- *Corresponding Author:
- Cong Nie
Department of Bioengineering, Qilu University of Technology, Jinan, Shandong 250353, China
E-mail: congnie2009@163.com
| This article was originally published in a special issue, “Transformative Discoveries in Biomedical and Pharmaceutical Research” |
| Indian J Pharm Sci 2023:85(4) Spl Issue “114-127” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Lactic acid bacteria are widely found in nature, have extensive biodiversity and are widely applied to food, feed and medicine. Currently, most probiotics in the market are lactic acid bacteria. In recent years, antibiotic technology has been actively developed, and the development and application of lactic acid bacteria greatly promoted the development of resources. All genome information based on lactic acid bacteria can systematically fully understand the metabolic properties of the strains, potential probiotic functions and application directions and can further study the genetics, evolution, and classification of lactic acid bacteria. The safety evaluation plan for lactic acid bacteria requires determining whether the genes are resistant, virulent, or virulence-related genes and whether or not the related genes can propagate horizontally. Study of 18 strains of Streptococcus from different regions and milk sources, including myriad bacteria, Streptococcus, Enterococcus and Streptococcus pyogenes. 26S ribosomal ribonucleic acid analysis of gene sequences was performed; amino glycosides, tetracyclines, fluoroquinolones, glycosides and macro lactones using the minimum inhibitory concentration method, the authors examined the resistance and sensitivity of 15 kinds of general antibiotics of phenyl propylene glycols and amphotericin to different lactic acid strains. As a result, three kinds of Staphylococcus aureus showed different resistance to five kinds of antibiotics. The results of this experiment provide guidance to the use of human antibiotics and lactic acid bacteria and make them more rational and secure. Antibiotic sequencing techniques are used to compare the gene sequence and the structural differences of antibiotics in different genomes. Based on whole genome information, the reconstruction of the genome scale metabolic network model can simulate and predict the behavior of bacterial cells in a particular environment of antibiotics, and to systematically control the metabolic engineering strategy of the strain.
Keywords
Lactococcus genus, antibiotic, antibacterial concentration, metabolic network model, epidemiology
Antibiotics have important physiological functions in the human body and are necessary substances which constitute synthetic hormone and nerve cells in the human body. In the body, substances such as steroid and bile acid can be converted[1]. Epidemiological and clinical studies showed that serum antibiotic levels were positively correlated with the incidence of cardiovascular disease. Any normal level serum antibiotic increases about 35 % of cardiovascular disease risk daily[2]. Nutritionists suggest taking 250-300 mg of food antibiotics a day. Excess intake causes increased serum antibiotics and causes coronary atherosclerosis[3]. Therefore, the development of drugs and health foods that reduce antibiotics is the focus of the present study. Lactic acid bacteria refers to a group of bacteria producing a large amount of lactic acid by fermenting sugar[4]. This is considered to be a safe microorganism day. In recent years, studies have shown that beneficial lactic acid bacteria planted in the gut have a variety of healthy benefits; maintaining microbiological balance and intestinal function, reducing serum antibiotics, alleviating lactose resistance, improving liver function and enhancing immune function and antitumor effect[5]. In vitro experiments revealed that lactic acid bacteria isolated from traditional fermented millet were potential application of probiotics to the fermentation industry[6]. The phylogenetic tree of lactic acid bacteria is located on the Clostridium bifurcation of gram positive bacteria. Many members of the lactic acid bacteria family are currently responsible for maintaining the balance of the intestinal tract, reproductive tract, oral and skin microbes as an important component of probiotics. Production of extracellular polysaccharides and flavors, the lactic acid bacteria play an important role in fermented dairy products and it is widely used as a natural biological preservative[7]. China's traditional fermentation products have long history and contain abundant lactic acid bacteria resources. Genomics can collaborate, quantify and compare different genomes of all genes in microbes[8]. The whole genome sequencing technique of lactic acid bacteria analyzes the metabolic property and the genetic background of the strain in whole country systematically and clarifies the mechanism of the probiotic function[9]. Rapid development of lactic acid economy accelerated the development and industrial application of excellent lactic acid bacteria. Lactic acid bacteria are aggregates of lactic acid and produce a large amount of lactic acid bacteria based on the fermentation of carbohydrates[10]. They have no inhibitory concentration in the human body. The probiotic family belongs to the most important bacterial community in the digestive tract[11]. From the angle of the digestive tract, there are many differences in the function of probiotics, such as remarkable reaction and the suppression of the growth and the reproduction of the enteric pathogen[12]. Among them, the antimicrobial concentration of lactic acid bacteria in the stomach is relatively low and gastric acid can kill lactic acid bacteria[13].
In this experiment, lactic acid bacteria in traditional fermented foods were separated by transport and culture, and different types of lactic acid bacteria were screened[14]. The specific changes in the inhibitory concentration of six different strains of lactobacilli in the mouse feces before and after Gavir in a mixed bacterial solution were measured using a quantitative real-time fluorescence Polymerase Chain Reaction (PCR) technique. Antibiotics are widely applied in the body of human and animal, and it is a basic active component with heavy cells, and it can prevent hyperlipidemia and it can reduce the incidence of cardiovascular disease in the body[15]. The excess amount of antibiotics in the body has a serious effect. Edible lactic acid bacteria cause arterial fat deposition and hyperlipidemia, which can increase the incidence of cardiovascular disease[16,17]. Therefore, the serum antibiotic content of the food microorganism is lowered and the bacteria in the intestine, and the secretion of the bile salt hydrolyzing enzyme are utilized, and the serum antibiotic can be lowered, and it acts on the prevention and treatment of the hyperlipidemia[18]. Bile Salt Hydrolase (BSH) can decompose complex bile salt water in the body into free bile salts and promote the formation of excretion between free bile salts and antibiotics[19]. Excretion is difficult to be absorbed in the intestine and is discharged to the outside with feces and the inhibitory concentration of antibiotics is the lowest. In the course of growth, lactic acid bacteria produce naturally occurring functional, protein rich amino acids, i.e., Y-amino butyric acid, which have the ability to lower blood lipids and blood pressure and serve to treat cardiovascular diseases. Lactic acid bacteria have certain antioxidant activity, remove radicals in the body, improve oxidative stress, balance lipid metabolism, and reduce the appearance of hyperlipidemia[20]. Therefore, lactic acid bacteria have important potential as functional health food[21].
Based on the good fortune of three kinds of lactic acid bacteria, this research was selected and preserved several strains from the laboratory of the resource microbiology research center of Seinan Jiaotong University. By studying the beneficial properties of each strain, we acquired cheese Lactobacillus S1, Enterococcus S4 and have Bifidobacterium S6 with better antibiotic reducibility and antioxidant activity, prepared it to triple lactic acid bacteria, studied adaptability to the gastrointestinal environment and Gamma-Aminobutyric Acid (GABA) content and antioxidant activity we must provide some technical support for the development of functional probiotics or health foods.
Materials and Methods
Samples and reagents:
Test strains: Cheese Lactobacillus S1, Enterococcus S4 and lotus lactose S6 can be selected from commercial yogurt and lactic acid bacteria beverages, cooled for 2 y and stably transported five generations. To produce and count lactic acid bacteria, triticale media and deMan Rogosa Sharpe (MRS) medium are produced from artificial gastrointestinal fluid. Antibiotic lowering ability of lactic acid bacteria was detected using a high antibiotic medium (0.1 mg/ml).
Artificial gastric juice: 0.20 % sodium chloride, 0.30 % pepsin and pH 2.5.
Artificial intestinal juice: Trypsin 0.10 %, bile salt 0.30 %, pH value is set to 8.0 % and filtered to disinfect, and prepare for later use. Samples were collected from intestinal contents. After aseptic sampling, the sample was sent to our laboratory and stored in a refrigerator at -20°.
