- *Corresponding Author:
- Man Guo
Department of Obstetrics and Gynecology, The Second Affiliated Hospital of Harbin Medical University, Harbin, Heilongjiang 150001, China
E-mail: 51864118@qq.com
| This article was originally published in a special issue, “Recent Progression in Pharmacological and Health Sciences” |
| Indian J Pharm Sci 2024:86(2) Spl Issue “101-113” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Preeclampsia is a disease associated with developmental defects in the placenta. However, the pathogenesis of preeclampsia has not been precisely elucidated. Extensive studies have found that noncoding ribonucleic acid is involved in transcription and post-transcriptional translation. In the present study, we expounded on the pathogenic mechanism and diagnostic potential of noncoding ribonucleic acid in preeclampsia. We obtained the differentially expressed long noncoding ribonucleic acid urothelial carcinoma-associated 1 from the gene expression omnibus dataset. Urothelial carcinoma-associated 1 participated as a core gene in the construction of the competing endogenous ribonucleic acid network. The receiver operating characteristic curves were used to construct a diagnostic model for preeclampsia. Besides, the biological processes that urothelial carcinoma-associated 1 may influence in preeclampsia development were explored. Then, we detected urothelial carcinoma-associated 1, microRNA-18a, and hypoxia-inducible factor-1 alpha expressions by real-time quantitative polymer chain reaction and Western blotting. Based on the HTR-8/SVneo cell hypoxia model, we altered the expression of urothelial carcinoma-associated 1 or microRNA-18a by transfection. Cell counting kit-8, transwell, and wound healing assay were applied to treated cells to detect changes in their biological behaviors. Clinical samples confirmed that urothelial carcinoma-associated 1 and hypoxia-inducible factor-1 alpha messenger ribonucleic acid were more abundant in the placenta of patients with preeclampsia, while microRNA-18a was deficient. Urothelial carcinoma-associated 1 regulated the expression of hypoxia-inducible factor-1 alpha via microRNA-18a, promoting trophoblastic cell migration, invasion, and proliferation. Aspirin treatment significantly repaired the invasive function of drug-induced hypoxic trophoblasts, and the expression levels of these three genes were altered. Thus, we concluded that urothelial carcinoma-associated 1/microRNA-18a/hypoxia-inducible factor-1 alpha forms a molecular pathway that affects the biological behaviors of trophoblast cells and participates in the pathogenesis of preeclampsia.
Keywords
Preeclampsia, urothelial carcinoma-associated 1, microRNA-18a, hypoxia-inducible factor-1 alpha, aspirin
Preeclampsia (PE), a clinical syndrome appearing in pregnancy after 20 gestational weeks, is characterized by hypertension and proteinuria and can cause multisystem organ dysfunction in the mother. It can produce short-term adverse pregnancy outcomes such as fetal distress, fetal growth restriction, and stillbirth, and long-term negative effects such as cardiovascular diseases, autism spectrum disorder, and attention deficit hyperactivity disorder[1-3]. PE affects 2 % to 4 % of worldwide pregnancies and is associated with approximately 46 000 maternal deaths and 500 000 fetal and neonatal deaths each year[4,5]. PE brings severe disease and economic burdens to pregnant women around the world. Terminating the pregnancy is currently the only way to stop the disease from progressing. Despite having been probed for many years, the precise etiology and pathogenesis of PE have not yet been elucidated. One internationally recognized theory is that in PE, the capacity of extravillous trophoblast infiltration is impaired, and uterine spiral arterioles remodeling is insufficient[6]. These pathological changes contribute to reduced placental perfusion and placental hypoxia, releasing a variety of placental factors. Ultimately, a range of adverse clinical symptoms that threaten the mother and fetus occur. Overall, it is vital to elucidate the etiology and pathogenesis of PE as soon as possible and to find more sensitive and specific methods for early diagnosis, treatment, and prognosis in clinical practice.
With the development of gene sequencing technology and biological information mining technology, multiple noncoding RNAs (ncRNAs) differentially expressed in placental tissues, peripheral blood, and exosomes have been identified[7-9]. MicroRNAs (miRNAs) and long noncoding RNAs (lncRNAs) are the main research objectives. ncRNAs regulate multiple cellular functions and are therefore widely used as potential biomarkers for various diseases. Urothelial Carcinoma-Associated 1 (UCA1) was the first tumorigenic lncRNA detected in bladder cancer[10]. Numerous works have found that UCA1 is aberrantly expressed in various diseases and plays a vital role in the pathophysiological formation of cancer and pregnancy complications through crosstalk with miRNAs[11]. UCA1 affects cell invasion and proliferation in diverse tumor types, including cervical, colorectal, and pancreatic cancers[12,13]. Furthermore, UCA1 is highly expressed in the placental tissues of pregnancies with PE. Silencing of UCA1 enhanced trophoblast proliferation and metastasis but inhibited apoptosis[14]. Hence, the downstream-specific mechanisms and pathways of UCA1 involved in PE occurrence need to be further elucidated.
Aspirin belongs to the family of non-steroidal antiinflammatory drugs that inhibit the cyclooxygenase pathway and is widely used to prevent a variety of diseases including cardiovascular complications and tumors. Clinical trials and systematic reviews demonstrated that Low-Dose Aspirin (LDA) in early pregnancy may decrease the risk of PE[15,16]. Nevertheless, the molecular mechanisms by which LDA benefits PE prevention remain enigmatic. One of the most recognized mechanisms is associated with the anticoagulant effect of aspirin. However, this mechanism is too simple to explain all the mechanisms involved in the prevention of PE by LDA. A recent study has shown that aspirin protects the migration and invasive functions of hypoxic trophoblasts and inhibits apoptosis through the c-Jun NH2-Terminal Kinase (JNK)/Activator Protein-1 (AP-1)/sFlt1 pathway[17]. Aspirin has also been demonstrated to regulate trophoblast invasive function through the miR-200 family and Transforming Growth Factor Beta 1 (TGFβ1)[18,19]. There is still a large knowledge gap about the mechanistic effects of aspirin and the pathogenesis of PE that needs to be further explored.
The progress of ncRNAs in pathological mechanism, marker prediction, diagnosis, and drug-targeted treatment of tumor diseases is at least 10 y ahead of PE research. Through such findings, it has been identified that ncRNAs have excellent prospects in the noninvasive prediction, diagnosis of diseases, and targeted precision therapy. Building on existing research, here we used databases prediction and in vitro experimental verification to dig deeper into the impact of UCA1/miR-18a/Hypoxia-Inducible Factor-1 Alpha (HIF-1α) on the biological behavior of PE trophoblast cells and to discover new molecular pathological mechanisms of PE. In addition, the pathway was found to be involved in the prevention of PE by aspirin. We ultimately aimed to uncover effective biomarkers for the early prediction of PE that would benefit pregnant women worldwide.
Materials and Methods
Bioinformatics analysis:
The maternal placental tissue dataset GSE75010 was derived from the Gene Expression Omnibus (GEO), (https://www.ncbi.nlm.nih.gov/geo/) database including 80 PE samples and 77 normal samples. Data variance analysis was performed using the limma package in the R software (version 4.2.2). Genes meeting |log2FC>0.6| and p<0.05 (Benjamini & Hochberg reconciliation) were used as significantly differentially expressed genes. miRNAs that lncRNA could target for binding were predicted in the miRcode (http://www.mircode.org/index.php) database. Possible target-binding mRNAs for miRNAs were predicted using miRNA Database (miRDB) (https:// mirdb.org/), TargetScan (https://www.targetscan. org/vert_80/), miRTarBase (https://mirtarbase.cuhk. edu.cn/~miRTarBase/miRTarBase_2022/php/index. php) databases. miRNAs target-binding messenger Ribonucleic Acid (mRNA) supported by two and more databases were selected. Protein-Protein Interaction (PPI) networks were performed through the String (https://cn.string-db.org/) database and node relationships were displayed. Using cytoscape software (version 3.8.2), we constructed a competing endogenous RNAs (ceRNAs) network of lncRNAs, miRNAs, and key mRNAs.
