- *Corresponding Author:
- Xianghai Zhou
Department of Endocrinology and Metabolism, Peking University People's Hospital, Xicheng, Beijing 100044, China
E-mail: xianghaizhou1@163.com
| This article was originally published in a special issue, “Innovations in Biomedical Research and Drug Development” |
| Indian J Pharm Sci 2023:85(3) Spl Issue “211-222” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Non-alcoholic fatty liver disease is a high-risk factor for the morbidity and mortality of type 2 diabetes mellitus. Thus, we conducted a meta-analysis to investigate whether the microRNAs associated with non-alcoholic fatty liver disease are associated with diabetes and their potential to predict diabetes in non-alcoholic fatty liver disease individuals. We conducted a systematic literature search using PubMed, Cochrane library and Embase databases. The literature about circulating microRNA levels in nonalcoholic fatty liver disease individuals was screened according to the inclusion and exclusion criteria. The sensitivity, specificity and area under curve of different microRNAs were summarized. These microRNAs with higher diagnostic efficacy and stability were selected for further analysis in people with diabetes. In all, 10 of 1950 studies determining microRNA expression profiles in non-alcoholic fatty liver disease were included in the final analysis. Using area under curve ≥0.7 as the criterion, we screened out 10 microRNAs that have moderate diagnostic efficacy in non-alcoholic fatty liver disease, including microRNA-122, microRNN-34a, microRNA-99a, microRNA-16, microRNA-379, microRNA-197, microRNA-181d, microRNA-146b, microRNA-375 and microRNA-12. The pooled area under curve of microRNA-122 and microRNA-34a in the diagnosis of non-alcoholic fatty liver disease was 0.88 (95 % confidence interval: 0.81–0.96) and 0.86 (95 % confidence interval: 0.82–0.91), respectively. Further, 30 of 2674 studies related to diabetes and the above microRNA were included in the final analysis. The results showed that microRNA-122 and microRNA-34a showed higher stability in non-alcoholic fatty liver disease and diabetes. In conclusion, microRNA-122 and microRNA-34a are common biomarkers of both non-alcoholic fatty liver disease and type 2 diabetes mellitus. However, whether they can be used as effective biomarkers for the early prediction of diabetes in patients with non-alcoholic fatty liver disease lacks valid evidence.
Keywords
Meta-analysis, microRNA, non-alcoholic fatty liver disease, type 2 diabetes mellitus
Pathologically, Nonalcoholic Fatty Liver Disease (NAFLD) is characterized by >5 % hepatic fat accumulation without other recognized causes of fatty liver, such as alcohol, viruses, drugs and autoimmunity[1]. Currently, it is the most common liver disorder worldwide and the leading cause of chronic liver diseases[2]. By 2030, it is predicted to be the predominant cause of orthotropic liver transplantation[3]. Diabetes is a metabolic disorder with rapidly rising prevalence worldwide and is characterized by chronically elevated blood glucose levels due to pancreatic beta (β)-cell dysfunction or insulin resistance[4,5]. Although the underlying relationship between NAFLD and Type 2 Diabetes Mellitus (T2DM) is complicated and controversial, there is a consensus that NAFLD increases the risk of T2DM[1,6-8]. Retrospective analysis of 2920 participants revealed that the non-obese NAFLD individuals had a significantly higher risk of approximately twofold for DM than the non-obese controls[9]. In a cohort study of 129 adults with biopsy-proven NAFLD, researchers found that the prevalence rate of T2DM increased from 8.5% at baseline to 78 % in 13 y, including patients who developed either T2DM (58 %) or impaired glucose tolerance (20 %)[10]. A meta-analysis of a pooled population of 117 020 patients summarized that NAFLD was associated with an increased risk of incidence of T2DM with a pooled relative risk of approximately twofold[11]. Furthermore, diabetic patients with NAFLD frequently have poorer glycemic control compared with patients without NAFLD and NAFLD was also associated with worse all-cause and cardiovascular mortality[12-14]. An investigation of 167 621 diabetic individuals revealed that they have an approximately threefold higher risk of dying due to chronic liver diseases, which is largely attributed to NAFLD[15]. In a cohort study of 2103 patients with T2DM, the prevalence of Chronic Kidney Disease (CKD) was almost twice in those with ultrasonography-diagnosed NAFLD than in those without NAFLD[16]. Considering the high risk and poor clinical outcomes associated with NAFLD in diabetic patients, it is imperative to identify biomarkers that can predict the risk of diabetes before the onset of glycemic abnormalities. Few studies have examined the common markers of diabetes and NAFLD and even fewer have investigated the appropriate markers for predicting diabetes in patients with NAFLD.
MicroRNAs (miRNAs) are small endogenous non- coding Ribonucleic Acid (RNA) with a length of about 22–25 nucleotides that regulate target gene expression at the post-transcriptional level[17,18]. Mostly, a single miRNA targets various messenger RNA (mRNAs) and influences the expression of several genes that are often involved in an interconnected pathway[19]. Owing to the remarkable stability that protects them from degradation by endogenous ribonucleases, circulating miRNAs have greater application compared with other molecular markers[20,21]. Researchers found that parts of specific miRNAs can influence various pathophysiological processes, such as the metabolism of lipids and glucose, chronic inflammation and liver function[22,23], and have used them as biomarkers to diagnose diseases, including NAFLD and other metabolic diseases[24-27]. Therefore, we conducted a meta- analysis to investigate whether miRNAs associated with NAFLD are associated with diabetes. This will eventually aid in identifying valid biomarkers to predict diabetes in patients with NAFLD.
Materials and Methods
Search strategies:
We comprehensively retrieved data from the PUBMED, EMBASE and Cochrane databases. Firstly, we explored studies concerning miRNA expression profiling in NAFLD using the following MeSH terms and keywords; “miRNA”, “NAFLD” and a combination of these terms. All studies published from inception to 7th November 2021 were included. The first round of literature screening and analysis identified target miRNAs for the second round of research. In the second step, studies relevant to specific miRNA expression profiling in patients with T2DM were searched using the following MeSH terms; “miRNA-12”, “miRNA-122”, “miRNA-146”, “miRNA-16”, “miRNA-181”, “miRNA-197”, “miRNA-34a”, “miRNA-375”, “miRNA-379” and “miRNA-99a” in combination with “T2DM” and “noninsulin-dependent diabetes mellitus” in December 2021. The language of searching records was not limited and all retrieved items were managed with Endnote X9.
Study selection:
Two independent investigators, namely Yu and Sun, manually screened the studies based on the following inclusion and exclusion criteria. In case of a discrepancy, it was resolved by the corresponding author Zhou. We included studies that reported miRNA expression profiles in adult patients with NAFLD or T2DM, age range 18-65 y. The exclusion criteria were as follows; duplicate reports; studies conducted on animals or cell lines; conference abstracts, case reports, comments, letters to the editors, systematic reviews and meta- analyses; studies that used tissue samples other than serum, plasma or blood; studies without data proving diagnostic efficacy in retrieval results of NAFLD and studies without relevant data about target miRNAs in retrieval results of T2DM.