Culture medium:
MRS medium 1 l contains 10.0 g protein, 10.0 g beef extract, 5.0 g yeast extract, 2.0 g ammonium bicarbonate, 5.0 g sodium acetate, 2.0 g potassium hydrogen phosphate, 0.58 g magnesium sulfate, 0.2 g manganese sulfate, 1.0 ml Tween 80, 20 g glucose and distilled water up to 1 l. Adjust the pH to around 6.4 and sterilize at 121° for 20 min. Then mix at a ratio of 1:1:1 to obtain triple lactic acid bacteria. Take the commercially available Bifidobacterium preparation as the control group and adjust the number of viable bacteria in its bacterial suspension to the same level according to the above method. Tolerance of artificial gastrointestinal fluid by inoculating it in MRS medium and incubate it at 37° for 24 h. Mix 1 ml of fermentation broth with 9 ml of artificial gastric fluid and place it in a shaker at 37°. Then transfer 1 ml of artificial gastric juice containing bacteria for 2 h and mixes it with 9 ml of artificial intestinal juice with pH 6.8. Take samples at 0 h, 2 h, 6 h, 10 h and 24 h and measure the number of viable bacteria using the plate counting method. Calculate the survival rate according to equation (1).
N1/N0×100 % (1)
In the formula, Nl is the number of viable bacteria after N h of artificial gastrointestinal fluid treatment and N0 is the number of viable bacteria after 0 h.
Test method:
Inoculate it in a high antibiotic MRS medium and incubate it in a shaker at 37° for 24 h. Use the ammonium iron sulfate method to determine the minimum inhibitory concentration of antibiotics in its supernatant[22]. BSH-specific activity determination was performed using colorimetric method using benzotriazole. It was vaccinated in MRS medium and cultivated at 37° for 24 h. The fermentation broth was centrifuged and precipitated twice with phosphate buffer at pH 7.0. Then the concentration of the regulator was set to 1-2 at OD6oo. 25 μl and 20 mmol lysozyme were added to break the wall and a water bath at 37° was used for 1 h, 0.1 ml raw solution 1+8 ml (pH). Buffer solution+0.1 ml of sodium taurocholate solution 0.2 mmol/l, water bath at 37° for 30 min, add 1.5 ml 15 % trichloroacetic acid to finish the reaction, centrifuge the bacterial solution, take 1.0 ml of the supernatant+2.0 ml of the chromogenic solution benzotriazole and boil in water for 15 min. Determine the minimum inhibitory concentration of antibiotics at OD6oo. Determination of raw enzyme protein; take 0.l ml of raw enzyme supernatant, add water to 1 ml, add 5 ml to Coomassie Brilliant Blue G-250 solution and determine the minimum inhibitory concentration of antibiotics at OD6oo.
Definition of enzyme activity: The number of μmol of taurine produces per ml enzyme dissolution reaction per minute. As defined in formula (2), enzyme-specific activity (U/mg) is the ratio of enzyme activity to protein content (mg) per ml of enzyme solution.
(U/ml)=A×4000/30 (2)
Antibiotic co-precipitation, bacterial absorption and determination of the content of cell membrane incorporation were vaccinated in highly antibiotic MRS medium with and without bile salt, cultivated at 37° for 24 h. After centrifugation of the fermentation broth, the minimum inhibitory concentration of antibiotics in the supernatant was determined. Take 1 ml resuspended bacterial solution, add 20 μ/l, 50 mmol/l lysozyme, 50 μl, 10 % Sodium Dodecyl Sulfate (SDS) and water bath at 37° for 1 h. Then ultrasonic fragmentation is used to determine the minimum inhibitory concentration of antibiotics in the supernatant[23]. The broken bacterial body is washed with phosphate buffer, and the precipitation is resuspended and centrifuged with 1.5 ml anhydrous ethanol for 10 min. The minimum inhibitory concentration of antibiotics in the supernatant is determined. The unbroken spent bacterial solution is centrifuged to determine the minimum concentration of antibiotics in 10 min.
F=(I-A2)-(I-B3) (3)
H=2D+3E-6C (4)
J=(2I-3B)-5H (5)
Expressions: A is the lowest inhibitory concentration of antibiotics in the supernatant of the bile salts and antibiotics added to the medium; B is the minimum inhibitory concentration of antibiotics in the media on the medium containing antibiotics, but the minimum inhibitory concentration of antibiotics in the stents on the bacteria cleaners; D is the lowest inhibitory concentration of antibiotics in the supernatant after cell wall rupture; E is the minimum inhibitory concentration of antibiotics in supernatants after cell wall rupture, salt and co absorption of antibiotics; H is the amount of antibiotics removed by bacterial absorption; 1 is the minimum inhibitory concentration of antibiotics in inoculated medium containing bile salts and j is the cell membrane binding quantity.
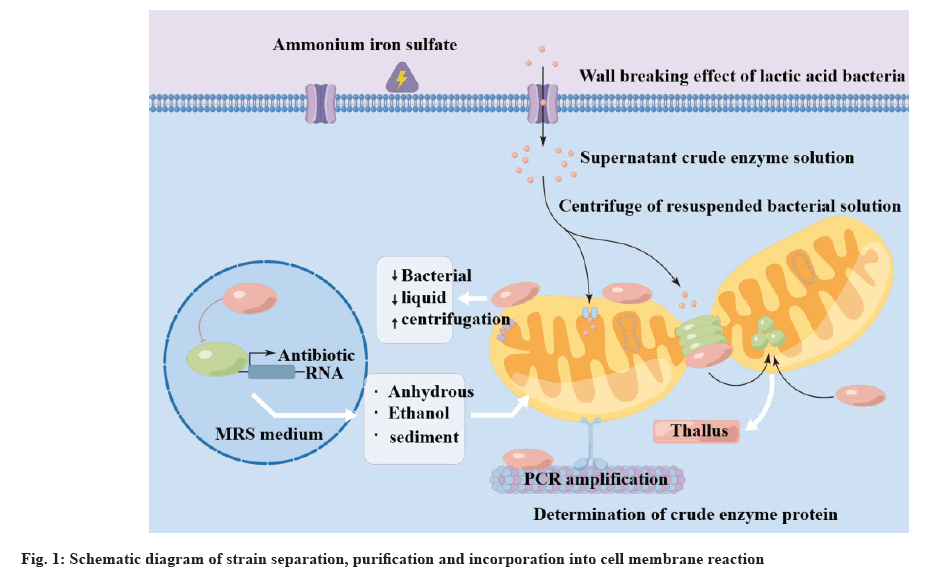
PCR Deoxyribonucleic Acid (DNA) amplification was performed using 16S Ribosomal DNA (16S rDNA) pathway. An enhanced product was placed in the agarose gel to obtain a distinct strip. The reaction of strain separation, purification and incorporation into the cell membrane is shown in fig. 1. The sequencing results of GenBank standard strain sequences differing from the PCR products of the strain were compared and the molecular identification of lactic acid bacteria was completed.