Receiver Operating Characteristic curves (ROC) was calculated using the pROC package, and a ROC PE diagnostic model was acquired by the Glmnet package in R software. The placental dataset GSE114691 was gained from the GEO database as the ROC model validation group. This dataset was composed of 20 PE samples and 21 normal samples. Principal Component Analysis (PCA) was applied to test whether UCA1 and downstream mRNA could discriminate between PE and normal samples in GSE114691. Gene Ontology (GO) (http://www. geneontology.org/) database was used to perform functional enrichment analysis of key mRNAs in three areas; Biological Process (BP), Cellular Component (CC), and Molecular Function (MF). Pathway enrichment analysis was also carried out utilizing Kyoto Encyclopedia of Genes and Genomes (KEGG) (https://www.kegg.jp/) database.
Clinical samples collection:
In this study, placental tissue collection from all patients was approved by the Medical Ethics Committee of the Second Affiliated Hospital of Harbin Medical University. 15 cases of placentas clinically diagnosed PE and 15 cases of placental tissue without other chronic diseases and obstetric complications were collected. PE patients exhibited systolic blood pressure ≥140 mmHg or diastolic blood pressure ≥90 mmHg on two or more occasions, accompanied by proteinuria after gestational 20 w. In exclusion criteria, patients with previous chronic diseases (such as hypertension, kidney disease, heart disease, rheumatism, and immune disease), complications during pregnancy (such as gestational diabetes mellitus and premature rupture of membrane), personal history of poor pregnancy (such as spontaneous miscarriage, fetal malformation, fetal chromosomal abnormalities, stillbirth), and multiple pregnancies have been excluded. Clinical characteristics of all patients are summarized in Table 1. Placenta samples of approximately 2 cm in diameter were obtained from the middle of villous lobules. The specimen was rinsed with pre-cooled phosphate-buffered saline to remove blood and then rapidly frozen in liquid nitrogen. Placental tissue was stored in a -80° refrigerator.
| Variable | Normal, n=151 | PE, n=151 | p value2 |
|---|---|---|---|
| Maternal age (years) | 30.0±4.1 | 29.0±6.5 | 0.7 |
| Before pregnancy BMI (kg/m2) | 21.1±3.1 | 25.0±3.6 | 0.013 |
| Delivery BMI (kg/m2) | 28.3±2.8 | 31.2±5.2 | 0.051 |
| Systolic blood pressure (mmHg) | 113±11 | 157±23 | <0.001 |
| Diastolic blood pressure (mmHg) | 79±7 | 105±13 | <0.001 |
| Proteinuria (+) | 0±0.5 | 3±1.2 | <0.001 |
| Newborn weight (g) | 3,650±428 | 2,350±891 | <0.001 |
| Newborn length (cm) | 50.0±1.8 | 44.0±6.6 | <0.001 |
| Apgar score | 9±0 | 8±2 | <0.001 |
| Gestational week (weeks) | 39.7±7.5 | 36.0±4.2 | <0.001 |
Note: BMI: Body Mass Index; (1) median±SD, n (%) and (2) Wilcoxon rank sum test
Table 1: Statistical analysis of clinical characteristics Of Patients with Pe ond normal pregnancy
Cells culture and treatment:
The trophoblast cell line HTR-8/SVneo was purchased from Zhong Qiao Xin Zhou Biotechnology (Shanghai, China) and verified by Short Tandem Repeat (STR) analysis before usage. Cells were fully cultured in a mixture of 90 % Roswell Park Memorial Institute (RPMI)-1640 (Corning, United States of America (USA)), 10 % fetal bovine serum (Procell, Wuhan, China), and 1 % penicillin (100 U/ ml) and streptomycin (100 mg/ml) (Procell, Wuhan, China). All cells were maintained at 37° and 5 % carbon dioxide. In this study, Cobalt dichloride (CoCl2) (Sigma, USA) was used to induce HTR-8/ SVneo to establish a hypoxic trophoblast cell model of PE. A study has demonstrated the above method's effect was similar to the traditional hypoxia method, which could induce the stable expression of HIF- 1α and is more convenient to study[20]. In aspirin experiments, the hypoxic HTR-8/SVneo cell model was treated with aspirin (0.5 mM; acetylsalicylic acid, MedChemExpress, USA), and exposed for 48 h before further analysis[19].
Cells transfection:
miR-18a mimic, miR-18a inhibitor, si-UCA1, and related Negative Control (NC) were purchased from Genepharma (Suzhou, China). RNA Oligo was transfected into HTR-8/SVneo using GP-transfect- Mate (Genepharma, Suzhou, China) transfection reagent. We replaced the old medium with a freshly complete medium after 5 h of transfection. The RNA expression was detected after (1-3) d of culturing cells, and the protein expression was examined after (2-4) d of culturing cells.
Real Time-quantitative Polymerase Chain Reaction (RT-qPCR):
The total RNA of placental tissue and trophoblast cells was extracted with Trizol Reagent (HaiGene, Haerbin, China). The ABScript One-Step SYBR Green RT-qPCR kit (ABclonal, Wuhan, China) was used to detect the expression of UCA1 and HIF- 1α mRNA according to its instruction. The internal reference gene was β-actin. Besides, the total RNA was reverse transcribed by transcriptor first strand cDNA synthesis kit (Roche, Switzerland), and FastStart Universal SYRB Green Master (ROX) (Roche, Switzerland) was used to detect miR-18a expression. The internal reference gene was RNU6. The gene primer sequences are shown in Table 2.
| Gene | Primer sequence (5'-3') |
|---|---|
| HIF-1α | Forward CGTTCCTTCGATCAGTTGTC |
| Reverse TCAGTGGTGGCAGTGGTAGT | |
| UCA1 | Forward CTCTCCATTGGGTTCACCATTC |
| Reverse GCGGCAGGTCTTAAGAGATGAG | |
| β-actin | Forward TGGAATCCTGTGGCATCCATGAAAC |
| Reverse TAAAACGCAGCTCAGTAACAGTCCG | |
| miR-18a | Forward GCCGCTAAGGTGCATCTAGT |
| Reverse CAGAGCAGGGTCCGAGGTA | |
| RNU6 | Forward CTCGCTTCGGCAGCACA |
| Reverse AACGCTTCACGAATTTGCGT |
Table 2: Primer sequences of target genes and internal reference genes
Cell protein extraction and Western Blotting (WB):
Radioimmunoprecipitation Assay (RIPA) lysis buffer and protease inhibitor were mixed at 1:100. Protein concentration was detected by the Bicinchoninic Acid Assay (BCA) method. Prepare 10 % Polyacrylamide Gel Electrophoresis (PAGE) gel according to the instruction of the PAGE gel rapid preparation kit (YaMei, Shanghai, China). All primary antibodies (Wanleibio, Shenyang, China) were derived from rabbits, and secondary antibodies (Wanleibio, Shenyang, China) were goat anti-rabbit antibodies. Glyceraldehyde 3-Phosphate Dehydrogenase (GAPDH) protein was used as the internal reference protein. The gray value of the target protein/the gray value of the inner reference protein was used as the relative expression level of the target protein.
Cell proliferation assay:
The proliferation of HTR-8/SVneo was detected after transfection or (and) hypoxic treatment. HTR- 8/SVneo cells were seeded in 96-well plates at 3000 cells/well. Cell Counting Kit-8 (CCK8) (Seven, Beijing, China) was added to the 96-well plate 1 d, 2 d, and 3 d after cell seeding, and the absorbance was measured at 450 nm wavelength with a microplate reader after 1 h.
Wound healing assay:
Transfected or (and) hypoxia-treated trophoblasts were cultured in 6-well plates to near 100 % density. Cells are cultivated in a medium without fetal bovine serum. Photographs were captured at 0 h, 1 d, 2 d, and 3 d after scratching, and the experimental results were processed with ImageJ software (version 1.53a).
Transwell invasion assay:
In the experiment, a layer of Matrigel was pre-laid on the upper layer of the transwell chamber. The cell suspension was added after it solidified. The upper layer of each chamber was filled with 200 μl (3×105 cells/ml) of the resuspended cell suspension with medium without fetal bovine serum, and 700 μl complete medium was added in the lower chamber. After 2 d, the chambers were taken out. The bottom of the chambers was immersed with 4 % paraformaldehyde for 30 min. Afterward; the chambers were gently washed three times with Phosphate Buffer Solution (PBS). They were stained with crystal violet for 30 min. 6 fields were randomly photographed in each experimental group, and the number of cells in each visual area was counted.