Data extraction and literature quality assessment:
We extracted the following information and data from the full text and supplementary materials of each study. First author, journal, year of publication, country of the study, sample type, miRNA expression assay type, sample size, the direction of expression difference, cut-off criteria of deregulated miRNAs and fold-change from miRNA expression profile studies. Moreover, we obtained relevant data proving diagnostic efficacy, including Sensitivity (SEN), Specificity (SPE), Positive Likelihood (PLR), Negative Likelihood (NLR), Diagnostic Odds Ratio (DOR) and the area under the Receiver Operating Characteristic (ROC) and Area Under the Curve (AUC). In addition, for articles without clear data reports, we extracted data from the graphical plots to calculate the mean fold changes using Web Plot Digitizer (version 4.5)[24]. Different miRNA names used in various studies were standardized according to miRBase version 22.1[28]. We evaluated the methodological quality of the included studies using the Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) criteria[29]. It assesses the research bias based on four domains; patient selection, index test, reference standard and flow timing. Furthermore, the first three domains were also used to evaluate the applicability of the articles[29].
Statistical analysis:
This study was mainly conducted in two stages. First, we selected all efficient miRNAs in the diagnosis of NAFLD followed by identifying their roles in T2DM. For selecting miRNAs in the first step, we ranked them according to the vote-counting strategy[30,31]. According to this strategy, all miRNAs were ranked in order of importance based on the following criteria; number of studies reporting a miRNA in the same expression direction, total sample size of the case and control in the studies, and mean fold changes. We used AUC as the dominant parameter of diagnostic power. For miRNAs with more than one study reporting AUC, we built the diagnostic 2×2 contingency table based on the extracted data and calculated the pooled AUC. In addition, we assessed the heterogeneity among studies using I2 statistics. If I2>50 %, heterogeneity was considered statistically significant and a random effects model was used. After identifying all useful miRNAs in diagnosing NAFLD, the second round of retrieving, screening and extracting was performed to confirm their role in T2DM. All the statistical analyses were displayed using Review manager version 5.4 and Meta-DiSc version 1.4. p<0.05 was considered statistically significant.
Results and Discussion
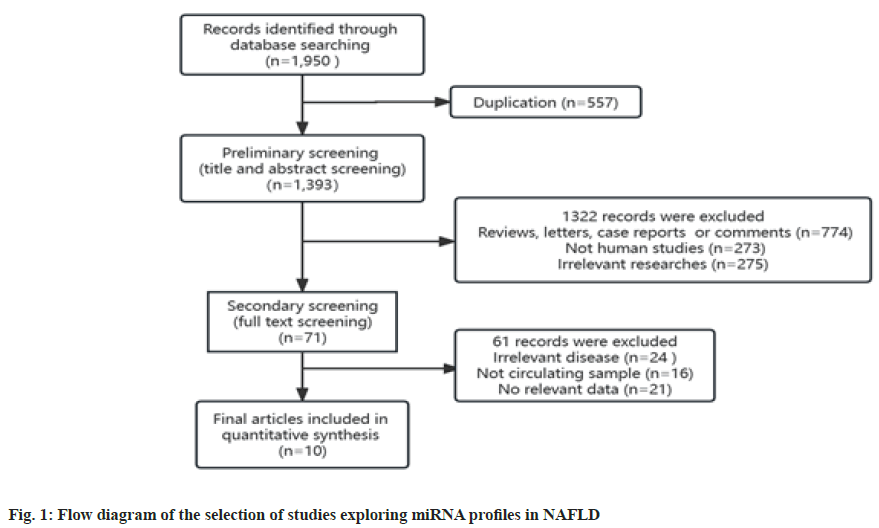
In all, we obtained 1950 studies that explored miRNA expression profiles in NAFLD. Of these, 557 duplicate records were recognized and removed using EndnoteX9. Further, during the primary screening of the titles and abstracts, 1322 records were excluded for unfit literary forms (n=774), animal or cellular studies (n=273) and irrelevant studies (n=275). Next, we conducted a full-text review of the remaining 71 records, among which 61 were excluded for irrelevant diseases (n=24), non-circulating samples except serum, plasma, and blood (n=16), and irrelevant data (n=21). Ultimately, 10 studies were included in the analysis[32-42]. Fig. 1 shows the flow diagram of the process of literature screening. The primary characteristics of the studies included are summarized in Table 1, and they are arranged by publication year, from 2011-2020. The included studies provided information on 700 patients with NAFLD and 417 healthy individuals. Two studies were conducted in China, two in the United States of America (USA), and others were conducted in Turkey, Philippines, Spain, Thailand, Egypt and Japan. Serum was used as the circulating sample and miRNA expression was measured by Real-Time quantitative Polymerase Chine Reaction (RT-qPCR) in all studies. Researchers typically detected more than one miRNA in their studies. We included only miRNAs with clear reports of diagnostic performance (SEN, SPE or AUC) (Table 1).
| Study | Country | Sample type | Measurement methods | Sample size | No. of miRNAs | miRNAs | |
|---|---|---|---|---|---|---|---|
| NAFLD | Control | ||||||
| Cermelli[32] | USA | Serum | RT-qPCR | 34 | 19 | 4 | miR-122, miR-16, miR-34a and miR-21 |
| Tan[34] | China | Serum | RT-qPCR | 152 | 90 | 6 | miR-122-5p, miR-1290, miR-27b-3p, miR-192-5p, miR-99a-5p and miR-148a-3p |
| Celikbilek[41] | Turkey | Serum | RT-qPCR | 20 | 20 | 4 | miR-197, miR-146b, miR-181d, miR-99a |
| Salvoza[35] | Philippines | Serum | RT-qPCR | 28 | 36 | 2 | miR-34a and miR-122 |
| Liu[38] | China | Serum | RT-qPCR | 48 | 37 | 2 | miR-34a |
| Auguet[42] | Spain | Serum | RT-qPCR | 61 | 61 | 1 | miR-122 |
| Jampoka[39] | Thailand | Serum | RT-qPCR | 58 | 34 | 3 | miR-29a, miR-29c and miR-122 |
| Hendy[40] | Egypt | Serum | RT-qPCR | 210 | 90 | 3 | miR-122, miR-34a and miR-99a |
| Pillai[36] | USA | Serum | RT-qPCR | 10 | 20 | 5 | miR-122, miR-34a, miR-375, miR-16 and miR-12 |
| Okamoto[37] | Japan | Serum | RT-qPCR | 79 | 10 | 1 | miR-379 |
Table 1: Characteristics of the Studies Included In the miRNA Profiles of NAFLD
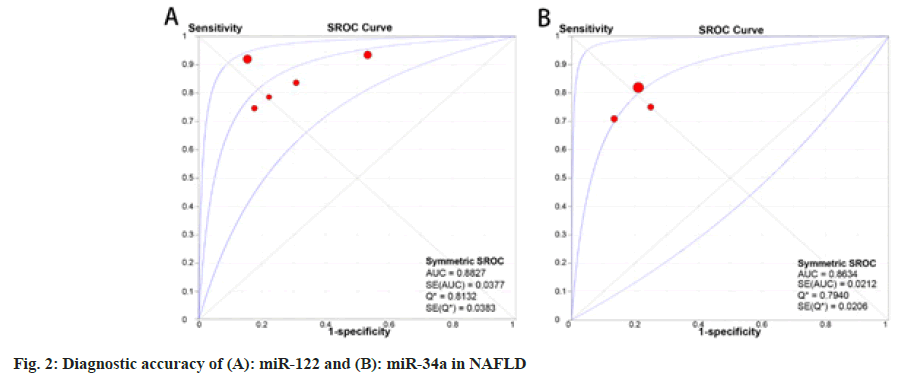
The 10 articles studied in this meta-analysis included 18 circulating miRNAs, including miR-122, miR- 34a, miR-99a, miR-16, miR-99a-5p, miR-27b-3p, miR-192-5p, miR-148a-3p, miR-1290, miR-122-5p, miR-29c, miR-29a, miR-379, miR-197, miR-181d, miR-146b, miR-375 and miR-12, ranked according to the vote-counting strategy (Table 2)[30,31]. Of these circulating miRNAs, six studies reported miR-122 to be significantly up regulated with a total sample size of 661 and a mean fold-change ranging from 3.76-7.20 in patients with NAFLD compared with healthy controls. The pooled AUC of miR-122 in diagnosing NAFLD was 0.88 (95 % Confidence Interval (CI): 0.81–0.96) and the pooled DOR was 18.05 (95 % CI: 8.68–37.54) (fig. 2A). In addition, four studies reported up regulation of miR-34a with a total sample size of 479 and a mean fold-change of 1.93–2.8. The pooled AUC of miR-34a was 0.86 (95 % CI: 0.82– 0.91) and DOR was 14.83 (95 % CI: 9.09–24.17) (fig. 2B). Down regulation of miR-99a expression was reported in two studies with a combined sample size of 340. The other two studies with a combined sample size of 83 revealed significantly higher levels of miR-16 expression and both AUC was higher than 0.85. Other miRNAs were reported only once, which indicates low credibility. Further, taking AUC as a reference for diagnostic power, we selected 10 miRNAs with an AUC ≥0.7, which is regarded as a moderate diagnostic value for distinguishing patients with NAFLD. These miRNAs were, namely, miR- 122, miR-34a, miR-99a, miR-16, miR-379, miR- 197, miR-181d, miR-146b, miR-375 and miR-12.