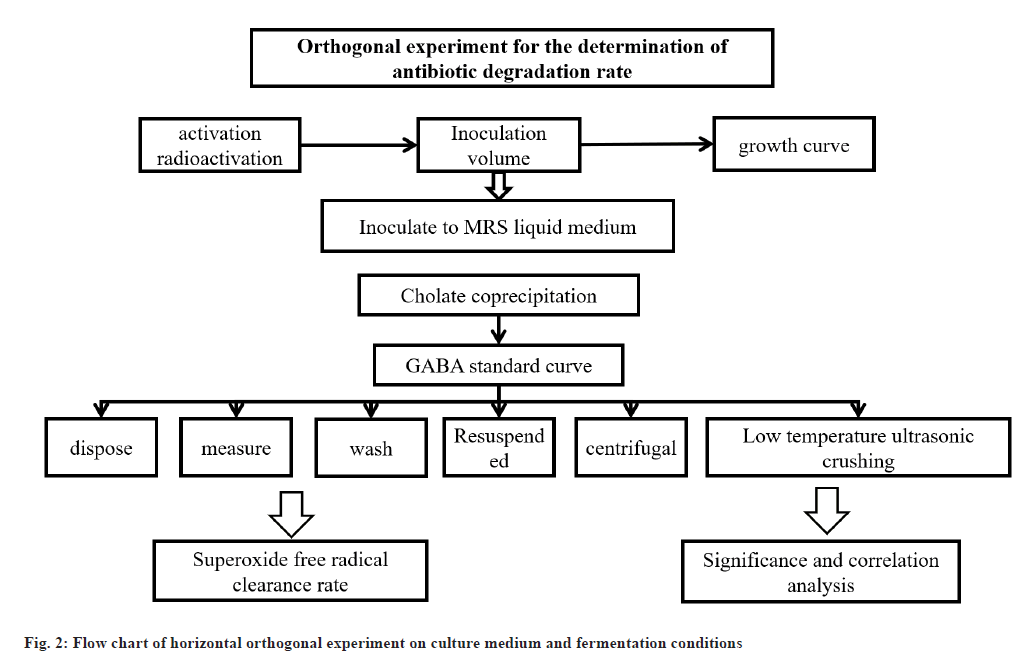
Orthogonal experiment for the determination of growth characteristics and antibiotic degradation rate:
Inoculate into MRS liquid culture medium under sterile conditions, incubate at 37° for 24 h, and then activate twice at a 2 % inoculation rate to obtain a Lactobacillus suspension. Transfer the activated strain HJS2 to 500 ml MRS liquid culture medium with a 2 % inoculation amount and incubate it in a shaker at 37°. Continuously measure for 48 h and draw the growth curve and pH change of strain HJ-S2. Correlation analysis of specific activity of BSH and antibiotic reducing ability take specific activity of BSH (U/mg) as Y, and the content of degrading antibiotics as Xi (μg/ml), where X represents the minimum inhibitory concentration (μg/ml) of antibiotics co-precipitated with bile salt, X2 represents the minimum inhibitory concentration (μg/ml) of antibiotics absorbed by bacteria, X. The minimum inhibitory concentration (μg/ml) of antibiotics that represent cell membrane binding. GABA standard curve; take 0, 0.2 ml, 0.4 ml, 0.6 ml, 0.8 ml and 1.0 ml of the prepared 1 mg/ ml and GABA content determination by taking 1.0 ml of fermentation broth and filter it, place it in a test tube, measure its absorbance according to the above method, and calculate its content through a standard curve. The activated strain culture solution was centrifuged for 10 min, and the cell and fermentation supernatant were collected respectively; the cells were washed with pH 7.0 phosphate buffer solutions and then the cells were resuspended with it, and then crushed with low-temperature ultrasound and then centrifuged for 20 min, and the supernatant was collected as cell-free extract. Reducibility using potassium ferro cyanide reduction method. Superoxide radical clearance rate using pyrogallol autoxidation method. Hydroxyl radical clearance rate by using Fenton reaction method.
Process the experimental data and repeat the measurement three times (n=3), with the result expressed as x±s. Use Statistical Package for the Social Sciences (SPSS) 22.0 statistical software for significance and correlation analysis. p<0.01 indicates that the difference is statistically significant, and the error line represents the effect of conditions on antibiotic degradation rate using standard error. The pancreatic protein gland, soybean protein gland and interstitial protein gland were used to replace the MRS-CHOL medium with equal amounts of protein. The strain HJ-S2 was inoculated with 2 % of the inoculation amount in MRS-CHOL medium containing different nitrogen sources, and incubated in a constant temperature shaking at 37° for 24 h. The effect of different types of nitrogen sources on the antibiotic degradation rate of strain HJ-S2 was measured using the same method. After selecting the determined nitrogen source, further investigate the effect of nitrogen source addition amount on degradation rate.
Sucrose, maltose, soluble starch, lactose and inulin were used to replace glucose in MRS-CHOL medium respectively. The measurement method was the same as above and the effect of different carbon sources and additions on the degradation rate was investigated.
Inoculate strain HJ-S2 with a 2 % inoculum in MRSCHOL medium containing antibiotics with minimum inhibitory concentrations of 0.5, 1.0, 1.5, 2.0, 2.5, 3.0 and 3.5 mg/ml and shake at 37° for 24 h determine the effect of different minimum inhibitory concentrations of antibiotics on the antibiotic degradation rate of strain HJ-S2.
Inoculate strain HJ-S2 with a 2 % inoculum in MRSCHOL medium containing 1.0, 2.0, 3.0, 4.0 and 5.0 mg/ml of bovine bile salt and shake at 37° for 24 h to determine the effect of different bovine bile salt contents on the degradation rate of HJ-S2 antibiotics. The activated strain HJS2 was inoculated into the MRS-CHOL medium with a 2 % inoculum based on the content of various components. The strain was incubated in a constant temperature shaker at 37° for 54 h and the culture medium was taken every 6 h to determine the effect of different cultivation times on the antibiotic degradation rate of strain HJ-S2.
After activation, strain HJ-S2 was inoculated into MRS-CHOL medium with a 2 % inoculation amount. The strain was incubated in a shaker at 37°, 38°, 40°, 41° and 42° for 24 h. The effect of different cultivation temperatures on the antibiotic degradation rate of strain HJ-S2 was measured. Strain HJ-S2 was inoculated in MRS-CHOL medium with initial pH values of 2, 3, 4, 5, 6, 7, 8, 9 and 10, and incubated in a constant temperature shaker at 37° for 24 h to determine the effect of different initial pH on the antibiotic degradation rate of strain HJ-S2. Strain HJ-S2 was inoculated in MRS-CHOL medium at doses of 1 %, 2 %, 3 %, 4 %, 5 % and 6 %, and incubated in a constant temperature shaker at 37° for 24 h to determine the effect of different inoculation doses on the antibiotic degradation rate of strain HJ-S2. The orthogonal experimental process of culture medium and fermentation conditions is shown in fig. 2. Orthogonal experiment of fermentation conditions; in order to determine the optimal liquid culture medium formula, based on the experimental results of the influence of single factor conditions on the antibiotic degradation rate of strain HJ-S2 in the early stage and the basic principles of nutrient elements required for bacterial growth, using the antibiotic degradation rate as an indicator, four factors, including glucose, protein, antibiotics and bile salt content, were designed in a 3-level orthogonal experiment, as well as three factors, pH, cultivation time and inoculation amount.
Results and Discussion
A total of 30 individual colonies were isolated and purified. Physiological and morphological observations revealed that 30 strains were gram positive bacteria, the colony morphology was clear, white or yellow and the edge rule. Under the oil mirror, the bacterial form is spherical or rod shaped, and the hydrogen peroxide enzyme detection is negative, and it is consistent with Lactobacillus. The obtained lactic acid bacteria detect the ability to reduce the minimum inhibitory concentration of antibiotics. Among them, the decomposition rate of 6 strains was less than 10 %, the decomposition rate of 8 stocks was 10 %-20 %, 8 stocks were 20 %-30 %, 5 stocks were 30 %-40 %, and the decomposition rate of 3 stocks exceeded 40 % (HJ-W33, HJ-W64, HJ-S2), and the decomposition rate was 40.69 % and 40.78 %, respectively. It was 48.82 %, and the study value of HJ-GC was the highest. The distribution of experimental data on lactic acid bacteria colony antagonism is shown in fig. 3; there is no bacterial inhibitory region between two kinds of lactic acid bacteria on the same flat plate. Separation allows different types of lactic acid bacteria to be obtained from various fermented foods. The 16S rDNA sequence analysis and blast comparison were carried out for isolated strains and each genus was determined. The results are shown isolated rhamnose lactate and Bulgaria Lactobacillus from commercial yogurt and separated cheese Lactobacillus and Swiss Lactobacillus from Chen vinegar and vinegar cheese.