Statistical analysis:
Clinical patient’s characteristics were analyzed using the R software rank sum test, and the data were shown with mean±Standard Deviation (SD). GraphPad Prism 9 software performed an unpaired t-test to analyze the experimental data of two groups. Statistical analysis of experimental data of multiple groups was performed using conventional one-way Analysis of Variance (ANOVA). All experiments were repeated at least three times. p<0.05 indicated statistical significance (*p<0.05, **p<0.01, ***p<0.001 or ****p<0.0001).
Results and Discussion
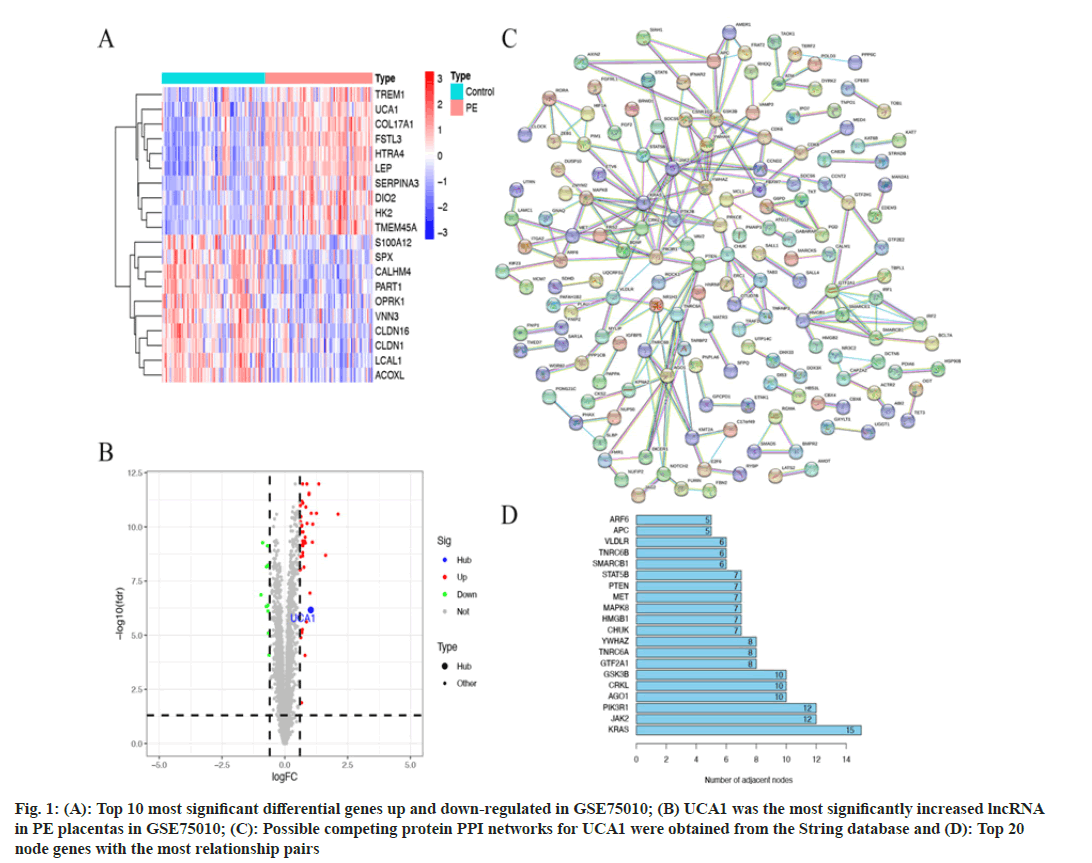
53 differentially expressed genes were generated by GSE75010 data differential analysis, including 10 down-regulated genes and 43 up-regulated genes (|log2FC>0.6| and p<0.05). We showed the top 10 most significant differences in up-regulated and down-regulated genes separately (fig. 1A). UCA1 was identified as the single most significant lncRNA with increased expression in PE placental samples (fig. 1B). The trend of UCA1 differential expression was consistent with the results of previous work[14]. The miRcode database predicted 44 miRNAs that can bind directly to UCA1. 401 mRNAs supported by two and more prediction databases are the target genes for 44 miRNAs. Notably, miR-18a was only predicted to bind to HIF-1α in the TargetScan database. 401 predicted mRNAs were subjected to PPI analysis using the String database (fig. 1C). There were 222 relationship pairs and 162 node genes in the PPI networks, and the top 20 node genes with the most relationship pairs were selected for presentation, including Kirsten Rat Sarcoma Virus (KRAS), Janus Kinase 2 (JAK2), Phosphoinositide- 3-Kinase Regulatory 1 (PIK3R1), Argonaute1 (AGO1), CRKL, and others (fig. 1D).
Fig. 1: (A): Top 10 most significant differential genes up and down-regulated in GSE75010; (B) UCA1 was the most significantly increased lncRNA in PE placentas in GSE75010; (C): Possible competing protein PPI networks for UCA1 were obtained from the String database and (D): Top 20 node genes with the most relationship pairs
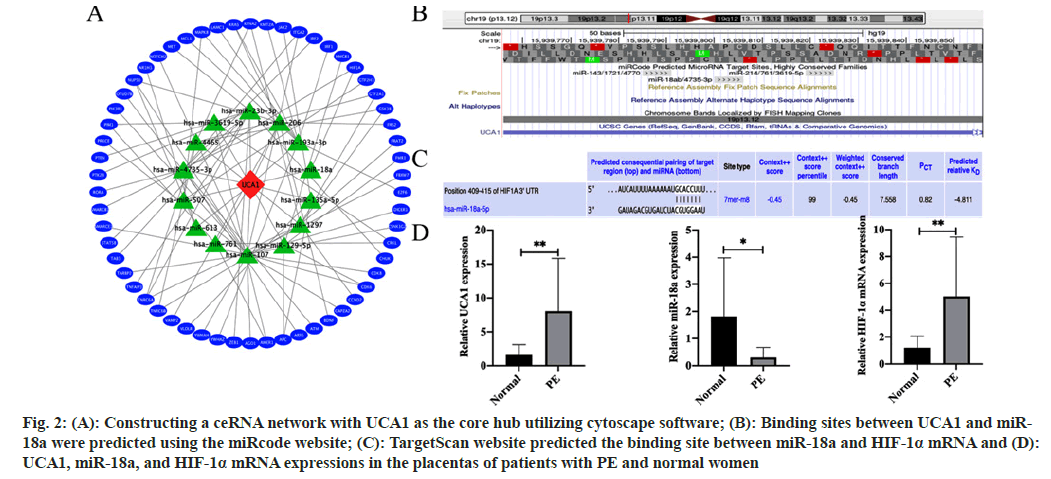
We picked node genes with relationship pairs equal to or >3 pairs as key mRNAs and used Cytoscape software to construct a ceRNA network with UCA1 as the core hub (fig. 2A). The ceRNA network consisted of UCA1, 14 miRNAs including miR-18a, and 58 mRNAs including HIF-1α. Subsequently, we explored and examined in depth the evidence that UCA1/miR-18a/HIF-1a constituted a ceRNA mechanism involved in PE pathogenesis. Based on the prediction from the miRcode website, UCA1 had a highly conserved binding site that binds directly to miR-18a (fig. 2B). The possible targeted binding sites of miR-18a and HIF-1α mRNA were predicted utilizing the TargetScan website (fig. 2C). Further confirmation of clinical samples by RT-qPCR showed that UCA1 was more abundant in the placenta of PE patients than in normal placentas, indicating that it might play an essential role in the pathogenesis of PE. Furthermore, the content of miR-18a in the placenta of PE patients was lacking compared with that of normal women. At the same time, HIF-1α mRNA expression was increased (fig. 2D).