| miRNA | Study | Sample size | Expression direction | Mean | p | Diagnostic power | ||
|---|---|---|---|---|---|---|---|---|
| NAFLD/Control | Sen (%) | Spe (%) | AUC | |||||
| miR-122 | Cermelli | 34/19 | Up regulated | 7.2 | <0.0001 | - | - | 0.927 |
| Salvoza | 28/36 | Up regulated | - | <0.0001 | 78.6 | 77.8 | 0.858 | |
| Auguet | 61/61 | Up regulated | - | - | 83.1 | 69.8 | 0.82 | |
| Jampoka | 58/34 | Up regulated | 3.76 | <0.001 | 75 | 82.4 | 0.831 | |
| Hendy | 210/90 | Up regulated | 5.06 | <0.001 | 92 | 85 | 0.92 | |
| Pillai | 10/20 | Up regulated | 30.3 | <0.05 | - | - | 0.85 | |
| miR-34a | Salvoza | 28/36 | Up regulated | - | <0.0001 | 75 | 75 | 0.781 |
| Liu | 48/37 | Up regulated | 2.8 | <0.05 | 70.4 | 87.5 | 0.811 | |
| Hendy | 210/90 | Up regulated | 1.93 | <0.05 | 82 | 79 | 0.77 | |
| Pillai | 10/20 | Up regulated | 8.3 | <0.01 | - | - | 0.86 | |
| miR-99a | Celikbilek | 20/20 | Down regulated | 0.26 | 0.0262 | 65 | 95 | 0.76 |
| Hendy | 210/90 | Down regulated | 0.82 | <0.05 | 78 | 76 | 0.73 | |
| miR-16 | Cermelli | 34/19 | Up regulated | 5.5 | <0.0001 | - | - | 0.962 |
| Pillai | 10/20 | Up regulated | 23 | <0.01 | - | - | 0.86 | |
| miR-99a-5p | Tan | 152/90 | Up regulated | 2.38 | <0.01 | 69.1 | 41.1 | 0.559 |
| miR-27b-3p | Tan | 152/90 | Up regulated | 2.71 | <0.01 | 59.9 | 72.7 | 0,693 |
| miR-192-5p | Tan | 152/90 | Up regulated | 2.61 | <0.01 | 34.9 | 93.3 | 0.652 |
| miR-148a-3p | Tan | 152/90 | Up regulated | 2.04 | <0.01 | 25.7 | 90 | 0.54 |
| miR-1290 | Tan | 152/90 | Up regulated | 4.05 | <0.01 | 0.586 | 0.656 | 0.629 |
| miR-122-5p | Tan | 152/90 | Up regulated | 9.27 | <0.01 | 0.934 | 0.467 | 0.729 |
| miR-29c | Jampoka | 58/34 | Down regulated | 0.37 | 0.3 | 0.434 | 0.6129 | 0.529 |
| miR-29a | Jampoka | 58/34 | Down regulated | 0.08 | 0.006 | 0.6087 | 0.8235 | 0.679 |
| miR-379 | Okamoto | 79/10 | Up regulated | - | 0.026 | - | - | 0.72 |
| miR-197 | Celikbilek | 20/20 | Down regulated | 0.33 | 0.0109 | 0.6 | 0.95 | 0.77 |
| miR-181d | Celikbilek | 20/20 | Down regulated | 0.18 | <0.0001 | 0.7 | 0.85 | 0.86 |
| miR-146b | Celikbilek | 20/20 | Down regulated | 0.35 | 0.0133 | 0.55 | 1 | 0.75 |
| miR-375 | Pillai | 10/20 | Up regulated | 34.2 | <0.01 | - | - | 0.88 |
| miR-12 | Pillai | 10/20 | Up regulated | 9.9 | <0.01 | - | - | 0.83 |
Table 2: A Systematic Review of the Diagnostic Performance of miRNAs in Patients with NAFLD
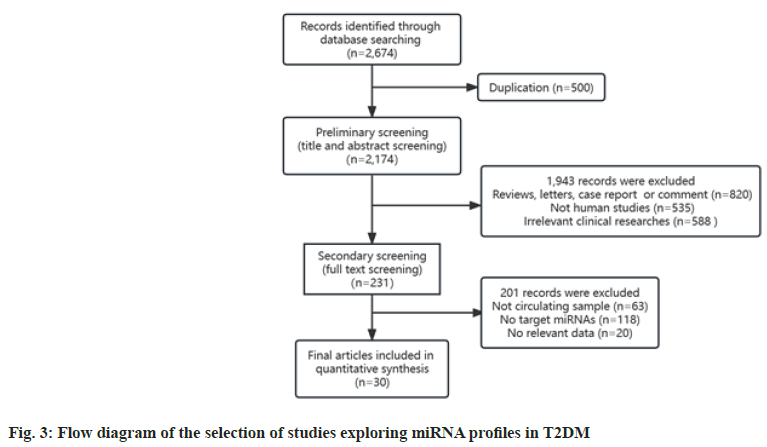
Overall, we retrieved 2674 studies that explored miRNA expression profiles in T2DM. Of these, 500 duplicate records were removed. Next, we excluded 1943 reports in the initial screening for the following reasons, unfit literary forms (n=820), animal or cellular studies (n=535) and irrelevant studies (n=588). Further, we conducted a full-text review of the remaining 231 records, among which 201 were excluded for the following reasons; non-circulating samples (n=63), no information about target miRNAs (n=118) and lack of valid data verified by RT-qPCR (n=20). Finally, 30 studies were included in the analysis[43-72]. Fig. 3 exhibits the flow chart of the process of literature screening. Table 3 summarizes the main characteristics of the included studies, in order of the publication year, ranging from 2010-2022. In all, 1229 patients with T2DM, 409 pre-diabetic patients and 1659 healthy controls were included. Of the 30 studies, ten were conducted in China, five in the USA and other studies were conducted in different countries. Most studies used serum as the test sample, while plasma and whole blood were also used. The miRNA expression levels were determined by RT-qPCR in all studies. Notably, the majority of studies detected a greater number of miRNAs than those listed here. Considering the purpose of this study, we extracted and listed data of only target miRNAs and only one or two target miRNAs were identified per research.