The samples were sampled before and after injection in the experimental mouse. Using soil fast DNA rotation kits, you can extract corresponding genomic DNA from feces samples. Using the above genomic DNA as a template, quantitative PCR was amplified with 6 pairs of Lactobacillus specific primers. The standard curves of different lactobacilli were converted into copy numbers to obtain 6 kinds of Lactobacillus species per gram of fecal samples. We compared the actual contents of Lactobacilli in the mouse feces before and after treatment with Lactobacilli. The specific planting amount of each strain can be continuously observed and compared. The results based on the content of 6 kinds of lactobacillus were performed on the 4th d of the completion period of the adjustment period and the gastric irrigation stomach. The reaction system parameters used for detecting trace antibiotic concentrations are shown in fig. 4. After cultivation, a small amount of a variety is selected to perform the gram coloring, thereby identifying the purity by Cetyltrimethyl Ammonium Bromide (CTAB) liquid nitrogen freezing. All DNA in the stem genome was extracted using the method. Put 1 ml of active stalk into the Eppendorf (EP) tube, add 500 μl pH 8.0 test buffer solution, and shake it uniformly. The degradation rate changes of different sample groups are shown in Table 1. After shaking, put in liquid nitrogen and dew defrosted. A 200 g/ μl protease K10 μl and 10 % SDS 80 μl were added to the EP tube and cultured in a pot at 37° for 3 h. After water bath at 65° for 30 min, the same volume of phenol chloroform isoamyl alcohol was added and extracted. Mix well and leave for 2 min. The slurry was centrifuged and repeatedly extracted, followed by a suspension of 50 μl and 3 mol/l acetate, stirred uniformly for 30 min on the ice, centrifuged, washed with 70 % ethanol, centrifuged and dried naturally. DNA concentration can be used as a template for subsequent PCR amplification reactions. Agarose gel electrophoresis was performed. Without obvious specific binding, the DNA is pure. The concentration of genomic DNA was diluted to about 100 ng/ μl using the buffer solution. In this experiment, the 50 μl reaction system was used to amplify the Streptococcus pneumoniae gene.
| Sample number | Degradation rate (%) | Sample number | Degradation rate (%) | Sample number | Degradation rate (%) |
|---|---|---|---|---|---|
| HJ-W01 | 14.917 | HJ-W08 | 9.042 | HJ-W15 | 2.426 |
| HJ-W02 | 13.913 | HJ-W09 | 13.095 | HJ-W16 | 11.724 |
| HJ-W03 | 4.845 | HJ-W10 | 12.903 | HJ-W17 | 6.895 |
| HJ-W04 | 9.348 | HJ-W11 | 6.348 | HJ-W18 | 11.141 |
| HJ-W05 | 3.857 | HJ-W12 | 5.712 | HJ-W19 | 5.968 |
| HJ-W06 | 5.136 | HJ-W13 | 6.476 | HJ-W20 | 7.859 |
| HJ-W07 | 7.596 | HJ-W14 | 3.518 | HJ-W21 | 13.074 |
Table 1: Change in Degradation Rate Of Different Sample Groups
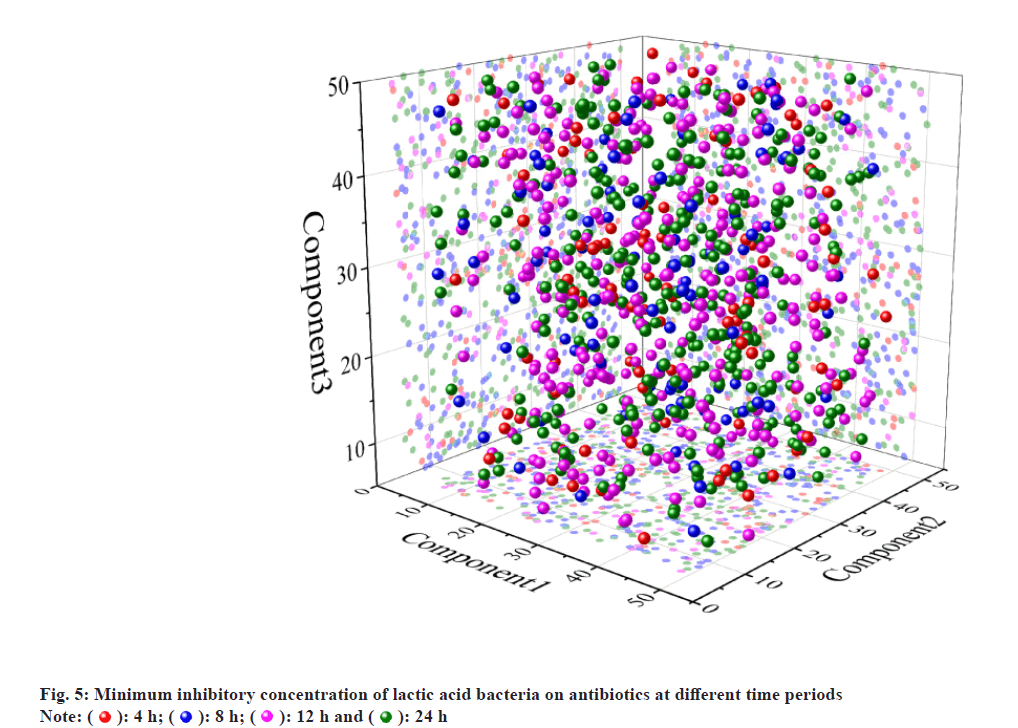
The minimum inhibitory concentration of lactic acid bacteria against antibiotics at different time periods is shown in fig. 5. By developing a fluorescent quantitative PCR with specific PCR primers, it is possible to quantitatively determine different lactobacilli in the fecal samples. Through a series of tests, such primers have good specificity and can resist interference by homologous lactobacilli. The standard curves of the six strains of Lactobacillus are relatively high, and the conditions for PCR are very appropriate, and the primer specificity is relatively good and this method can be applied to the related quantitative examination of fecal samples. As an experimental animal model, the ability of 6 strains of Lactobacillus was determined in the intestine. As a result, the difference in the planting ability of different strains is comparatively clear and it is necessary to study deeply whether there is a difference between the species of which the size of planting ability is different. In the early stage of the growth period, the strain was slow in the incubation period, the growth was slow, the increase in the quantity was small, and OD6oo increased gradually from 0.25 to 0.38, and the pH value decreased from 6.10 to 5.63 in the initial medium. 3 h to 24 h was the growth logarithmic phase of the strain. In this stage, the metabolism and the propagation of bacteria rapidly increased, the growth was rapid, the number of bacteria increased in the formula and OD6oo rapidly increased from 0.38 to 1.65 and the pH value rapidly decreased from 5.63 to 3.72. 24-34 h was the stable stage of the growth of the strain, and the number of viable bacteria in the meantime reached the maximum value and the biomass metabolism and reproduction balance, the metabolite accumulated continuously, and Odeo was maintained at around 1.65 and the acid metabolite was stabilized, and the system pH value was stabilized about 3.70. 34-48 h was in the stage of the collapse of the strain. In this stage, the number of active bacteria was slightly decreased, and some strains died, and the strain activity was lowered, and the OD6oo was lowered from 1.65 to 1.50, and it continued to slow gradually, and the apoptosis of the strain decreased some acid metabolite and the pH value recovered a little. It recovered from 3.70° to 3.90° degrees. Therefore, the growth rate of the strain HJ-S2 is relatively high and the acid production ability is strong and it is suitable for the production of the functional pharmaceutical by the fermentation method. By this, the inoculation quantity in the orthogonal test was determined to be 1 %, 2 % and 3 %. Based on the experimental results of the single factor conditions in the early stage, orthogonal test of the medium and fermentation conditions is designed according the test results are shown the difference in the degree of the effect of the medium condition is C>D>B>A, i.e. it is the content of the minimum antibiotic concentration of the substrate antibiotic which affects the decomposition rate of the antibiotic minimum static bacteria and the remainder is the bile salt content, the glucose content and the protein stand content. As a result of the orthogonal optimization, the decomposition effect of the strain on antibiotic minimum inhibitory concentration is the strongest and the optimum medium condition is prescribed under the A, B, C and D conditions. The antibiotic minimum static bacteria concentration was 56.95 %, and the optimal medium conditions for lowering the antibiotic minimum sterile concentration were 56.95 l, 25 g/l of glucose, 1.0 g/l of antibiotics and 2.0 g/l of bile salt.