Fig. 2: (A): Constructing a ceRNA network with UCA1 as the core hub utilizing cytoscape software; (B): Binding sites between UCA1 and miR- 18a were predicted using the miRcode website; (C): TargetScan website predicted the binding site between miR-18a and HIF-1α mRNA and (D): UCA1, miR-18a, and HIF-1α mRNA expressions in the placentas of patients with PE and normal women
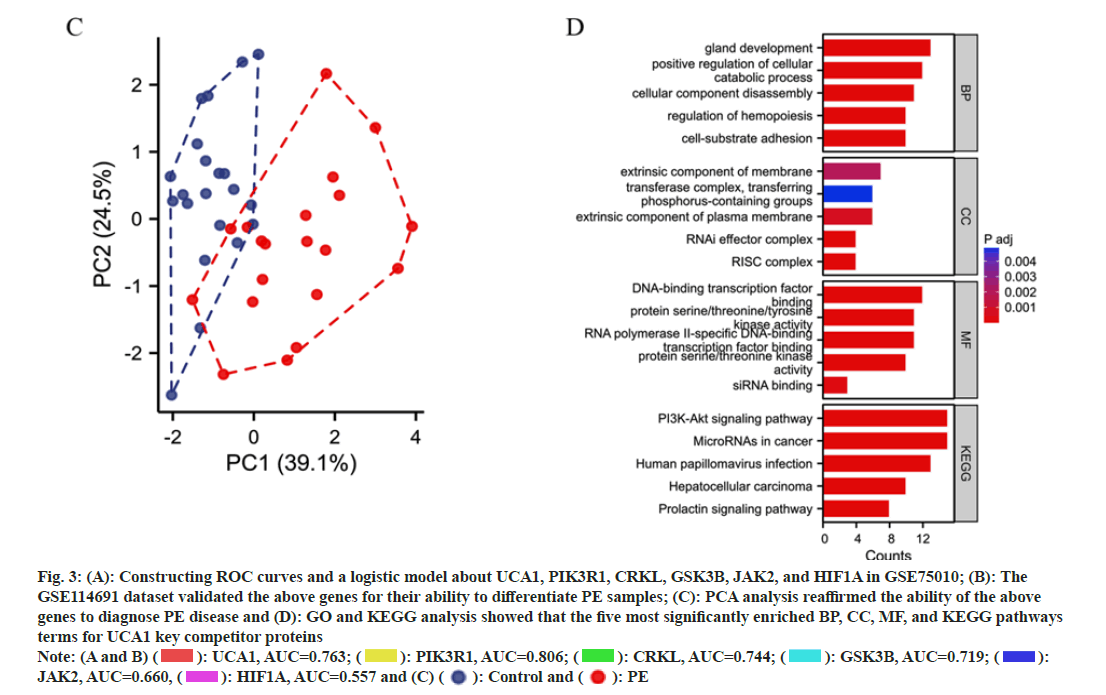
We constructed ROC curves and a logistic model about UCA1 and its competing mRNAs PIK3R1, CRKL, GSK3B, JAK2, and HIF1A (i.e., HIF-1α) in GSE75010. This model's area under the ROC curve (Area under the Curve (AUC)) was 0.861 (95 % Confidence Interval (CI); 0.799-0.918) (fig. 3A). The GSE114691 dataset validated the above genes for their ability to differentiate PE samples. The validation group model exhibited AUC of 0.990 (95 % CI; 0.964-1.000), indicating that the above genes in combination were more capable of predicting PE (fig. 3B). PCA analysis revealed only a small overlap of regions between the control and PE groups in GSE114691, reaffirming the ability of the above genes to diagnose PE disease (fig. 3C). GO and KEGG analysis revealed that the five most significantly enriched BP, CC, MF, and KEGG pathways terms for UCA1 key competitor mRNAs (fig. 3D). UCA1 mainly affected BP such as intracellular protein catabolism, cellular adhesion, apoptosis, and migration. Transcription-related CC that may be potentially influenced by UCA1 include the RISC complex and RNAi effector complex. UCA1 may have roles in such as RNA polymerase IIspecific DNA-binding, transcription factor binding, and DNA-binding transcription factor binding to interfere with protein transcription. UCA1 is engaged in the Phosphoinositide 3-Kinase (PI3K)-Protein Kinase B (Akt) signaling pathway, microRNAs in multiple cancers, human papillomavirus infection, and other pathways affecting disease progression.
Fig. 3: (A): Constructing ROC curves and a logistic model about UCA1, PIK3R1, CRKL, GSK3B, JAK2, and HIF1A in GSE75010; (B): The
GSE114691 dataset validated the above genes for their ability to differentiate PE samples; (C): PCA analysis reaffirmed the ability of the above
genes to diagnose PE disease and (D): GO and KEGG analysis showed that the five most significantly enriched BP, CC, MF, and KEGG pathways
terms for UCA1 key competitor proteins
Note: (A and B) ( ): UCA1, AUC=0.763; (
): UCA1, AUC=0.763; (  ): PIK3R1, AUC=0.806; (
): PIK3R1, AUC=0.806; ( ): CRKL, AUC=0.744; (
): CRKL, AUC=0.744; (  ): GSK3B, AUC=0.719; (
): GSK3B, AUC=0.719; ( ):
JAK2, AUC=0.660, (
):
JAK2, AUC=0.660, ( ): HIF1A, AUC=0.557 and (C) (
): HIF1A, AUC=0.557 and (C) ( ): Control and (
): Control and (  ): PE
): PE
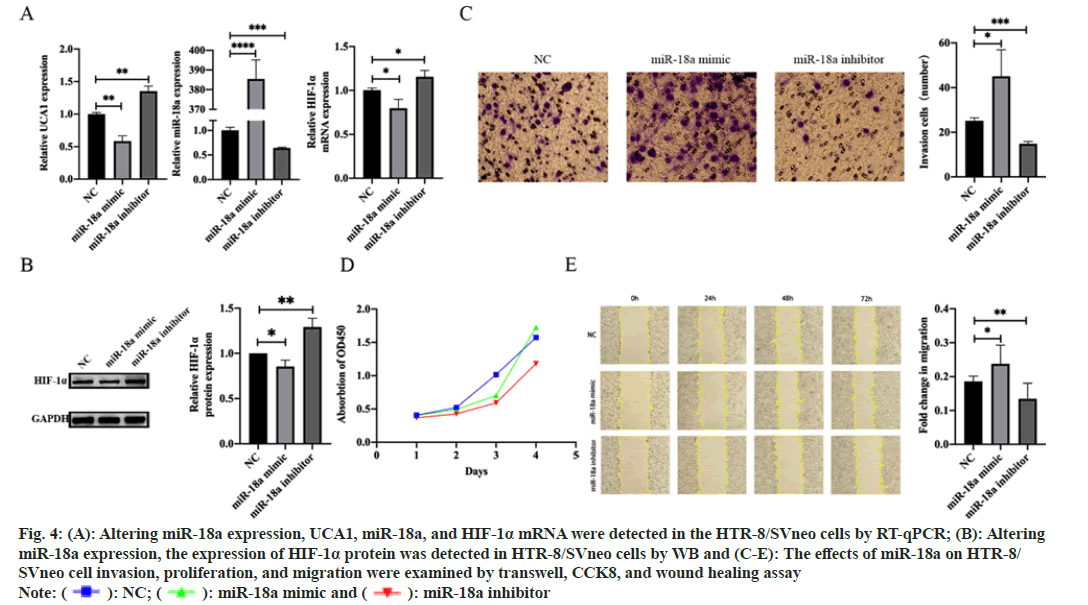
HTR-8/SVneo cells were transfected with NC, miR- 18a mimic, and miR-18a inhibitor, respectively, to investigate the effect of miR-18a on the biological behavior of trophoblast cells. After transfection, the transfection efficiency of cells and the expression levels of UCA1, HIF-1α mRNA, and HIF-1α protein were measured by RT-qPCR and WB. We found that overexpression of miR-18a, which induced UCA1 and HIF-1α decreased expression at the RNA and protein levels. Meanwhile, the knockdown of miR-18a resulted in UCA1 and HIF-1α increased expression at both RNA and protein levels (fig. 4A and fig. 4B). CCK8, transwell, and wound healing assay were performed. The results demonstrated that overexpression of miR-18a promoted migration, invasion, and proliferation of trophoblast cells, while knockdown of miR-18a produced the opposite effects (fig. 4C-fig. 4E).