| Study | Country | Sample type | Measurement methods | Sample size | No. of miRNAs | miRNAs | ||
|---|---|---|---|---|---|---|---|---|
| DM | Pre-DM | Control | ||||||
| Zampetaki[71] | USA | Plasma | RT-qPCR | 80 | 80 | 1 | miR-197 | |
| Kong[57] | China | Serum | RT-qPCR | 18 | 18 | 19 | 2 | miR-34a and miR-375 |
| Sun[65] | China | Serum | RT-qPCR | 100 | 100 | 1 | miR-375 | |
| Wang (1)[66] | China | Plasma | RT-qPCR | 54 | 44 | 53 | 1 | miR-375 |
| Wang (2)[67] | China | Plasma | RT-qPCR | 33 | 119 | 1 | miR-197 | |
| Flowers[49] | USA | Plasma | RT-qPCR | 72 | 55 | 2 | miR-122-5p and miR-197-3p | |
| Rojas[45] | Ecuador | Serum | RT-qPCR | 56 | 40 | 1 | miR-122 | |
| Lopez[62] | USA | Serum | RT-qPCR | 17 | 20 | 1 | miR-34a | |
| Seyhan[64] | USA | Serum | RT-qPCR | 31 | 12 | 27 | 2 | miR-375 and miR-34a |
| Candia[47] | Italy | Serum | RT-qPCR | 9 | 9 | 9 | 2 | miR-99a-5p and miR-122-5p |
| Dias[48] | South Africa | Serum | RT-qPCR | 4 | 4 | 1 | miR-379 | |
| Jones[55] | New Zealand | Plasma | RT-qPCR | 15 | 12 | 1 | miR-34a | |
| Krauskopf[58] | USA | Serum | RT-qPCR | 7 | 22 | 1 | miR-375 | |
| Yin[70] | China | Plasma | RT-qPCR | 100 | 100 | 1 | miR-375 | |
| Al-Muhtaresh[43] | Bahrain | Serum | RT-qPCR | 30 | 30 | 30 | 1 | miR-375 |
| Fomison-Nurse[50] | New Zealand | Plasma | RT-qPCR | 39 | 32 | 1 | miR-34a | |
| Jaeger[54] | Switzerland | Serum | RT-qPCR | 43 | 43 | 43 | 1 | miR-122 |
| Ashoori[44] | Iran | Serum | RT-qPCR | 50 | 50 | 1 | miR-375 | |
| García-Jacobo[51] | Mexico | Serum | RT-qPCR | 54 | 16 | 35 | 2 | miR-34a and miR-375 |
| Gok[52] | Turkey | Serum | RT-qPCR | 25 | 25 | 1 | miR-34a | |
| Kokkinopoulou[56] | Greece | Blood | RT-qPCR | 40 | 37 | 1 | miR-34a | |
| Meerson[59] | Israel | Serum | RT-qPCR | 29 | 30 | 1 | miR-122-5p | |
| Mononen[60] | Finland | Blood | RT-qPCR | 19 | 207 | 427 | 1 | miR-122-5p |
| Regmi[63] | China | Serum | RT-qPCR | 50 | 25 | 1 | miR-122-5p | |
| Banerjee[46] | India | Plasma | RT-qPCR | 30 | 30 | 1 | miR-34a | |
| Hu[53] | China | Serum | RT-qPCR | 50 | 50 | 1 | miR-197-3p | |
| Wu[68] | China | Serum | RT-qPCR | 10 | 21 | 1 | miR-375 | |
| Ye[69] | China | Plasma | RT-qPCR | 40 | 40 | 1 | miR-16-5p | |
| Zeinali[72] | Iran | Serum | RT-qPCR | 30 | 30 | 30 | 1 | miR-122 |
| Nie[61] | China | Plasma | RT-qPCR | 94 | 94 | 1 | miR-122-5p | |
Table 3: Characteristics of the Studies Included in the Target miRNA Expression Profiles of T2DM
From 30 included articles, we acquired data on seven target circulating miRNAs, including miR-375, miR-34a, miR-122, miR-197, miR-16-5p, miR-99a-5P and miR-379. We divided the data into two groups based on the type of disease as follows; T2DM vs. healthy controls (Table 4) and prediabetes vs. healthy controls (Table 5). The miRNAs in both groups were ranked according to the vote-counting strategy[30,31]. miR- 375, which was reported only once in the diagnosis of NAFLD, had the greatest number of 10 studies in T2DM. It was consistently up regulated in seven of the ten studies with a total sample size of 735 and a mean fold-change ranging from 1.72–5.9. miR-34a also showed a consistent up regulation in six studies with a total sample size of 290 and a mean fold-change ranging from 2.04–11.3. For miR-122, five of the nine studies reported a consistent up regulation with a total sample size of 787 and a mean fold-change ranging from 1.53–8.61, while the other four studies revealed no significant change in its expression. miR-197 displayed no significant dysregulation in most studies. The remaining three miRNAs, miR-16- 5p, miR-99a-5P, miR-379, have only been reported by a single study, with lower accuracy and reliability. In the prediabetic group, miR-122 showed consistent up regulation with a sample size of 712, consistent with the T2DM group. However, the direction of expression change was not always the same in the diabetic and prediabetic patients. miR-34a showed no significant dysregulation in all three prediabetic studies and miR-375 was reported with inconsistent directions, implying low credibility and reference value.
