH and AMHR2 in PCOS granulosa cells. Increased AMH and AMHR2 expression was consistent with the observed increase in FF AMH level in PCOS patients, suggesting that there is increased AMH secretion from granulosa cells of PCOS patients. The elevated AMH level observed in our study is in accordance with the results from other studies[3,16]. Given the physiological roles of AMH binding to AMHR2, upregulation of AMH and AMHR2 expression in PCOS granulosa cells support their synergistic cooperation in promoting abnormal follicular growth and development and consequent PCOS follicular developmental disorders.
The higher LH/FSH level in PCOS patients observed in this study promotes anovulation in PCOS and supports previous findings[17]. We observed no significant change in E2 level in the PCOS group, likely due to the conversion to estrogen in the presence of elevated androgen levels. The E2/T and E2/A ratios, which are established indicators of P450 aromatase activity, were lower in the PCOS group due to elevated T and A levels, indicating low aromatase activity. The negative correlation between CYP19A1 DNA methylation level and follicular E2/A ratio suggests that CYP19A1 DNA methylation inhibits aromatase activity and increases A synthesis in the ovary, supporting the role of DNA methylation in regulating androgen metabolism.
The inverse correlation between AMH and CYP19A1 gene expression in this study indicates the inhibitory role of AMH in aromatase activity in granulosa cells. Indeed, AMH inhibits aromatase activity and contributes to PCOS pathogenesis[1,14,18]. AMH-mediated inhibition of aromatase may occur via FSH inhibition, which could impact antral follicle growth and differentiation[19,20]. Further research is warranted to clarify the exact mechanism of AMH modulation of aromatase activity and consequently antral follicular maturation and oocyte quality. Pearson correlation analysis revealed that FF AMH level is not correlated with relative AMH promoter DNA methylation level in ovarian granulosa cells (data not shown). This appears to contradict our results of the correlation between AMH DNA hypomethylation and elevated AMH mRNA and protein levels. AMH production varies over the menstrual cycle, with relatively high secretion by the granulosa cells of preantral and small antral follicles during the antral stage and tapering off after the dominant follicle has been selected[12]. Because we used mature follicles in this study, AMH level may be relatively lower in this study. Moreover, a myriad of factors including smoking[21] and vitamin deficiency[22] may represent confounding factors that could influence accuracy of the analysis. Thus, more precise patient selection and stratification to account for these factors may be required to obtain more reliable results[23], for which a larger sample size is necessary.
Our results suggest the correlation between DNA methylation and downstream mRNA and protein expression of three genes that modulate androgen metabolism and ovarian follicular growth and development in PCOS pathogenesis. Further studies are warranted to fully confirm and understand the role of DNA methylation in PCOS pathogenesis, and to elucidate the mechanistic relationships among CYP19A1, AMH, and AMHR2 in PCOS patients, for deeper insights and potential application in PCOS diagnosis and treatment.
Funding support:
We thank the National Natural Science Foundation of China (Nos. 81701402) for the funding support.
Conflicts of interest:
The authors declare no conflicts of interest.
References
- Tata B, Mimouni NE, Barbotin AL, Malone SA, Loyens A, Pigny P, et al. Elevated prenatal anti-Müllerian hormone reprograms the fetus and induces polycystic ovary syndrome in adulthood. Nat Med 2018;24(6):834-46.
- Owens LA, Kristensen SG, Lerner A, Christopoulos G, Lavery S, Hanyaloglu AC, et al. Gene expression in granulosa cells from small antral follicles from women with or without polycystic ovaries. J Clin Endocrinol Metab 2019;104(12):6182-92.
- Abbara A, Eng PC, Phylactou M, Clarke SA, Hunjan T, Roberts R, et al. Anti-Müllerian hormone (AMH) in the diagnosis of menstrual disturbance due to polycystic ovarian syndrome. Front Endocrinol 2019;10:656.
- De Leo V, Musacchio MC, Cappelli V, Massaro MG, Morgante G, Petraglia F. Genetic, hormonal and metabolic aspects of PCOS: an update. Reproduc Biol Endocrinol 2016;14(1):1-7.
- Lanzo E, Monge M, Trent M. Diagnosis and management of polycystic ovary syndrome in adolescent girls. Pediatr Annal 2015;44(9):e223-30.