Fig. 4: (A): Altering miR-18a expression, UCA1, miR-18a, and HIF-1α mRNA were detected in the HTR-8/SVneo cells by RT-qPCR; (B): Altering
miR-18a expression, the expression of HIF-1α protein was detected in HTR-8/SVneo cells by WB and (C-E): The effects of miR-18a on HTR-8/
SVneo cell invasion, proliferation, and migration were examined by transwell, CCK8, and wound healing assay
Note: (  ): NC; (
): NC; (  ): miR-18a mimic and (
): miR-18a mimic and (  ): miR-18a inhibitor
): miR-18a inhibitor
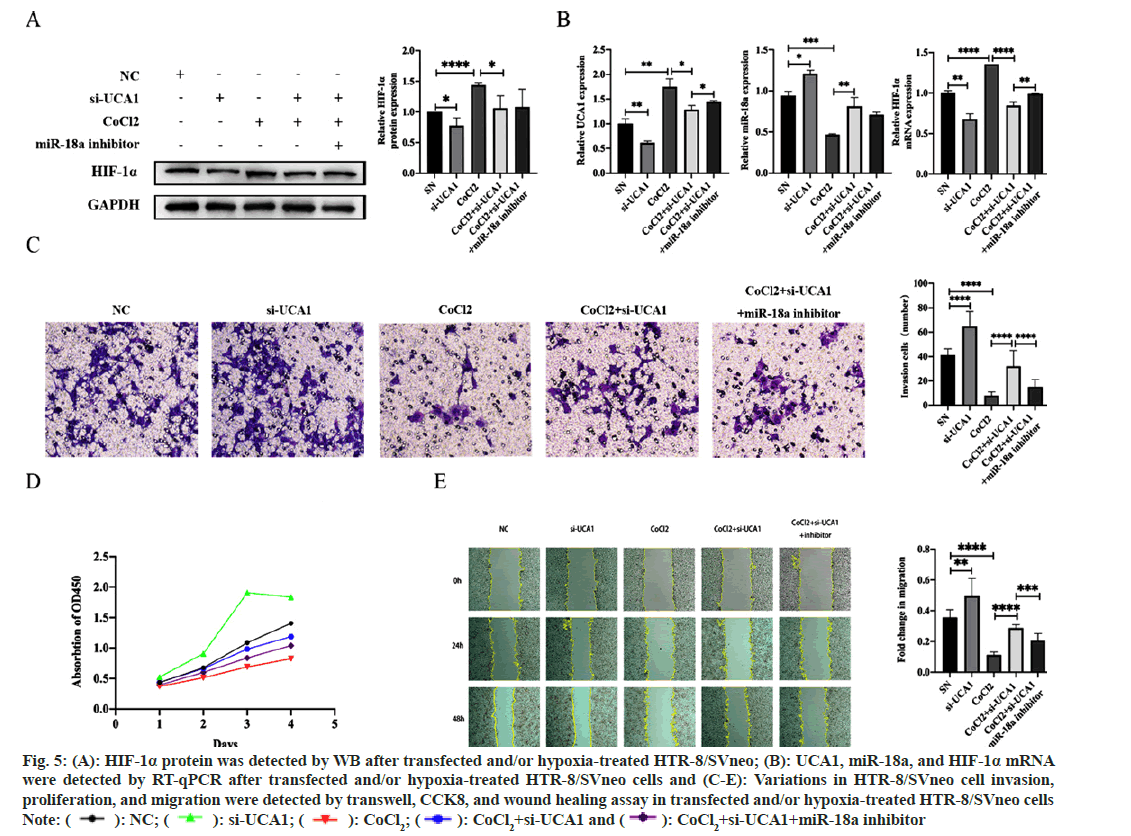
To exploit the pathological and physiological roles of UCA1 in PE and investigate the possibility that miR-18a mediates UCA1 to affect HIF-1α involved in PE, HTR-8/SVneo cells without other treatments were transfected with ncRNA or si-UCA1, respectively. The HTR-8/SVneo hypoxic model established by CoCl2 induction was transfected with si-UCA1 and simultaneously transfected with si-UCA1 and a miR-18a inhibitor. We had the following findings; compared with trophoblast cells under normoxic conditions, the expression of HIF-1α mRNA and protein were increased in the hypoxia model, indicating that this model was successfully established. Additionally, when treated with si-UCA1, miR-18a expression was upregulated, and HIF-1α, mRNA and protein expressions were downregulated (fig. 5A and fig. 5B). Therefore, we considered that the UCA1/miR-18a/HIF-1α axis is likely to participate in the pathogenesis of PE.
We further explored the effect of UCA1 on the biological functions of trophoblast cells. The results displayed that si-UCA1 transfection enhanced such cells’ migration, invasion, and proliferation. Thus, the knockdown of UCA1 reduced the decline of cell migration, invasion, and proliferation capacity affected by hypoxia, while inhibiting miR-18a expression reversed this circumstance to a certain extent (fig. 5C-fig. 5E). Therefore, we concluded that the UCA1/miR-18a/HIF-1α axis interfered with the invasion, proliferation, and migration of trophoblast cells in the hypoxia model and is involved in the pathophysiology of PE.
Fig. 5: (A): HIF-1α protein was detected by WB after transfected and/or hypoxia-treated HTR-8/SVneo; (B): UCA1, miR-18a, and HIF-1α mRNA
were detected by RT-qPCR after transfected and/or hypoxia-treated HTR-8/SVneo cells and (C-E): Variations in HTR-8/SVneo cell invasion,
proliferation, and migration were detected by transwell, CCK8, and wound healing assay in transfected and/or hypoxia-treated HTR-8/SVneo cells
Note: ( ): NC; (
): NC; ( ): si-UCA1; (
): si-UCA1; ( ): CoCl2; (
): CoCl2; ( ): CoCl2+si-UCA1 and (
): CoCl2+si-UCA1 and ( ): CoCl2+si-UCA1+miR-18a inhibitor
): CoCl2+si-UCA1+miR-18a inhibitor
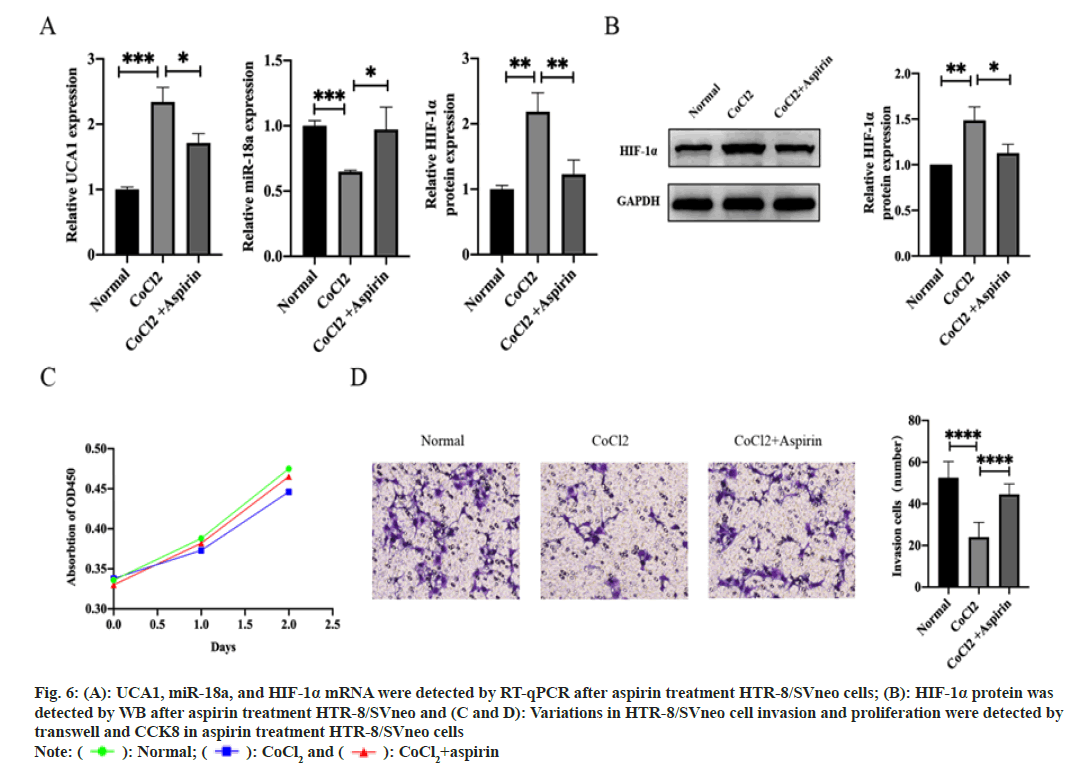
We sought to dig out whether the UCA1/miR-18a/ HIF-1α pathway is one of the mechanisms by which aspirin prevents the development of PE. Aspirin treatment altered the expression levels of UCA1, miR-18a, and HIF-1α in hypoxic HTR-8/SVneo cells. UCA1 and HIF-1α expression was decreased, while miR-18a expression was increased (fig. 6A and fig. 6B). Importantly, the invasive function was repaired after exposing in aspirin, although the proliferative function of trophoblast cells was not significantly changed (fig. 6C and fig. 6D). Thus, we considered that UCA1/miR-18a/HIF-1α was involved in the pharmacological mechanism of aspirin in controlling the pathogenesis of PE.