| miRNA | Study | Sample size | Expression direction | Mean | p | |
|---|---|---|---|---|---|---|
| DM | Control | |||||
| miR-375 | Kong | 18 | 19 | Up regulated | - | 0.004 |
| Sun | 100 | 100 | Up regulated | - | 0.0313 | |
| Wang | 54 | 53 | Up regulated | 1.72 | <0.05 | |
| Seyhan | 31 | 27 | n.c. | 1.77 | n.s. | |
| Krauskopf | 7 | 22 | Down regulated | - | <0.05 | |
| Yin | 100 | 100 | Up regulated | 3 | 0.026 | |
| Al-Muhtaresh | 30 | 30 | Up regulated | 5.9 | <0.001 | |
| Ashoori | 50 | 50 | Up regulated | 2.1 | 0.02 | |
| García-Jacobo | 54 | 35 | n.c. | - | n.s. | |
| Wu | 10 | 21 | Up regulated | - | <0.001 | |
| miR-34a | Kong | 18 | 19 | Up regulated | - | <0.0001 |
| Lopez | 17 | 20 | Up regulated | 2.04 | 0.011 | |
| Seyhan | 31 | 27 | Up regulated | 3.3 | 0.0184 | |
| Jones | 15 | 12 | Up regulated | 11.3 | 0.0049 | |
| Fomison-Nurse | 39 | 32 | Up regulated | - | <0.01 | |
| García-Jacobo | 54 | 35 | n.c. | - | n.s. | |
| Gok | 25 | 25 | n.c. | - | n.s. | |
| Kokkinopoulou | 40 | 37 | Down regulated | - | < 0.0001 | |
| Banerjee | 30 | 30 | Up regulated | >2 | 0.045 | |
| miR-122 | Flowers | 72 | 55 | n.c. | 1.12 | n.s. |
| Rojas | 56 | 40 | n.c. | 0.86 | n.s. | |
| Jaeger | 43 | 43 | n.c. | - | n.s. | |
| Zeinali | 30 | 30 | Up regulated | 8.61 | <0.001 | |
| Candia | 9 | 9 | Up regulated | 1.75 | >0.05 | |
| Meerson | 29 | 30 | n.c. | - | n.s. | |
| Mononen | 19 | 427 | Up regulated | 1.53 | <0.0001 | |
| Nie | 94 | 94 | Up regulated | 2.82 | 0.001 | |
| Regmi | 50 | 25 | Up regulated | - | <0.05 | |
| miR-197 | Flowers | 72 | 55 | n.c. | 1.09 | n.s. |
| Zampetaki | 80 | 80 | Down regulated | 0.456 | 0.0013 | |
| Wang | 33 | 119 | n.c. | - | n.s. | |
| Hu | 50 | 50 | n.c. | - | n.s. | |
| miR-16-5p | Ye | 40 | 40 | n.c. | 1.711 | n.s. |
| miR-99a-5P | Candia | 9 | 9 | Up regulated | 1.35 | >0.05 |
| miR-379 | Dias | 4 | 4 | n.c. | 1.64 | n.s. |
Note: n.c.: not changed and n.s.: not significant
Table 4: Summary of the Target miRNA Expression Profiles in T2DM NAFLD
| miRNA | Study | Sample size | Expression direction | Mean | p | |
|---|---|---|---|---|---|---|
| Pre-DM | Control | |||||
| miR-122 | Jaeger | 43 | 43 | n.c. | - | n.s. |
| Zeinali | 30 | 30 | Up regulated | 3.73 | <0.001 | |
| Candia | 9 | 9 | Up regulated | 2.58 | <0.01 | |
| Mononen | 207 | 427 | Up regulated | 1.14 | 0.006 | |
| miR-34a | Kong | 18 | 19 | n.c. | - | n.s. |
| Seyhan | 12 | 27 | n.c. | 0.86 | n.s. | |
| García-Jacobo | 16 | 35 | n.c. | - | n.s. | |
| miR-375 | Kong | 18 | 19 | n.c. | - | n.s. |
| Wang | 44 | 53 | Down regulated | 0.88 | <0.05 | |
| Seyhan | 12 | 27 | Down regulated | 0.65 | 0.4004 | |
| Al-Muhtaresh | 30 | 30 | Up regulated | >3 | <0.05 | |
| García-Jacobo | 16 | 35 | n.c. | - | n.s. | |
| miR-99a-5P | Candia | 9 | 9 | Up regulated | 1.74 | <0.01 |
Note: n.c.: not changed and n.s.: not significant
Table 5: Summary of the Target miRNA Expression Profiles in Pre-Diabetes
According to previous data and reports, miRNAs can be used as diagnostic markers of NAFLD. This study conducted a meta-analysis of the diagnostic miRNAs of NAFLD to determine whether they are associated with diabetes. Since different miRNAs have varying diagnostic power, we selected 10 miRNAs with a pooled AUC >0.7, which suggests moderate diagnostic accuracy. Of these, miR-122 and miR-34a were the most frequently reported miRNAs with consistent up regulation and both had a superior diagnostic power with a pooled AUC >0.85 in NAFLD. Next, we figured out whether these miRNAs can function as biomarkers of T2DM as well. The results of this study indicate that miR-122, miR-34a and miR-375 are the most promising biomarkers of both NAFLD and T2DM. miR-122 is up regulated in NAFLD, pre-diabetic and diabetic patients, and has greater potential than other miRNAs. miRNA-34a was up regulated in patients with NAFLD and diabetes, but not altered in those with prediabetes. Notably, its expression level is found to be associated with NAFLD severity[35] and it can distinguish patients with Non-Alcoholic Steatohepatitis (NASH) from those with NAFLD[24]. Several studies acknowledge that miR-375 is up regulated in diabetic individuals; however, reported data on NAFLD and pre-diabetic individuals are insufficient and inconsistent.
The functions of the target genes of these three miRNAs can partly explain our conclusion. We searched and analyzed the target genes of miR-122, miR-34a and miR-375, and their mediated biological processes in the Gene Ontology database. The target genes of these miRNAs are involved in various processes, such as cell proliferation, pancreatic β-cell development, fatty acid transport, oxidative stress and other processes. Except for clinical research, two recently published reports revealed that miR-122 and miR-34a regulate the development of NAFLD by inducing lipid absorption, adipogenesis, inflammation and apoptosis while inhibiting fatty acid oxidation in mice[73,74]. Additionally, a series of animal experiments support the modulating roles of miR-122, miR-34a and miR-375 in the pathogenesis and development of diabetes[75-77].
Although we identified miR-122, miR-34a and miR- 375 as common biomarkers of NAFLD and T2DM, they exhibit limited effectiveness in identifying diabetics among patients with NAFLD. The primary reason is that only a few studies focus on the diagnostic effectiveness of miRNA in diabetes, which makes it difficult to reach a firm and reliable conclusion. Even though the mean fold-changes in different researches differ considerably, we observe that the expression levels of miR-122 and miR-34a are lower in patients with diabetes than in patients with NAFLD, which may be associated with different stages of the metabolic syndrome. Several studies suggest that miR-375 plays a crucial role in diabetic pathophysiology; however, there are limited data concerning its role in NAFLD. Besides, a study of 90 individuals found that miR-375 had an AUC >0.7 in separating diabetic and pre-diabetic conditions from healthy controls[43]. Interestingly, numerous studies have found that miRNA panels exhibit better diagnostic performance than individual miRNAs, which provides a further studying direction for this topic[34,36,57].
The inconsistency in miRNA expression profiling studies is one of its major drawbacks[28] and the included studies had conflicting results. For example, Krauskopf et al.[58] found that miR-375 was down regulated in T2DM while other studies claimed that it was up regulated. Kokkinopoulou et al.[56] also found that miR-34a was down regulated in T2DM while other studies claimed that it was up regulated. This inconsistency may be attributed to a lack of representativeness caused by a small sample size and an unknown bias in patient selection. Considering that the few anomalous data in one or two studies shall have limited effect on the final results, we take the most consistent results as final results. The studies on miR-375 expression in pre-diabetic patients varied greatly, indicating a low level of credibility and requiring further evaluation.
Inevitably, our study has several limitations. First, even though we limited the sample type, the results of miRNA expression profiling vary largely due to different expression profile platforms, methods of detection and normalization control types[78,79]. We ranked all target miRNAs based on a vote-counting strategy and discarded data not validated by RT- qPCR to reduce the concomitant bias. Still, the levels of circulating miRNAs vary under specific physiological and pathological conditions. Second, some articles about T2DM that met inclusion criteria were excluded for not reporting the target miRNAs, which may be because the target miRNAs were not statistically significant. Undoubtedly, publication bias might affect our results to some extent. However, due to the small amount of reference literature, we did not calculate the publication bias of each target miRNA. Finally, the strength of our conclusion might also be affected by the relatively small sample size of patients with NAFLD and T2DM. Nevertheless, our results may provide a new perspective on existing controversy and for future research. MiRNAs serve as potential biomarkers of both NAFLD and T2DM. MiR-122 and miR-34a can distinguish patients with NAFLD from healthy controls. Additionally, miR- 122, miR-34a and miR-375 are common biomarkers of both NAFLD and T2DM. However, whether these miRNAs can be used for early prediction of diabetes in patients with NAFLD needs further investigation.