- Chen J, Shen S, Tan Y, Xia D, Xia Y, Cao Y, et al. The correlation of aromatase activity and obesity in women with or without polycystic ovary syndrome. J Ovarian Res 2015;8(1):1-6.
- Khan MJ, Ullah A, Basit S. Genetic basis of polycystic ovary syndrome (PCOS): current perspectives. Appl Clin Genet 2019;12:249.
- Sagvekar P, Kumar P, Mangoli V, Desai S, Mukherjee S. DNA methylome profiling of granulosa cells reveals altered methylation in genes regulating vital ovarian functions in polycystic ovary syndrome. Clin Epigenet 2019;11(1):1-6.
- Yu YY, Sun CX, Liu YK, Li Y, Wang L, Zhang W. Genome-wide screen of ovary-specific DNA methylation in polycystic ovary syndrome. Fertil Steril 2015;104(1):145-53.
- Edson MA, Nagaraja AK, Matzuk MM. The mammalian ovary from genesis to revelation. Endocrine Rev 2009;30(6):624-712.
- Yu YY, Sun CX, Liu YK, Li Y, Wang L, Zhang W. Promoter methylation of CYP19A1 gene in Chinese polycystic ovary syndrome patients. Gynec Obstet Invest 2013;76(4):209-13.
- Xu J, Bishop CV, Lawson MS, Park BS, Xu F. Anti-Müllerian hormone promotes pre-antral follicle growth, but inhibits antral follicle maturation and dominant follicle selection in primates. Hum Reprod 2016;31(7):1522-30.
- Borges E, Braga DP, Setti A, de Cássia Figueira R, Iaconelli Jr A. The predictive value of serum concentrations of anti-Müllerian hormone for oocyte quality, fertilization, and implantation. JBRA Assisted Reprod 2017;21(3):176.
- Liu XY, Yang YJ, Tang CL, Wang K, Chen JJ, Teng XM, et al. Elevation of antimüllerian hormone in women with polycystic ovary syndrome undergoing assisted reproduction: effect of insulin. Fertil Steril 2019;111(1):157-67.
- Wang R, Mol BW. The Rotterdam criteria for polycystic ovary syndrome: evidence-based criteria?. Hum Reprod 2017;32(2):261-4.
- Tal R, Seifer CM, Khanimov M, Seifer DB, Tal O. High serum Antimullerian hormone levels are associated with lower live birth rates in women with polycystic ovarian syndrome undergoing assisted reproductive technology. Reprod Biol Endocrinol 2020;18(1):1-8.
- Dumitrescu R, Mehedintu C, Briceag I, Purcarea VL, Hudita D. The polycystic ovary syndrome: an update on metabolic and hormonal mechanisms. J Med Life 2015;8(2):142.
- Gorsic LK, Kosova G, Werstein B, Sisk R, Legro RS, Hayes MG, et al. Pathogenic anti-Müllerian hormone variants in polycystic ovary syndrome. J Clin Endocrinol Metab 2017;102(8):2862-72.
- Dewailly D, Robin G, Peigne M, Decanter C, Pigny P, Catteau-Jonard S. Interactions between androgens, FSH, anti-Müllerian hormone and estradiol during folliculogenesis in the human normal and polycystic ovary. Hum Reprod 2016;22(6):709-24.
- Homer MV, Rosencrantz MA, Shayya RF, Chang RJ. The effect of estradiol on granulosa cell responses to FSH in women with polycystic ovary syndrome. Reprod Biol Endocrinol 2017;15(1):1-6.
- White AJ, Sandler DP, D'Aloisio AA, Stanczyk F, Whitworth KW, Baird DD, et al. Antimüllerian hormone in relation to tobacco and marijuana use and sources of indoor heating/cooking. Fertil Steril 2016;106(3):723-30.
- Bednarska-Czerwińska A, Olszak-Wąsik K, Olejek A, Czerwiński M, Tukiendorf A. Vitamin D and anti-müllerian hormone levels in infertility treatment: the change-point problem. Nutrients 2019;11(5):1053.
- Thakre N, Homburg R. A review of IVF in PCOS patients at risk of ovarian hyperstimulation syndrome. Expert Rev Endocrinol Metab 2019;14(5):315-9.