The effect of each factor in the fermentation conditions on the degradation rate of antibiotic minimum bacteria is A>C>B, i.e. the most influential factor is the positive H value of the medium, followed by inoculation and incubation time. As a result of orthogonal optimization, the degradation of the antibiotic minimum static bacteria concentration is the strongest under the A, B and C conditions and the optimum fermentation condition are prescribed. The correlation coefficient between the minimum inhibitory concentrations of lactic acid bacteria in each experimental group of antibiotics is shown in fig. 6. The repeat test was carried out according to the culture conditions a, B, C, and the antibiotic minimum bacteria concentration was 69.81 %.
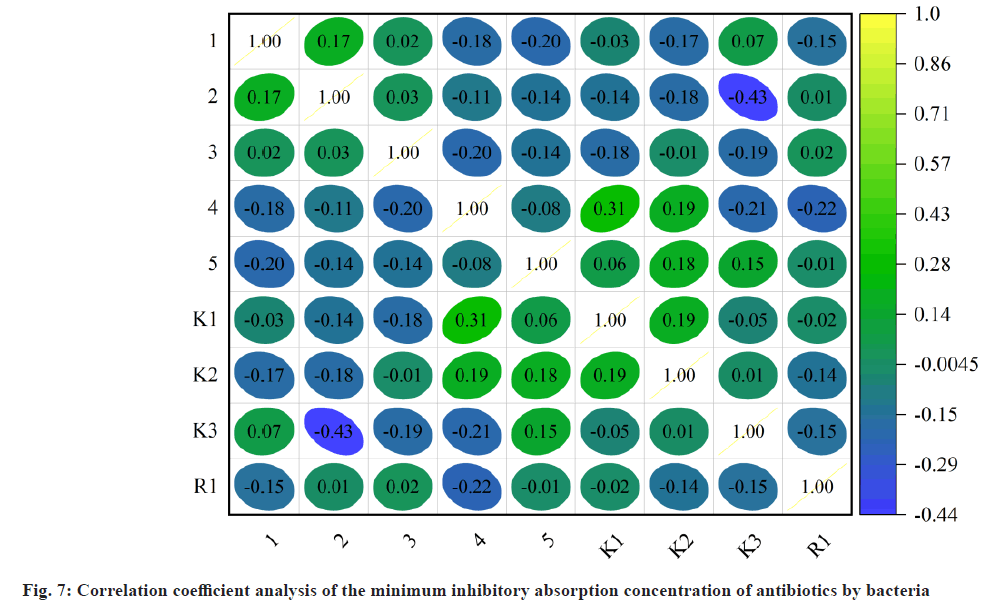
Fig. 7 shows the effect of different nitrogen sources on the rate of decomposing bacteria. The difference in the percentage of bacterial isolates is also considered that protein prices are relatively low and the protein is selected as a fermentative nitrogen source. In addition, the effect of different amounts of protein glands on the minimum inhibitory concentration degradation rate of strain HJ-S2 antibiotics is considered, the content of the nitrogen source has a certain effect on the minimum inhibitory concentration decomposition rate of the antibiotic. The protein content of the protein was 10 g/l, the decomposition rate was highest, 39.28 %, and the addition of protein glands continued, and the decomposition rate decreased, and it was maintained at around 25.80 %, and the orthogonal test level was decided to be 5, 10 and 15 g/l of the protein test level.
The effect of different carbon sources on the degradation of bacterial strains of strain HJ-S2 antibiotics; the difference in the percentage of bacterial isolates in the CHL medium containing different carbon sources was significant (p<0.05). The degradation rate of the antibiotic minimum bacteria was highest in the medium containing glucose as the carbon source, and the decomposition rate was lower in the case of lactose, maltose and Chrysanthemum and the decomposition rate was low when the soluble starch was carbon source. Glucose was selected as the fermentation carbon source because glucose was used as a single sugar, and the microorganism can be directly absorbed. The minimum concentrations of different lactobacilli under the action of antibiotics are shown in Table 2.
| Lactobacillus type | Adaptation period | 4 d | 6 d | 8 d | 10 d | 12 d | 14 d |
|---|---|---|---|---|---|---|---|
| Fermented Lactobacillus Z1 | 10.625 | 11.365 | 7.62 | 8.706 | 10.003 | 10.967 | 9.318 |
| Lactobacillus helveticus C36 | 9.038 | 8.369 | 9.283 | 9.284 | 7.705 | 10.794 | 8.871 |
| Lactobacillus bulgaricus Y3 | 7.167 | 9.728 | 10.635 | 11.113 | 8.514 | 9.194 | 9.488 |
| Lactobacillus plantarum H7 | 9.744 | 8.605 | 10.737 | 8.656 | 11.219 | 9.743 | 9.36 |
| Lactobacillus rhamnosus Y4 | 8.108 | 9.817 | 9.008 | 7.678 | 9.293 | 9.078 | 7.326 |
| Lactobacillus casei C6 | 9.351 | 11.516 | 11.867 | 10.727 | 11.356 | 11.367 | 7.035 |
Table 2: Minimum Concentrations Of Different Lactobacilli Under Antibiotic Action
In addition, the effect of different amounts of glucose on the decomposition rate of bacterial strains of HJ-S2 antibiotics is discussed. When the amount of glucose added was 20 g/l, the decomposition rate of the strain HJ-S2 was highest at 42.13 % and the rate of glucose addition continued to decrease by 29.30 %. The content of the carbon source is too high, the growth of the organism is too fast, and this occurs in the early aging and it affects the absorption of the minimal inhibitory concentration of the fungus. Thus, the glucose content in the orthogonal test was determined to be 15, 20 and 25 g/l.
Fig. 8 shows the effect of different bile salt content on the decomposition rate of the minimum static bacteria concentration of strain HJ-S2 antibiotics. The increase in the concentration of the entophytic salt in the strain of strain HJ-S2 decreased after the increase and the bile salt content was 1.0 g/l, the decomposition rate of the antibiotic minimum sterile concentration was low at 18.62 %, and when the bile salt content was 2.0 g/l, the maximum decomposition rate was 43.99 %, when the bile salt content reached 3.0 g/l, and the decomposition rate of the antibiotic minimum static bacteria reached about 38.62 % and the decomposition rate of the antibiotic minimum bacteria concentration decreased as the bile salt content increased, and it was maintained at around 37.51 %. Thus, the bile salt content in the orthogonal test was determined to be 1.5, 2.0 and 2.5 g/l.