Fig. 6: A): UCA1, miR-18a, and HIF-1α mRNA were detected by RT-qPCR after aspirin treatment HTR-8/SVneo cells; (B): HIF-1α protein was
detected by WB after aspirin treatment HTR-8/SVneo and (C and D): Variations in HTR-8/SVneo cell invasion and proliferation were detected by
transwell and CCK8 in aspirin treatment HTR-8/SVneo cells
Note: ( ): Normal; (
): Normal; (  ): CoCl2 and (
): CoCl2 and ( ): CoCl2+aspirin
): CoCl2+aspirin
PE affects >20 000 of pregnant women worldwide. The heterogeneity and complexity of PE make prediction, diagnosis, and treatment become difficult. Clarifying the molecular mechanisms of PE and searching for new PE biomarkers are necessary to reduce maternal and child harm and the global economic burden. With advances in precise molecular targeting methods, extensive studies have found that ncRNAs are involved in the occurrence and development of various diseases including multiple pregnancy complications[21-23]. In particular, lncRNAs and miRNAs play essential roles in disease diagnosis and treatment. For example, the lncRNA TUG1 promotes angiogenesis of human umbilical vein endothelial cells in PE by regulating the miR- 29a-3p/Vascular Endothelial Growth Factor A (VEGFA) pathway[24]. These findings prompted the exploration of the role of ncRNA in the pathology, diagnosis, and treatment of PE.
Herein, we aimed to broaden the molecular mechanisms by which combinations of lncRNA/ miRNA/mRNA are involved in the pathogenesis of PE. Utilizing bioinformatics mining, we identified the potential biological significance of UCA1 in the placenta of women with PE. Afterward, the ceRNA network we constructed illustrated that UCA1 can act as a core gene that competed with multiple mRNAs to bind miRNAs, influence protein expression, and engage in the pathogenesis of PE. Four miRNAs (miR-206, miR-18a, miR-135a-5p and miR-507) of the ceRNA network have been demonstrated to be abnormally abundant in PE placental tissue, affecting specific molecular mechanisms leading to PE[25-28]. Key mRNAs in the PPI network influenced by UCA1 included JAK2, PIK3R1, CRKL, GSK3B, and HIF-1α. They have been supported by several evidence to influence trophoblast function and participate in PE occurrence by activating JAK2/ STAT3, AKT signaling pathway, the RAS and JUN kinase signaling pathways, and inhibiting PI3K-Akt signaling pathway, respectively[29-34].
We selected the above genes to constitute a diagnostic model for PE and validated that the model is diagnostically sound, identifying that UCA1 and its competing mRNAs were diagnostic for PE. KEGG analysis displayed that UCA1 competing mRNAs were significantly enriched in the PI3KAkt signaling pathway and microRNAs in cancer. PI3K/Akt/endothelial Nitric Oxide Synthase (eNOS) and PI3K/Akt/mammalian Target of Rapamycin (mTOR) pathways activation has been shown to affect trophoblast angiogenesis, oxidative stress, and invasion leading to PE in pregnant women[35-37]. There are similarities in the alteration of certain functions of cancer and PE placental cells, such as proliferation, invasion, and angiogenesis. Fully evaluating the GO and KEGG analytic result, we considered that UCA1 may disrupt transcription and post-translationally affect protein expression, leading to impaired BP such as cell metabolism, migration, invasion, apoptosis, and adhesion, thereby inducing PE.
UCA1/miR-18a/HIF-1α was identified in the ceRNA network. Through searching extensive literature, we discovered that UCA1 was responsible for the onset and development of diverse diseases[38,39]. For instance, UCA1 could target miR-18a to strengthen tamoxifen resistance in breast cancer cells by impacting cell proliferation and the cell cycle[40]. And miR-18a also restrained HIF-1α expression through targeted binding and regulated gastric cancer cell apoptosis and invasion[41]. HIF-1α induced UCA1 upregulation facilitated osteosarcoma cell proliferation[42]. Based on the above discoveries, we hypothesized that UCA1/miR-18a/HIF-1α might form a pathway; affect the biological function of trophoblast cells, and participate in the pathophysiology of PE.
HIF-1 has been broadly researched as a major regulator of the cellular response to hypoxic tension. A study has shown that HIF-1 is a crucial link in the pathogenesis of PE[43]. As a component of HIF-1, HIF- 1α regulates multiple pathways mediating abnormal PE placental vascular remodeling[44,45]. Primarily, HIF-1α can activate anti-angiogenic and inhibit pro-angiogenic factors, such as soluble FMS-Like Tyrosine kinase-1 (FLT1), VEGFA, Notch homolog 1[46]. HIF-1α also enhances placental inflammation and dysfunction by increasing the production of p38MAPK and NLRP3 inflammatory vesicles[46]. Moreover, serum HIF-1α levels with or without uterine artery Doppler ultrasonography at 11-13+6 w of gestation have been proven to effectively predict PE[47].
miR-18a is one of the most conserved and functional miRNAs in the miR-17-92 cluster[48]. The altered miR-18a expression has been found in various physiological and pathological processes, including cell proliferation, invasion, apoptosis, epithelial-mesenchymal transition, and metastasis[49]. Interestingly, miR-18a has dual functional roles in promoting or inhibiting tumorigenesis in different human cancers.
We looked at the UCA1/miR-18a/HIF-1 pathway as a focused research target to further explore. Clinical samples verified by RT-qPCR showed that the expressions of UCA1 and HIF-1α in PE placentas were higher than those in normal pregnancy. In contrast, miR-18a expression was decreased, consistent with previous research results. Using different databases, we predicted possible binding sites among UCA1, miR-18a, and HIF-1α mRNA. Subsequently, we overexpressed miR-18a trophoblast cells and exhibited that UCA1 and HIF- 1α were decreased at both RNA and protein levels. Meanwhile, reducing the expression of miR-18a in vitro had the opposite result. It is well known that the ability of trophoblast cells to migrate, invade and proliferate is critical for a successful pregnancy. Therefore, we performed gain-and-loss studies of functions focusing on the above biological functions. They revealed that overexpression of miR-18a fostered the migration, invasion, and proliferation of trophoblast cells. Taken together, these experiments have suggested that overexpression or knockdown of miR-18a transformed the expressed levels of UCA1 and HIF-1α, interfering with trophoblast cell behaviors. Xu et al.[28] exhibited that diminished miR-18a enhanced TGF-β Signaling by reinforcing Mothers Against Decapentaplegic Homolog 2 (SMAD2) (FL) expression, eliciting PE in pregnant women. Similarly, this work sourced that miR-18a had a facilitative effect on trophoblast invasion. In addition, they observed no effect of miR-18a on the trophoblast cell cycle. A prior study assessed that miR-18a was highly expressed in PE maternal plasma and correlated with PE severity[50]. It can be evident that miR-18a is a promising molecule for non-invasive liquid biopsy detection in PE.
Based on the successful establishment of the PE cell hypoxia model, by interfering with UCA1 expression, miR-18a expression was upregulated, and the expressions of HIF-1α mRNA and protein were downregulated, demonstrating that UCA1 transformed the expression of HIF-1α in this model via miR-18a. Cell functional experiments were performed; depicting that knockdown of UCA1 attenuated hypoxia-affected cell migration, invasion, and proliferation while inhibiting miR- 18a expression reversed this phenomenon to some extent. In light of the above studies, we pinpointed that UCA1/miR-18a/HIF-1α affected the invasion, proliferation, and migration of trophoblasts in the PE hypoxia model and was involved in the development of PE. However, Wu et al.[51] implemented a dual luciferase reporter gene assay to demonstrate that HIF-1α mRNA bound directly to the UCA1 promoter to facilitate UCA1 expression. Hence, we had doubts about whether the UCA1/miR-18a/ HIF-1α relationship acted as ceRNAs or a feedback pathway. Resolving this question is imperative for understanding the pathogenesis of PE. Their data demonstrated that UCA1 recruited ubiquitin-specific peptidase 14 to profilin 1, inactivated ubiquitinated degradation of PFN1, and further stimulated the RhoA/Rho-kinase pathway to trigger reactive oxygen species production in endothelial cells. Through this molecular mechanism, UCA1 functioned in the adaptive pathogenesis of HIF-1α in response to PE and damaged maternal endothelial cells. The team also detected a rise in serum exosome UCA1 expression in PE mothers. Consequently, UCA1 holds promise as a target for the detection and therapy of PE in the coming years. Furthermore, we realized that aspirin improved the invasive function of trophoblast cells under hypoxia-induced and shifted the expression levels of UCA1, miR-18a, and HIF-1α. This result provides a theoretical basis for aspirin to attenuate the risk of developing PE.