Conflict of interests:
The authors declared no conflict of interests.
References
- Byrne CD, Targher G. NAFLD: A multisystem disease. J Hepatol 2015;62(1):S47-64.
[Crossref] [Google Scholar] [PubMed]
- Masuoka HC, Chalasani N. Nonalcoholic fatty liver disease: An emerging threat to obese and diabetic individuals. Ann N Y Acad Sci 2013;1281(1):106-22.
[Crossref] [Google Scholar] [PubMed]
- Charlton MR, Burns JM, Pedersen RA, Watt KD, Heimbach JK, Dierkhising RA. Frequency and outcomes of liver transplantation for nonalcoholic steatohepatitis in the United States. Gastroenterology 2011;141(4):1249-53.
- Yang W, Lu J, Weng J, Jia W, Ji L, Xiao J, et al. Prevalence of diabetes among men and women in China. N Engl J Med 2010;362(12):1090-101.
[Crossref] [Google Scholar] [PubMed]
- Mostafa T, Abdel-Hamid IA. Ejaculatory dysfunction in men with diabetes mellitus. World J Diabetes 2021;12(7):954.
[Crossref] [Google Scholar] [PubMed]
- Anstee QM, Targher G, Day CP. Progression of NAFLD to diabetes mellitus, cardiovascular disease or cirrhosis. Nat Rev Gastroenterol Hepatol 2013;10(6):330-44.
[Crossref] [Google Scholar] [PubMed]
- Scorletti E, Byrne CD. Extrahepatic diseases and NAFLD: The triangular relationship between NAFLD, type 2-diabetes and dysbiosis. Dig Dis 2016;34(1):11-8.
[Crossref] [Google Scholar] [PubMed]
- Bilson J, Sethi JK, Byrne CD. Non-alcoholic fatty liver disease: A multi-system disease influenced by ageing and sex, and affected by adipose tissue and intestinal function. Proc Nutr Soc 2022;81(2):146-61.
[Crossref] [Google Scholar] [PubMed]
- Kim SS, Cho HJ, Kim HJ, Kang DR, Berry JR, Kim JH, et al. Nonalcoholic fatty liver disease as a sentinel marker for the development of diabetes mellitus in non-obese subjects. Dig Liver Dis 2018;50(4):370-7.
[Crossref] [Google Scholar] [PubMed]
- Ekstedt M, Franzén LE, Mathiesen UL, Thorelius L, Holmqvist M, Bodemar G, et al. Long-term follow-up of patients with NAFLD and elevated liver enzymes. Hepatology 2006;44(4):865-73.
[Crossref] [Google Scholar] [PubMed]
- Ballestri S, Zona S, Targher G, Romagnoli D, Baldelli E, Nascimbeni F, et al. Nonalcoholic fatty liver disease is associated with an almost twofold increased risk of incident type 2 diabetes and metabolic syndrome. Evidence from a systematic review and meta-analysis. J Gastroenterol Hepatol 2016;31(5):936-44.
[Crossref] [Google Scholar] [PubMed]
- Williams CD, Stengel J, Asike MI, Torres DM, Shaw J, Contreras M, et al. Prevalence of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis among a largely middle-aged population utilizing ultrasound and liver biopsy: A prospective study. Gastroenterology 2011;140(1):124-31.
[Crossref] [Google Scholar] [PubMed]
- Williamson RM, Price JF, Glancy S, Perry E, Nee LD, Hayes PC, et al. Prevalence of and risk factors for hepatic steatosis and nonalcoholic fatty liver disease in people with type 2 diabetes: The Edinburgh type 2 diabetes study. Diabetes Care 2011;34(5):1139-44.
[Crossref] [Google Scholar] [PubMed]
- Wu W, Xiang J, Chen X. Association between diabetes mellitus and all-cause and cardiovascular mortality among individuals with ultrasound-defined non-alcoholic fatty liver disease. Front Endocrinol 2021;12:773342.
[Crossref] [Google Scholar] [PubMed]
- Zoppini G, Fedeli U, Gennaro N, Saugo M, Targher G, Bonora E. Mortality from chronic liver diseases in diabetes. Am J Coll Gastroenterol 2014;109(7):1020-5.
[Crossref] [Google Scholar] [PubMed]
- Targher G, Bertolini L, Rodella S, Zoppini G, Lippi G, Day C, et al. Non-alcoholic fatty liver disease is independently associated with an increased prevalence of chronic kidney disease and proliferative/laser-treated retinopathy in type 2 diabetic patients. Diabetologia 2008;51(3):444-50.
[Crossref] [Google Scholar] [PubMed]
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Ann Rev Biochem 2010;79:351-79.
[Crossref] [Google Scholar] [PubMed]
- Dongiovanni P, Meroni M, Longo M, Fargion S, Fracanzani AL. miRNA signature in NAFLD: A turning point for a non-invasive diagnosis. Int J Mol Sci 2018;19(12):3966.
[Crossref] [Google Scholar] [PubMed]
- Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol 2018;141(4):1202-7.
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci 2008;105(30):10513-8.
[Crossref] [Google Scholar] [PubMed]
- Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008;18(10):997-1006.
[Crossref] [Google Scholar] [PubMed]
- Marques-Rocha JL, Samblas M, Milagro FI, Bressan J, Martínez JA, Marti A. Noncoding RNAs, cytokines, and inflammation-related diseases. FASEB J 2015;29(9):3595-611.
[Google Scholar] [PubMed]
- Suksangrat T, Phannasil P, Jitrapakdee S. miRNA regulation of glucose and lipid metabolism in relation to diabetes and non-alcoholic fatty liver disease. Adv Exp Med Biol 2019;1134:129-48.
[Crossref] [Google Scholar] [PubMed]
- Liu CH, Ampuero J, Gil-Gómez A, Montero-Vallejo R, Rojas Á, Muñoz-Hernández R, et al. miRNAs in patients with non-alcoholic fatty liver disease: A systematic review and meta-analysis. J Hepatol 2018;69(6):1335-48.
[Crossref] [Google Scholar] [PubMed]
- Musso G, Gambino R, Cassader M, Pagano G. Meta-analysis: Natural history of non-alcoholic fatty liver disease (NAFLD) and diagnostic accuracy of non-invasive tests for liver disease severity. Ann Med 2011;43(8):617-49.
[Crossref] [Google Scholar] [PubMed]
- Guo J, Lin Y, Wei J, Zhang X, Yang C, Sun J, et al. Diagnostic value of serum miR-587 in patients with metabolic syndrome. Clin Lab 2019;65(7).
[Crossref] [Google Scholar] [PubMed]
- Xin S, Zhan Q, Chen X, Xu J, Yu Y. Efficacy of serum miRNA test as a non-invasive method to diagnose nonalcoholic steatohepatitis: A systematic review and meta-analysis. BMC Gastroenterol 2020;20(1):186.
[Crossref] [Google Scholar] [PubMed]
- Gholaminejad A, Abdul Tehrani H, Gholami Fesharaki M. Identification of candidate microRNA biomarkers in diabetic nephropathy: A meta-analysis of profiling studies. J Nephrol 2018;31:813-31.