The number of bacteria in this stage was exponentially increased, and the number of bacteria in this stage increased exponentially and the bacterial growth rate of bacteria rapidly increased, and the content of the antibiotic minimum bacteria suppression concentration rapidly decreased, and when it cultivated to 24 h, the decomposition rate of the antibiotic minimum bacteria suppressing concentration reached the maximum of 42.18 % and 24~34 h was the stable stage of the growth of lactic bacteria. The lowering rate of antibiotic minimum bacteria inhibition concentration was no longer increased, and it decreased slightly and occupied 42.18 %. 34-54 h was the decline stage of the strain, and some bacteria died and the lowering rate of the antibiotic minimum static bacteria concentration gradually stabilized around 32.15 %.
The effect of different culture temperatures on the decomposition rate of the minimum bactericidal bacteria of strain HJ-S2 antibiotics. The different culture temperatures had a certain effect on the minimum inhibitory concentration drop capacity of the strains, and the minimum inhibitory concentration drop rate of the antibiotic was minimum 25.82 % when the incubation temperature was 34°, and the decomposition rate of the bacterial strain HJ-S2 reached the highest bacteria concentration of 41.39 %, and the difference in the decomposition rate was not large if it was when the temperature reaches 40° or more, the activity of the bacteria is reduced, and the absorption of the bacteria is absorbed to the minimum static bacteria concentration, and the decomposition rate of the antibiotic concentration is lowered a little. Therefore, the following experiment is carried out at 37°, and the optimum temperature of the organism is 37°, and if the temperature is too low or too high, it affects the biological activity of the organism. The effect of different culture temperatures on the minimum inhibitory concentration degradation rate of strain HJ-S2 antibiotics is as shown in the case of the incubation temperature of 34°, the degradation rate of the antibiotic minimum inhibitory concentration was about 25.82 %, and the decomposition rate of the bacterial strain HJ-S2 reached the highest bacteria concentration of when the temperature reaches 40° or more, the bacterial activity is reduced, and the absorption of the bacteria to the antibiotic minimum bacteria concentration is absorbed, and the decomposition rate of the antibiotic minimum bacteria concentration is lowered a little. Therefore, the following experiment is carried out at 37°, and the optimum temperature of the organism is 37°, and if the temperature is too low or too high, it affects the biological activity of the organism.
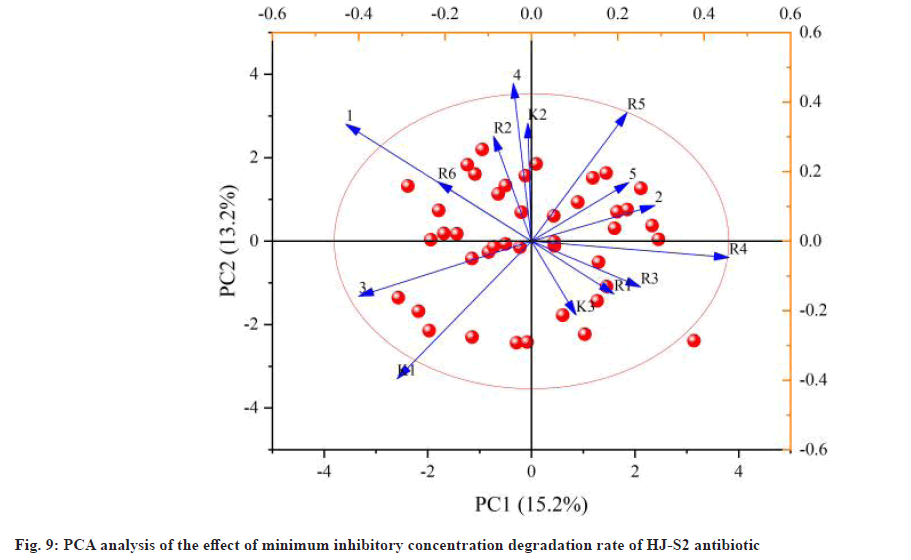
Fig. 9 shows the PCA effect of different pH values on the minimum inhibitory concentration decomposition rate of antibiotic HJ-S2 strain. The initial pH value of the medium markedly affects the rate of antibiotic degradation of the bacterial strains of strain HJ-S2. If the pH is less than 5, the degradation rate of the minimal inhibitory concentration of antibiotics is low because the strain has low activity under low pH conditions, which affects the effectiveness of the strain lowering the minimum inhibitory concentration of antibiotics. However, the strain of HJ-S2 was lower than the initial pH 2, and the degradation rate of the antibiotic was reduced to 5.41 %, indicating that the strain was resistant to acidic environment. The decomposition rate increased with increasing pH value and reached the highest point when the pH value was 6. In this environment, the activity of lactic acid bacteria was high, and the decomposition rate reached the maximum value of 42.24 %. When the pH value exceeds 6, the decomposition rate of the minimum inhibitory concentration of antibiotics decreases gradually, and the p field value in the orthogonal experiment becomes 5, 6 and 7.
The effect of different doses on the antibiotic degradation rate of HJ-S2 strain in the antibiotic degradation rate indicated that the inoculation effect had a significant effect on the antibiotic degradation rate of the lowest bacteria control concentration. When the inoculation amount was 1 %, the decomposition rate of the lowest inhibitory concentration of the antibiotic was relatively low (18.21 %), and as the inoculation quantity increased, the decomposition rate of the minimum inhibitory concentration gradually increased, and if the inoculation quantity was 2 %, the decomposition rate of the antibiotic minimum inhibitory concentration was the maximum value (35.21 %), and the inoculation quantity was over The degradation rate of the minimum inhibitory concentration of antibiotics decreased slightly and was around 32.10 %.
The interference of different factors in complex gut environments poses significant challenges for the tracing and quantification of lactic acid bacteria. The resistance of bacteria to antibiotics has been known long, and our knowledge of the relevant mechanisms involved has also significantly increased in recent years; the progress in genomics, systems biology and structural biology has also accurately described the relevant foundations of drug resistance and will continue to be further analyzed in depth; the resistance mechanism between lactic acid bacteria and antibiotics will also be further deepened and understood by people with the help of cutting-edge scientific knowledge. The results of this experiment can provide a certain reference for people when using antibiotics and lactic acid bacteria, making people more reasonable and safe in the joint application process of probiotics and antibiotics.
During the process of drug resistance transfer, exposed DNA that may carry drug resistance genes will be integrated by bacteria; and during the process of conjugation transfer, the drug resistant genes will be transferred from the donor to the receptor through plasmids or conjugation transposons after forming a conjugation bridge between the two cells, thereby further spreading. Although bacteria have inherent resistance to certain antibiotics, resistance may also be achieved through chromosomal gene mutations and horizontal gene transfer. Lactic acid bacteria colonize specific parts of the intestine and play an indispensable role in maintaining the balance of the gut microbiota. The use of antibiotics has to some extent alleviated the spread of diseases, but at the same time, it has disrupted the unique living environment of beneficial bacteria in the gut microbiota, causing strains to develop varying degrees of resistance. The 26 strains of lactic acid bacteria used in this article come from different milk sources in different regions and have different drug resistance. The reasons for the different drug resistance of 26 strains of lactic acid bacteria may be related to the geographical environment in which they are raised, the feeding mode during feeding, the dietary habits of animals that isolate lactic acid bacteria, the dosage of antibiotics taken when animals are sick, and the varying degrees of residue and enrichment after antibiotic treatment.
Conflict of interests:
The authors declared no conflict of interests.
References
- Adnan M, Patel M, Hadi S. Functional and health promoting inherent attributes of Enterococcus hirae F2 as a novel probiotic isolated from the digestive tract of the freshwater fish Catla catla. PeerJ 2017;5:e3085.