A tremendous challenge that therapists face with PE disease is the time difference between fundamental pathological changes starting before the 20th w of pregnancy and clinical symptoms appearing after that in PE disease. In the present study, we combined several genes associated with UCA1 to establish a model for diagnosing PE, and the model validity has been verified. In addition, we discovered a new molecular mechanism in the pathological processes of PE. UCA1 regulated the expression of HIF-1α via miR-18a, affected the biological function of trophoblast cells, and contributed to the occurrence of PE. The role of UCA1 implies that UCA1 may form an effective prenatal monitoring system for PE in combination with other indicators to guide its prevention, diagnosis, treatment, and prognosis.
Ethical approval:
This study was performed in line with the principles of the ethical review of biomedical research involving human subjects and other relevant ethical regulations. The placental specimens in the medical records in this study were secondarily utilized, and the work met the original conditions for informed consent permission. Therefore, the study was approved by the Medical Ethics Committee of the Second Affiliated Hospital of Harbin Medical University (KY2021-309) and was conducted under the premise of waiving informed consent.
Author contributions:
Limin Song and Man Guo contributed to conception and design of the study. Xinying Zhao undertook the bioinformatics analysis. Limin Song and Jiaxi Chen performed experiments. Limin Song, Hang Yin, Hongyan Tang, Jiaxi Chen, Haijing Dong and Xinyue Li jointly collected clinical tissue samples and some part of the study experiments. Xinying Zhao, Xiaodan Chu, and Lianxiu Li provided technical assistance. Limin Song drafted the first manuscript. Man Guo supervised the whole study. All authors contributed to manuscript revision, read, and approved the submitted version. Limin Song and Xinying Zhao have contributed equally to this work and they both are considered as first authors.
Funding:
This research was supported by China Postdoctoral Science Foundation (No: 2018M641855).
Conflict of interest:
The authors declared no conflict of interests.
References
- Barron A, McCarthy CM, O’Keeffe GW. Preeclampsia and neurodevelopmental outcomes: Potential pathogenic roles for inflammation and oxidative stress? Mol Neurobiol 2021;58(6):2734-56.
[Crossref] [Google Scholar] [PubMed]
- Bokslag A, van Weissenbruch M, Mol BW, de Groot CJ. Preeclampsia; short and long-term consequences for mother and neonate. Early Hum Dev 2016;102:47-50.
[Crossref] [Google Scholar] [PubMed]
- Turbeville HR, Sasser JM. Preeclampsia beyond pregnancy: Long-term consequences for mother and child. Am J Physiol Renal Physiol 2020;318(6):F1315-26.
[Crossref] [Google Scholar] [PubMed]
- Wang H, Bhutta ZA, Coates MM, Coggeshall M, Dandona L, Diallo K, et al. Global, regional, national, and selected subnational levels of stillbirths, neonatal, infant, and under-5 mortality, 1980–2015: A systematic analysis for the global burden of disease study 2015. Lancet 2016;388(10053):1725-74.
[Crossref] [Google Scholar] [PubMed]
- Collaborators GB. Global, regional, and national levels of maternal mortality, 1990–2015: A systematic analysis for the global burden of disease study 2015. Lancet 2016;388:1775-812.
- Ives CW, Sinkey R, Rajapreyar I, Tita AT, Oparil S. Preeclampsia-pathophysiology and clinical presentations: JACC state-of-the-art review. J Am Coll Cardiol 2020;76(14):1690-702.
[Crossref] [Google Scholar] [PubMed]
- Demirer S, Hocaoglu M, Turgut A, Karateke A, Komurcu-Bayrak E. Expression profiles of candidate microRNAs in the peripheral blood leukocytes of patients with early-and late-onset preeclampsia vs. normal pregnancies. Pregnancy Hypertens 2020;19:239-45.
[Crossref] [Google Scholar] [PubMed]
- Li H, Ouyang Y, Sadovsky E, Parks WT, Chu T, Sadovsky Y. Unique microRNA signals in plasma exosomes from pregnancies complicated by preeclampsia. Hypertension 2020;75(3):762-71.
[Crossref] [Google Scholar] [PubMed]
- Lv Y, Lu C, Ji X, Miao Z, Long W, Ding H, et al. Roles of microRNAs in preeclampsia. J Cell Physiol 2019;234(2):1052-61.
[Crossref] [Google Scholar] [PubMed]
- Wang XS, Zhang Z, Wang HC, Cai JL, Xu QW, Li MQ, et al. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin Cancer Res 2006;12(16):4851-8.
[Crossref] [Google Scholar] [PubMed]
- Xuan W, Yu H, Zhang X, Song D. Crosstalk between the lnc RNA UCA 1 and micro RNAs in cancer. FEBS Lett 2019;593(15):1901-14.
[Crossref] [Google Scholar] [PubMed]
- Hosseini NF, Manoochehri H, Khoei SG, Sheykhhasan M. The functional role of long non-coding RNA UCA1 in human multiple cancers: A review study. Curr Mol Med 2021;21(2):96-110.
[Crossref] [Google Scholar] [PubMed]
- Yao F, Wang Q, Wu Q. The prognostic value and mechanisms of lncRNA UCA1 in human cancer. Cancer Manag Res 2019;11:7685-96.
[Crossref] [Google Scholar] [PubMed]
- Liu J, Luo C, Zhang C, Cai Q, Lin J, Zhu T, et al. Upregulated lncRNA UCA1 inhibits trophoblast cell invasion and proliferation by downregulating JAK2. J Cell Physiol 2020;235(10):7410-9.
[Crossref] [Google Scholar] [PubMed]
- Hoffman MK, Goudar SS, Kodkany BS, Metgud M, Somannavar M, Okitawutshu J, et al. Low-dose aspirin for the prevention of preterm delivery in nulliparous women with a singleton pregnancy (ASPIRIN): A randomised, double-blind, placebo-controlled trial. Lancet 2020;395(10220):285-93.
[Crossref] [Google Scholar] [PubMed]
- Roberge S, Nicolaides KH, Demers S, Villa P, Bujold E. Prevention of perinatal death and adverse perinatal outcome using low-dose aspirin: A meta-analysis. Ultrasound Obstetr Gynecol 2013;41(5):491-9.
[Crossref] [Google Scholar] [PubMed]
- Lin L, Li G, Zhang W, Wang YL, Yang H. Low-dose aspirin reduces hypoxia-induced sFlt1 release via the JNK/AP-1 pathway in human trophoblast and endothelial cells. J Cell Physiol 2019;234(10):18928-41.
[Crossref] [Google Scholar] [PubMed]
- Khaidakov M, Szwedo J, Mitra S, Mehta JL. Angiostatic effects of aspirin in hypoxia-reoxygenation are linked to modulation of TGFβ1 signaling. J Cardiovasc Pharmacol Ther 2011;16(1):105-10.
[Crossref] [Google Scholar] [PubMed]
- Su MT, Tsai PY, Wang CY, Tsai HL, Kuo PL. Aspirin facilitates trophoblast invasion and epithelial-mesenchymal transition by regulating the miR-200-ZEB1 axis in preeclampsia. Biomed Pharmacother 2021;139:111591.
[Crossref] [Google Scholar] [PubMed]
- Lee HR, Leslie F, Azarin SM. A facile in vitro platform to study cancer cell dormancy under hypoxic microenvironments using CoCl2. J Biolog Eng 2018;12(1):1-5.
- Mohapatra S, Pioppini C, Ozpolat B, Calin GA. Non-coding RNAs regulation of macrophage polarization in cancer. Mol Cancer 2021;20(1):24.