[Crossref] [Google Scholar] [PubMed]
- Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann Int Med 2011;155(8):529-36.
[Crossref] [Google Scholar] [PubMed]
- Griffith OL, Melck A, Jones SJ, Wiseman SM. Meta-analysis and meta-review of thyroid cancer gene expression profiling studies identifies important diagnostic biomarkers. J Clin Oncol 2006;24(31):5043-51.
[Crossref] [Google Scholar] [PubMed]
- Chan SK, Griffith OL, Tai IT, Jones SJ. Meta-analysis of colorectal cancer gene expression profiling studies identifies consistently reported candidate biomarkers. Cancer Epidemiol Biomarkers Prev 2008;17(3):543-52.
[Crossref] [Google Scholar] [PubMed]
- Cermelli S, Ruggieri A, Marrero JA, Ioannou GN, Beretta L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PloS One 2011;6(8):e23937.
[Crossref] [Google Scholar] [PubMed]
- Ye D, Zhang T, Lou G, Xu W, Dong F, Chen G, et al. Plasma miR-17, miR-20a, miR-20b and miR-122 as potential biomarkers for diagnosis of NAFLD in type 2 diabetes mellitus patients. Life Sci 2018;208:201-7.
[Crossref] [Google Scholar] [PubMed]
- Tan Y, Ge G, Pan T, Wen D, Gan J. A pilot study of serum microRNAs panel as potential biomarkers for diagnosis of nonalcoholic fatty liver disease. PloS One 2014;9(8):e105192.
[Crossref] [Google Scholar] [PubMed]
- Salvoza NC, Klinzing DC, Gopez-Cervantes J, Baclig MO. Association of circulating serum miR-34a and miR-122 with dyslipidemia among patients with non-alcoholic fatty liver disease. PloS One 2016;11(4):e0153497.
[Crossref] [Google Scholar] [PubMed]
- Pillai SS, Lakhani HV, Zehra M, Wang J, Dilip A, Puri N, et al. Predicting nonalcoholic fatty liver disease through a panel of plasma biomarkers and microRNAs in female West Virginia population. Int J Mol Sci 2020;21(18):6698.
[Google Scholar] [PubMed]
- Okamoto K, Koda M, Okamoto T, Onoyama T, Miyoshi K, Kishina M, et al. Serum miR-379 expression is related to the development and progression of hypercholesterolemia in non-alcoholic fatty liver disease. PLoS One 2020;15(2):e0219412.
[Crossref] [Google Scholar] [PubMed]
- Liu XL, Pan Q, Zhang RN, Shen F, Yan SY, Sun C, et al. Disease-specific miR-34a as diagnostic marker of non-alcoholic steatohepatitis in a Chinese population. World J Gastroenterol 2016;22(44):9844.
[Crossref] [Google Scholar] [PubMed]
- Jampoka K, Muangpaisarn P, Khongnomnan K, Treeprasertsuk S, Tangkijvanich P, Payungporn S. Serum miR-29a and miR-122 as potential biomarkers for non-alcoholic fatty liver disease (NAFLD). Microrna 2018;7(3):215-22.
[Crossref] [Google Scholar] [PubMed]
- Hendy OM, Rabie H, El Fouly A, Abdel-Samiee M, Abdelmotelb N, Elshormilisy AA, et al. The circulating micro-RNAs (− 122,− 34a and− 99a) as predictive biomarkers for non-alcoholic fatty liver diseases. Diabetes Metab Syndr Obes 2019;12(6):2715-23.
- Celikbilek M, Baskol M, Taheri S, Deniz K, Dogan S, Zararsiz G, et al. Circulating microRNAs in patients with non-alcoholic fatty liver disease. World J Hepatol 2014;6(8):613-20.
[Crossref] [Google Scholar] [PubMed]
- Auguet T, Aragonès G, Berlanga A, Guiu-Jurado E, Martí A, Martínez S, et al. miR33a/miR33b* and miR122 as possible contributors to hepatic lipid metabolism in obese women with nonalcoholic fatty liver disease. Int J Mol Sci 2016;17(10):1620.
[Crossref] [Google Scholar] [PubMed]
- Al-Muhtaresh HA, Al-Kafaji G. Evaluation of two-diabetes related microRNAs suitability as earlier blood biomarkers for detecting prediabetes and type 2 diabetes mellitus. J Clin Med 2018;7(2):12.
[Crossref] [Google Scholar] [PubMed]
- Ashoori MR, Rahmati-Yamchi M, Ostadrahimi A, Pahlavan-Gharebaba R, Mobasseri M, Bakhtiyari S, et al. Apelin-13 serum levels in type 2 diabetic obese women: Possible relations with microRNAs-107 and 375. Turkish J Biochem 2019;44(5):667-75.
- Baldeón Rojas L, Weigelt K, de Wit H, Ozcan B, van Oudenaren A, Sempértegui F, et al. Study on inflammation-related genes and microRNAs, with special emphasis on the vascular repair factor HGF and miR-574-3p, in monocytes and serum of patients with T2D. Diabetol Metab Syndr 2016;8(1):1-2.
[Crossref] [Google Scholar] [PubMed]
- Banerjee J, Roy S, Dhas Y, Mishra N. Senescence-associated miR-34a and miR-126 in middle-aged Indians with type 2 diabetes. Clin Exp Med 2020;20(1):149-58.
[Crossref] [Google Scholar] [PubMed]
- De Candia P, Spinetti G, Specchia C, Sangalli E, La Sala L, Uccellatore A, et al. A unique plasma microRNA profile defines type 2 diabetes progression. PloS One 2017;12(12):e0188980.
[Crossref] [Google Scholar] [PubMed]
- Dias S, Hemmings S, Muller C, Louw J, Pheiffer C. MicroRNA expression varies according to glucose tolerance, measurement platform and biological source. Biomed Res Int 2017;2017:1080157.
[Crossref] [Google Scholar] [PubMed]
- Flowers E, Gadgil M, Aouizerat BE, Kanaya AM. Circulating micrornas associated with glycemic impairment and progression in Asian Indians. Biomarker Res 2015;3(1):1-22.
[Crossref] [Google Scholar] [PubMed]
- Fomison-Nurse I, Saw EE, Gandhi S, Munasinghe PE, Van Hout I, Williams MJ, et al. Diabetes induces the activation of pro-ageing miR-34a in the heart, but has differential effects on cardiomyocytes and cardiac progenitor cells. Cell Death Differ 2018;25(7):1336-49.
[Crossref] [Google Scholar] [PubMed]
- García-Jacobo RE, Uresti-Rivera EE, Portales-Pérez DP, González-Amaro R, Lara-Ramírez EE, Enciso-Moreno JA, et al. Circulating miR-146a, miR-34a and miR-375 in type 2 diabetes patients, pre-diabetic and normal-glycaemic individuals in relation to β-cell function, insulin resistance and metabolic parameters. Clin Exp Pharmacol Physiol 2019;46(12):1092-1100.
[Crossref] [Google Scholar] [PubMed]
- Gok O, Karaali Z, Ergen A, Ekmekci SS, Abaci N. Serum sirtuin 1 protein as a potential biomarker for type 2 diabetes: Increased expression of sirtuin 1 and the correlation with microRNAs. J Res Med Sci 2019;24:56.
[Crossref] [Google Scholar] [PubMed]
- Hu Y, Li Q, Zhang L, Zhong L, Gu M, He B, et al. Serum miR-195-5p exhibits clinical significance in the diagnosis of essential hypertension with type 2 diabetes mellitus by targeting DRD1. Clinics 2021;76:e2502.