[Crossref] [Google Scholar] [PubMed]
- Aminnezhad S, Kermanshahi RK, Ranjbar R. Evaluation of synergistic interactions between cell-free supernatant of Lactobacillus strains and amikacin and genetamicin against Pseudomonas aeruginosa. Jundishapur J Microbiol 2015;8(4):e165092.
[Crossref] [Google Scholar] [PubMed]
- Andriani AD, Lokapirnasari WP, Karimah B, Hidanah S, Al-Arif MA. Potency of probiotic on broiler growth performance and economics analysis. Indian J Animal Sci 2020;90(8):1140-5.
- Arroyo-Moreno S, Cummings M, Corcoran DB, Coffey A, McCarthy RR. Identification and characterization of novel endolysins targeting Gardnerella vaginalis biofilms to treat bacterial vaginosis. NPJ Biofilms Microbiomes 2022;8(1):29.
[Crossref] [Google Scholar] [PubMed]
- Bang WY, Ban OH, Lee BS, Oh S, Park C, Park MK, et al. Genomic, phenotypic and toxicity based safety assessment and probiotic potency of Bacillus coagulans IDCC 1201 isolated from green malt. J Ind Microbiol Biotechnol 2021;48(5-6):26.
[Crossref] [Google Scholar] [PubMed]
- Brandi J, di Carlo C, Manfredi M, Federici F, Bazaj A, Rizzi E, et al. Investigating the proteomic profile of HT-29 colon cancer cells after Lactobacillus kefiri SGL 13 exposure using the SWATH method. J Am Soc Mass Spectrom 2019;30(9):1690-9.
[Crossref] [Google Scholar] [PubMed]
- Cartagena E, Alva M, Montanaro S, Bardón A. Natural sesquiterpene lactones enhance oxacillin and gentamicin effectiveness against pathogenic bacteria without antibacterial effects on beneficial lactobacilli. Phytother Res 2015;29(5):695-700.
[Crossref] [Google Scholar] [PubMed]
- Chang YC, Tsai CY, Lin CF, Wang YC, Wang IK, Chung TC. Characterization of tetracycline resistance lactobacilli isolated from swine intestines at western area of Taiwan. Anaerobe 2011;17(5):239-45.
[Crossref] [Google Scholar] [PubMed]
- Cherian PT, Wu X, Maddox MM, Singh AP, Lee RE, Hurdle JG. Chemical modulation of the biological activity of reutericyclin: A membrane-active antibiotic from Lactobacillus reuteri. Sci Rep 2014;4(1):4721.
[Crossref] [Google Scholar] [PubMed]
- Citron DM, Warren YA, Tyrrell KL, Merriam V, Goldstein EJ. Comparative in vitro activity of REP3123 against Clostridium difficile and other anaerobic intestinal bacteria. J Antimicrob Chemother 2009;63(5):972-6.
[Crossref] [Google Scholar] [PubMed]
- Gaiser RA, Ayerra Mangado J, Mechkarska M, Kaman WE, van Baarlen P, Conlon JM, et al. Selection of antimicrobial frog peptides and temporin-1 DR a analogues for treatment of bacterial infections based on their cytotoxicity and differential activity against pathogens. Chem Biol Drug Design 2020;96(4):1103-13.
[Crossref] [Google Scholar] [PubMed]
- Gomez-Sala B, Munoz-Atienza E, Diep DB, Feito J, del Campo R, Nes IF, et al. Biotechnological potential and in vitro safety assessment of Lactobacillus curvatus BCS35, a multibacteriocinogenic strain isolated from dry-salted cod (Gadus morhua). LWT 2019;112:108219.
- Gunaratnam S, Diarra C, Paquette PD, Ship N, Millette M, Lacroix M. The acid-dependent and independent effects of Lactobacillus acidophilus CL1285, Lacticaseibacillus casei LBC80R, and Lacticaseibacillus rhamnosus CLR2 on Clostridioides difficile R20291. Probiotics Antimicrob Proteins 2021;13(4):949-56.
[Crossref] [Google Scholar] [PubMed]
- Jadhav SR, Shandilya UK, Kansal VK. Exploring the ameliorative potential of probiotic dahi containing Lactobacillus acidophilus and Bifidobacterium bifidum on dextran sodium sulphate induced colitis in mice. J Dairy Res 2013;80(1):21-7.
[Crossref] [Google Scholar] [PubMed]
- Jepson RG, Williams G, Craig JC. Cranberries for preventing urinary tract infections. Cochrane Database Syst Rev 2012;10(10):CD001321.
[Crossref] [Google Scholar] [PubMed]
- Lin WC, Ptak CP, Chang CY, Ian MK, Chia MY, Chen TH, et al. Autochthonous lactic acid bacteria isolated from dairy cow feces exhibiting promising probiotic properties and in vitro antibacterial activity against foodborne pathogens in cattle. Front Vet Sci 2020;7:239.
[Crossref] [Google Scholar] [PubMed]
- Kalima K, Lehtoranta L, He L, Pitkäniemi J, Lundell R, Julkunen I, et al. Probiotics and respiratory and gastrointestinal tract infections in Finnish military conscripts–A randomised placebo-controlled double-blinded study. Benef Microbes 2016;7(4):463-71.
[Crossref] [Google Scholar] [PubMed]
- Kromann S, Hvidtfeldt A, Boye M, Sørensen DB, Jorgensen S, Nielsen JP, et al. In vitro synergy of sertraline and tetracycline cannot be reproduced in pigs orally challenged with a tetracycline resistant Escherichia coli. BMC Microbiol 2019;19(1):1-2.
[Crossref] [Google Scholar] [PubMed]
- Lokapirnasari WP, Al Arif MA, Maslachah L, Kirana AL, Suryandari A, Yulianto AB, et al. The potency of Lactobacillus acidophillus and L. lactis probiotics and Guazuma ulmifolia Lam. extract as feed additives with different application times to improve nutrient intake and feed efficiency in Coturnix coturnix japonica females. J Anim Feed Sci 2023;32(1):59-67.
- Meng F, Liu Y, Nie T, Tang C, Lyu F, Bie X, et al. Plantaricin A, derived from Lactiplantibacillus plantarum, reduces the intrinsic resistance of gram-negative bacteria to hydrophobic antibiotics. Appl Environ Microbiol 2022;88(10):e00317-22.
[Crossref] [Google Scholar] [PubMed]
- Rahayuningsih SR, Safitri R, Andayaningsih P. Potency of probiotic bacteria from noni fruit (Morinda citrifolia L.) as anti-Helicobacter pylori agent. AIP Conference Proc 2016;14(1744):020055.
- Yang X, Huang E, Yuan C, Zhang L, Yousef AE. Isolation and structural elucidation of brevibacillin, an antimicrobial lipopeptide from Brevibacillus laterosporus that combats drug-resistant gram-positive bacteria. Appl Environ Microbiol 2016;82(9):2763-72.
[Crossref] [Google Scholar] [PubMed]
- Zhang Q, Pan Y, Wang M, Sun L, Xi Y, Li M, et al. In vitro evaluation of probiotic properties of lactic acid bacteria isolated from the vagina of yak (Bos grunniens). PeerJ 2022;10:e13177.