[Crossref] [Google Scholar] [PubMed]
- Sun P, Hamblin MH, Yin KJ. Non-coding RNAs in the regulation of blood–brain barrier functions in central nervous system disorders. Fluids Barriers CNS 2022;19(1):1-39.
[Crossref] [Google Scholar] [PubMed]
- Zarkovic M, Hufsky F, Markert UR, Marz M. The role of non-coding RNAs in the human placenta. Cells 2022;11(9):1588.
[Crossref] [Google Scholar] [PubMed]
- Liu Z, Guo N, Zhang XJ. Long noncoding TUG1 promotes angiogenesis of HUVECs in PE via regulating the miR-29a-3p/VEGFA and Ang2/Tie2 pathways. Microvasc Res 2022;139:104231.
[Crossref] [Google Scholar] [PubMed]
- Li HQ, Fan JJ, Li XH, Bao D. miR-507 inhibits the growth and invasion of trophoblasts by targeting CAMK4. Eur Rev Med Pharmacol Sci 2020;24:5856-62.
[Crossref] [Google Scholar] [PubMed]
- Wu HY, Wang XH, Liu K, Zhang JL. LncRNA MALAT1 regulates trophoblast cells migration and invasion via miR-206/IGF-1 axis. Cell Cycle 2020;19(1):39-52.
[Crossref] [Google Scholar] [PubMed]
- Wu D, Shi L, Hong L, Chen X, Cen H. miR-135a-5p promotes the migration and invasion of trophoblast cells in preeclampsia by targeting β-TrCP. Placenta 2020;99:63-9.
[Crossref] [Google Scholar] [PubMed]
- Xu P, Li Z, Wang Y, Yu X, Shao X, Li YX, et al. miR-18a contributes to preeclampsia by downregulating Smad2 (full length) and reducing TGF-β signaling. Mol Ther Nucl Acids 2020;22:542-56.
[Crossref] [Google Scholar] [PubMed]
- Abdelzaher WY, Mostafa-Hedeab G, Bahaa HA, Mahran A, Atef Fawzy M, Abdel Hafez SM, et al. Leukotriene receptor antagonist, montelukast ameliorates L-Name-induced pre-eclampsia in rats through suppressing the IL-6/Jak2/STAT3 signaling pathway. Pharmaceuticals 2022;15(8):914.
[Crossref] [Google Scholar] [PubMed]
- Chiang MH, Liang FY, Chen CP, Chang CW, Cheong ML, Wang LJ, et al. Mechanism of hypoxia-induced GCM1 degradation: Implications for the pathogenesis of preeclampsia. J Biol Chem 2009;284(26):17411-9.
[Crossref] [Google Scholar] [PubMed]
- Li Y, Sun XL, Ma CL, Li C, Zhan Y, Li WT, et al. STX2 promotes trophoblast growth, migration, and invasion through activation of the PI3K-AKT pathway in preeclampsia. Front Cell Dev Biol 2021;9:615973.
[Crossref] [Google Scholar] [PubMed]
- Shah DM. The role of RAS in the pathogenesis of preeclampsia. Curr Hypertens Rep 2006;8(2):144-52.
[Crossref] [Google Scholar] [PubMed]
- Shi CS, Tuscano J, Kehrl JH. Adaptor proteins CRK and CRKL associate with the serine/threonine protein kinase GCKR promoting GCKR and SAPK activation. Blood 2000;95(3):776-82.
[Google Scholar] [PubMed]
- Yang HY. miR-133b regulates oxidative stress injury of trophoblasts in preeclampsia by mediating the JAK2/STAT3 signaling pathway. J Mol Histol 2021;52:1177-88.
[Crossref] [Google Scholar] [PubMed]
- Fisher SJ. Why is placentation abnormal in preeclampsia? Am J Obstetr Gynecol 2015;213(4):115-22.
[Crossref] [Google Scholar] [PubMed]
- Huang J, Zheng L, Wang F, Su Y, Kong H, Xin H. Mangiferin ameliorates placental oxidative stress and activates PI3K/Akt/mTOR pathway in mouse model of preeclampsia. Arch Pharm Res 2020;43:233-41.
[Crossref] [Google Scholar] [PubMed]
- Xu Y, Sui L, Qiu B, Yin X, Liu J, Zhang X. ANXA4 promotes trophoblast invasion via the PI3K/Akt/eNOS pathway in preeclampsia. Am J Physiol Cell Physiol 2019;316(4):481-91.
[Crossref] [Google Scholar] [PubMed]
- Luan Y, Li X, Luan Y, Zhao R, Li Y, Liu L, et al. Circulating lncRNA UCA1 promotes malignancy of colorectal cancer via the miR-143/MYO6 axis. Mol Ther Nucl Acids 2020;19:790-803.
[Crossref] [Google Scholar] [PubMed]
- Yang Z, Zhang H, Yin M, Cheng Z, Jiang P, Feng M, et al. TGF-β1/Smad3 upregulates UCA1 to promote liver fibrosis through DKK1 and miR18a. J Mol Med 2022;100(10):1465-78.
[Crossref] [Google Scholar] [PubMed]
- Li X, Wu Y, Liu A, Tang X. Long non-coding RNA UCA1 enhances tamoxifen resistance in breast cancer cells through a miR-18a-HIF1α feedback regulatory loop. Tumor Biol 2016;37:14733-43.
[Crossref] [Google Scholar] [PubMed]
- Wu F, Huang W, Wang X. MicroRNA-18a regulates gastric carcinoma cell apoptosis and invasion by suppressing hypoxia-inducible factor-1α expression. Exp Ther Med 2015;10(2):717-22.
[Crossref] [Google Scholar] [PubMed]
- Li T, Xiao Y, Huang T. HIF-1α-induced upregulation of lncRNA UCA1 promotes cell growth in osteosarcoma by inactivating the PTEN/AKT signaling pathway. Oncol Rep 2018;39(3):1072-80.
[Crossref] [Google Scholar] [PubMed]
- Tal R. The role of hypoxia and hypoxia-inducible factor-1alpha in preeclampsia pathogenesis. Biol Reprod 2012;87(6):134.
[Crossref] [Google Scholar] [PubMed]
- Shu L, Wang C, Ding Z, Tang J, Zhu Y, Wu L, et al. A novel regulated network mediated by downregulation HIF1A-AS2 lncRNA impairs placental angiogenesis by promoting ANGPTL4 expression in preeclampsia. Front Cell Dev Biol 2022;10:837000.
[Crossref] [Google Scholar] [PubMed]
- Yu N, Wu JL, Xiao J, Fan L, Chen SH, Li W. HIF-1α regulates angiogenesis via Notch1/STAT3/ETBR pathway in trophoblastic cells. Cell Cycle 2019;18(24):3502-12.
[Crossref] [Google Scholar] [PubMed]
- Albogami SM, Al-Kuraishy HM, Al-Maiahy TJ, Al-Buhadily AK, Al-Gareeb AI, Alorabi M, et al. Hypoxia-inducible factor 1 and preeclampsia: A new perspective. Curr Hypertens Rep 2022;24(12):687-92.
[Crossref] [Google Scholar] [PubMed]
- Tianthong W, Phupong V. Serum hypoxia-inducible factor-1α and uterine artery Doppler ultrasound during the first trimester for prediction of preeclampsia. Sci Rep 2021;11(1):6674.
- Kolenda T, Guglas K, Kopczynska M, Sobocinska J, Teresiak A, Blizniak R et al. Good or not good: Role of miR-18a in cancer biology. Rep Pract Oncol Radiother 2020;25(5):808-19.
[Crossref] [Google Scholar] [PubMed]
- Shen K, Cao Z, Zhu R, You L, Zhang T. The dual functional role of microRNA-18a (miR-18a) in cancer development. Clin Transl Med 2019;8(1):1-3.
- Jairajpuri DS, Malalla ZH, Mahmood N, Almawi WY. Circulating microRNA expression as predictor of preeclampsia and its severity. Gene 2017;627:543-8.
[Crossref] [Google Scholar] [PubMed]
- Wu S, Cui Y, Zhao H, Xiao X, Gong L, Xu H, et al. Trophoblast exosomal UCA1 induces endothelial injury through the PFN1-RhoA/ROCK pathway in preeclampsia: A human-specific adaptive pathogenic mechanism. Oxid Med Cell Longev 2022;2022.
[Crossref] [Google Scholar] [PubMed]