[Google Scholar] [PubMed]
- Jaeger A, Zollinger L, Saely CH, Muendlein A, Evangelakos I, Nasias D, et al. Circulating microRNAs-192 and-194 are associated with the presence and incidence of diabetes mellitus. Sci Rep 2018;8(1):14274.
- Jones A, Danielson KM, Benton MC, Ziegler O, Shah R, Stubbs RS, et al. miRNA signatures of insulin resistance in obesity. Obesity 2017;25(10):1734-44.
[Crossref] [Google Scholar] [PubMed]
- Kokkinopoulou I, Maratou E, Mitrou P, Boutati E, Sideris DC, Fragoulis EG, et al. Decreased expression of microRNAs targeting type-2 diabetes susceptibility genes in peripheral blood of patients and predisposed individuals. Endocrine 2019;66(2):226-39.
[Crossref] [Google Scholar] [PubMed]
- Kong L, Zhu J, Han W, Jiang X, Xu M, Zhao Y, et al. Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: A clinical study. Acta Diabetol 2011;48:61-9.
[Crossref] [Google Scholar] [PubMed]
- Krauskopf J, de Kok TM, Schomaker SJ, Gosink M, Burt DA, Chandler P, et al. Serum microRNA signatures as "liquid biopsies" for interrogating hepatotoxic mechanisms and liver pathogenesis in human. PloS One 2017;12(5):e0177928.
[Crossref] [Google Scholar] [PubMed]
- Meerson A, Najjar A, Saad E, Sbeit W, Barhoum M, Assy N. Sex differences in plasma microRNA biomarkers of early and complicated diabetes mellitus in Israeli Arab and Jewish patients. Noncoding RNA 2019;5(2):32.
[Crossref] [Google Scholar] [PubMed]
- Mononen N, Lyytikäinen LP, Seppälä I, Mishra PP, Juonala M, Waldenberger M, et al. Whole blood microRNA levels associate with glycemic status and correlate with target mRNAs in pathways important to type 2 diabetes. Sci Rep 2019;9(1):8887.
- Nie H, Hu H, Li Z, Wang R, He J, Li P, et al. Associations of plasma metal levels with type 2 diabetes and the mediating effects of microRNAs. Environ Pollut 2022;292:118452.
[Crossref] [Google Scholar] [PubMed]
- Lopez YO, Garufi G, Seyhan AA. Altered levels of circulating cytokines and microRNAs in lean and obese individuals with prediabetes and type 2 diabetes. Mol Biosyst 2017;13(1):106-21.
[Crossref] [Google Scholar] [PubMed]
- Regmi A, Liu G, Zhong X, Hu S, Ma R, Gou L, et al. Evaluation of serum microRNAs in patients with diabetic kidney disease: A nested case-controlled study and bioinformatics analysis. Med Sci Monit 2019;25:1699-1708.
[Crossref] [Google Scholar] [PubMed]
- Seyhan AA, Nunez Lopez YO, Xie H, Yi F, Mathews C, Pasarica M, et al. Pancreas-enriched miRNAs are altered in the circulation of subjects with diabetes: A pilot cross-sectional study. Sci Rep 2016;6(1):31479.
[Crossref] [Google Scholar] [PubMed]
- Sun K, Chang X, Yin L, Li J, Zhou T, Zhang C, et al. Expression and DNA methylation status of microRNA-375 in patients with type 2 diabetes mellitus. Mol Med Rep 2014;9(3):967-72.
[Crossref] [Google Scholar] [PubMed]
- Wang X, Chang X, Li J, Yin L, Sun K. DNA methylation of microRNA-375 in impaired glucose tolerance. Exp Ther Med 2014;8(3):775-80.
[Crossref] [Google Scholar] [PubMed]
- Wang X, Sundquist J, Zöller B, Memon AA, Palmér K, Sundquist K, et al. Determination of 14 circulating microRNAs in Swedes and Iraqis with and without diabetes mellitus type 2. PloS One 2014;9(1):e86792.
[Crossref] [Google Scholar] [PubMed]
- Wu X, Li Y, Man B, Li D. Assessing microRNA-375 levels in type 2 diabetes mellitus (T2DM) patients and their first-degree relatives with T2DM. Diabetes Metab Syndr Obes 2021;14:1445-51.
[Crossref] [Google Scholar] [PubMed]
- Ye Y, Liu X, Yan Y. Differential expression and bioinformatic analysis of microRNA in the plasma of patients with type 2 diabetes mellitus. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2021;38(6):536-40.
[Google Scholar] [PubMed]
- Yin L, Zhang T, Wei Y, Cai WJ, Feng G, Chang XY, et al. Epigenetic regulation of microRNA-375 and its role as DNA epigenetic marker of type 2 diabetes mellitus in Chinese Han population. Int J Clin Exp Pathol 2017;10(12):11986.
[Google Scholar] [PubMed]
- Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res 2010;107(6):810-7.
[Crossref] [Google Scholar] [PubMed]
- Zeinali F, Aghaei Zarch SM, Jahan-Mihan A, Kalantar SM, Vahidi Mehrjardi MY, Fallahzadeh H, et al. Circulating microRNA-122, microRNA-126-3p and microRNA-146a are associated with inflammation in patients with pre-diabetes and type 2 diabetes mellitus: A case control study. PloS One 2021;16(6):e0251697.
[Crossref] [Google Scholar] [PubMed]
- Xu Y, Zhu Y, Hu S, Pan X, Bawa FC, Wang HH, et al. Hepatocyte miR-34a is a key regulator in the development and progression of non-alcoholic fatty liver disease. Mol Metab 2021;51:101244.
[Crossref] [Google Scholar] [PubMed]
- Long JK, Dai W, Zheng YW, Zhao SP. miR-122 promotes hepatic lipogenesis via inhibiting the LKB1/AMPK pathway by targeting Sirt1 in non-alcoholic fatty liver disease. Mol Med 2019;25(1):26.
[Crossref] [Google Scholar] [PubMed]
- Erener S, Mojibian M, Fox JK, Denroche HC, Kieffer TJ. Circulating miR-375 as a biomarker of β-cell death and diabetes in mice. Endocrinology 2013;154(2):603-8.
[Crossref] [Google Scholar] [PubMed]
- Wei S, Zhang M, Yu Y, Xue H, Lan X, Liu S, et al. HNF-4α regulated miR-122 contributes to development of gluconeogenesis and lipid metabolism disorders in type 2 diabetic mice and in palmitate-treated HepG2 cells. Eur J Pharmacol 2016;791:254-63.
[Crossref] [Google Scholar] [PubMed]
- Su T, Hou J, Liu T, Dai P, Qin L, Ding L, et al. MiR-34a-5p and miR-452-5p: The novel regulators of pancreatic endocrine dysfunction in diabetic Zucker rats? Int J Med Sci 2021;18(14):3171.
[Crossref] [Google Scholar] [PubMed]
- Sato F, Tsuchiya S, Terasawa K, Tsujimoto G. Intra-platform repeatability and inter-platform comparability of microRNA microarray technology. PloS One 2009;4(5):e5540.
[Crossref] [Google Scholar] [PubMed]
- Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, et al. Serum microRNAs are promising novel biomarkers. PloS One 2008;3(9):e3148.
[Crossref] [Google Scholar] [PubMed]