[Crossref] [Google Scholar] [PubMed]
 ): A1; (
): A1; ( ): A2; (
): A2; ( ): A3; (
): A3; ( ): B1; (
): B1; ( ): B2; (
): B2; ( ): B3; (
): B3; ( ): C1 and (
): C1 and ( ): C2
): C2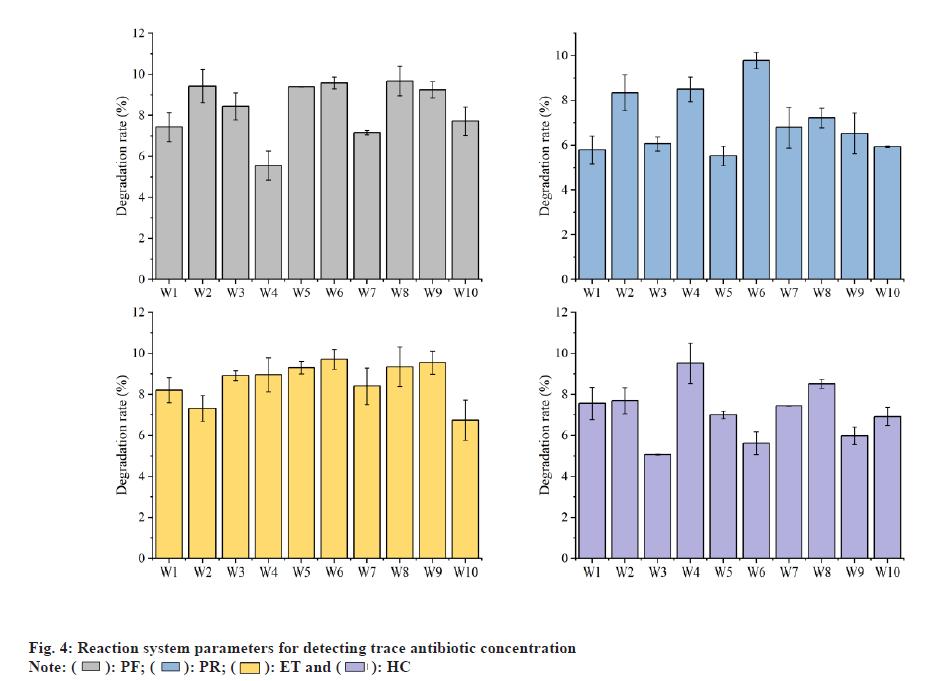
 ): 4 h; (
): 4 h; ( ): 8 h; (
): 8 h; ( ): 12 h and (
): 12 h and ( ): 24 h
): 24 h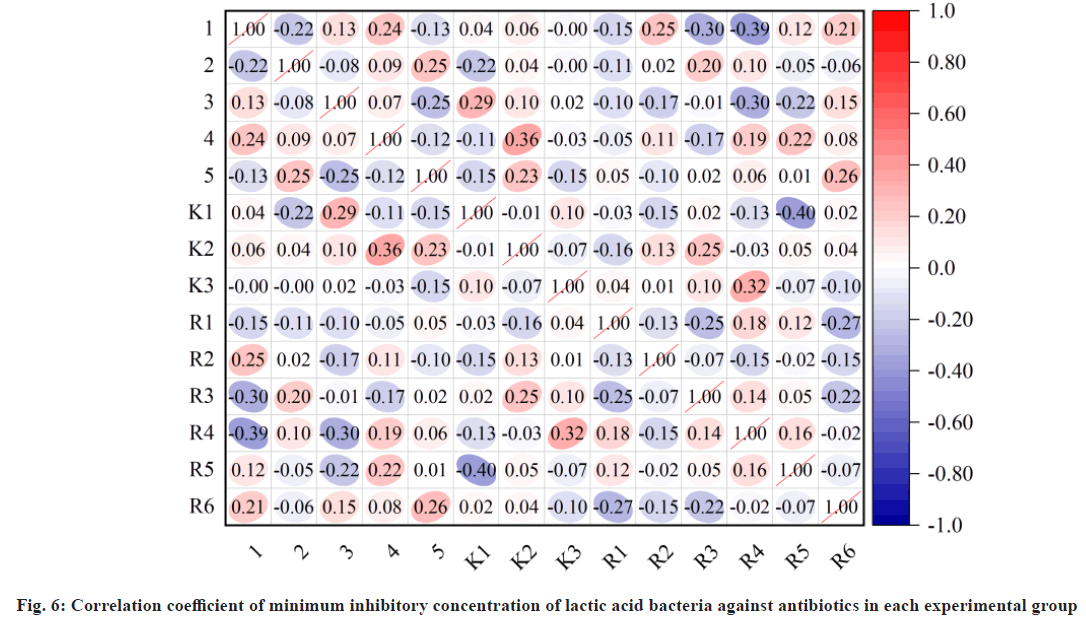
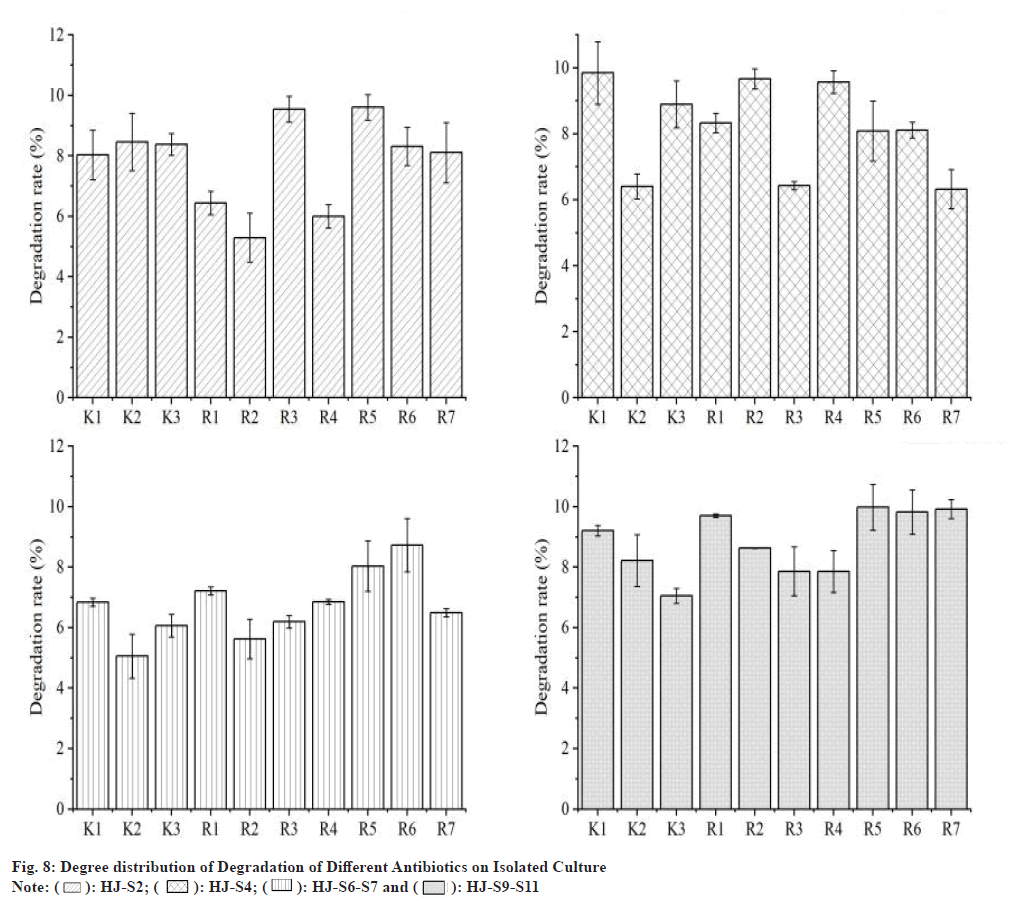
 ): HJ-S2; (
): HJ-S2; ( ): HJ-S4; (
): HJ-S4; ( ): HJ-S6-S7 and (
): HJ-S6-S7 and ( ): HJ-S9-S11
): HJ-S9-S11