- *Corresponding Author:
- Zhuandi Lin
Department of Respiratory Medicine, Intensive Care Unit, Panyu Central Hospital, Guangzhou, Guangdong Province 511400, China
E-mail: 13710911343@163.com
| This article was originally published in a special issue, “Innovations in Biomedical Research and Drug Development” |
| Indian J Pharm Sci 2023:85(3) Spl Issue “192-198” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Sepsis is associated with acute respiratory distress syndrome, which has a significant impact on the prognosis of patients. Several studies have shown that the microbiota plays a significant role in sepsisinduced acute respiratory distress syndrome, but the relationship between the microbiota and sepsisinduced acute respiratory distress syndrome is not fully understood. We conducted a case-control and single-center study in 19 sepsis-induced acute respiratory distress syndrome patients and 36 sepsis-noninduced acute respiratory distress syndrome patients to investigate the clinical features and microbiota expression. There were 55 subjects enrolled, 19 of whom suffered acute respiratory distress syndrome due to sepsis. A significant increase in the abundance of Pseudomonas aeruginosa, leptospiral virus, Cytomegalovirus, Klebsiella pneumoniae, Streptococcus pneumoniae, Candida albicans, Escherichia coli, Epstein-Barr virus and Staphylococcus aureus. Besides, expressions of peripheral T lymphocytes (cluster of differentiation 3+, cluster of differentiation 4+ and cluster of differentiation 3+, cluster of differentiation 8+) was much higher in the sepsis-induced acute respiratory distress syndrome group than that in the sepsis non-induced acute respiratory distress syndrome group. The acute physiology and chronic health evaluation II scores, duration of mechanical ventilation and mortality with 28 d and 90 d were much higher in the sepsis-induced acute respiratory distress syndrome group. Patients with sepsis-induced acute respiratory distress syndrome had worse clinical outcomes and a higher expression of peripheral T lymphocytes, as well as the relative abundance of microbiota dysbiosis.
Keywords
Microbiome, sepsis, acute respiratory distress syndrome, pulmonary infection
Acute Respiratory Distress Syndrome (ARDS) or Acute Lung Injury (ALI) is a life-threatening condition characterized by respiratory failure[1]. As one of the primary causes of death in critically ill patients, ALI/ARDS is one of the most common clinical critical illnesses with rapid onset and high mortality[2]. It is estimated that more than three million patients are diagnosed with ARDS worldwide every year, accounting for 10 % of the entire number of people who are admitted to Intensive Care Unit (ICU)[3]. While major advances have been made in the treatment of ARDS, including the use of Extracorporeal Membrane Oxygenation (ECMO), protective lung ventilation maneuvers and statins, the mortality from ARDS remains high[4]. A primary cause of ALI/ARDS in the ICU is sepsis and the lung is the most vulnerable organ during the course of sepsis[5]. The direct cause of sepsis-induced ALI/ARDS is pulmonary infection, while the indirect cause is extra pulmonary infection[6]. Although the mechanism by which sepsis induces ALI and ARDS is complex and multifactorial, we understand that gut microbiome plays a significant role[7].
A microbiome is a community of microorganisms including bacteria, viruses, fungi and archaea, that colonize the body[8]. A number of recent studies have demonstrated the close relationship between microbiota dysbiosis and multiple diseases, such as cardiovascular, digestive, and respiratory diseases[9]. It has also been discovered that the microbiota and its metabolites contribute to the occurrence, development, treatment and prognosis of sepsis[10]. A potential therapeutic target in sepsis is the microbiome, which modulates several responses to sepsis[11]. An individual's gut microbiome can be colonized with an entire donor microbiome by fecal microbiota transplant. As a result, fecal microbiota transplantation may enhance microbiome reconstitution and impact sepsis outcomes in several ways, including through the stimulation of short- chain fatty acid production and the modulation of the immune system[12]. It was found that healthy lungs also contain microbiomes and the microbiome of the lungs changes depending on the severity of the disease. After passing through the gut-lung axis, the intestinal and pulmonary microbiomes of severe patients exhibit obvious changes. These changes have an impact on sepsis-associated ARDS[13]. In spite of this, there is a lack of understanding regarding the relationship between microbiota and ARDS caused by sepsis.
In this study, we examined whether the sepsis induced ARDS group differed in clinical characteristics and microbiota at the time of diagnosis from the sepsis induced ARDS group, and to provide valuable evidence to help prevent and treat sepsis- induced ARDS.
Materials and Methods
General information:
We conducted a single-center case-control study at Panyu Central Hospital between September 1st 2020 and December 31st 2021. The study was conducted in accordance with the Declaration of Helsinki and was approved by the Hospital's Ethics Committee. Informed consent was obtained from all included patients before participation in the study.
Inclusion criteria: Age ≥18 y; a diagnosis of sepsis 3.0 criteria and ARDS was made.
Exclusion criteria: Terminal stage of chronic disease; acute pancreatitis but no definite infection; pregnant and lactating women; diagnosis of hematological, immunological diseases; treatment with immunosuppressant’s and chemotherapy agents.
The diagnostic criteria of sepsis associated ARDS:
Sepsis and ARDS were required criteria for enrollment[14]. According to the 3rd International Consensus Definitions for Sepsis and Septic Shock (sepsis-3), patients were diagnosed with sepsis[15]. ARDS is defined as acute onset of illness that meets all of the three criteria; an arterial partial Pressure of Oxygen (PaO2) to Fraction of inspired Oxygen (FiO2) ratio of not more than 300; the chest radiograph showed bilateral infiltrates consistent with pulmonary edema, without evidence of left atrial hypertension; and using an endotracheal tube to receive positive pressure mechanical ventilation[16]. To identify potential study participants and study researcher screen participating ICUs.
Collecting data:
Data were extracted as follows; demographics such as gender and age, Acute Physiology and Chronic Health Evaluation II (APACHE II) score, Sequential Organ Failure Assessment (SOFA) score, infection sites, history of smoking, history of drinking, tracheotomy, mechanical ventilation, duration of mechanical ventilation, comorbidity, infection sites, mortality within 28 d or 90 d medications. Expressions of peripheral T lymphocytes (Cluster of Differentiation (CD3+, CD4+, CD3+, CD8+ and ThTs) and metagenomic next-generation sequencing for the detection of pathogens in bronchoalveolar lavage fluid. APACHE II and SOFA score was calculated using variables obtained 24 h prior to admission.
Macrogenome extraction, preservation and sequencing:
Deoxyribonucleic Acid (DNA) was extracted from pretreated sputum samples using the FastDNA™ SPIN kit (see kit instructions for extraction steps). DNA was preserved by elution with 80 μl Drug- Eluting Stents (DES) and the extracted DNA was subjected to electrophoresis and DNA extraction was successful if bright bands were produced by electrophoresis. Store the extracted DNA at -20° in the refrigerator. The DNA stored at -20° is sequenced simultaneously: Primers are designed according to the conserved regions and the sequencing junction is added at the end of the primers, Polymerase Chain Reaction (PCR) amplification is performed and the products are purified, quantified and homogenized to form a sequencing library. The raw image data files obtained from high-throughput sequencing are transformed into raw sequencing sequences by base identification analysis, and the results are stored in FASTQ file format.
Statistics analysis:
Statistical Package for the Social Sciences (SPSS) 22.0 was used to process the statistics of all data. Frequencies and percentages (n, %) were used to express categorical variables, and a comparison between groups was performed using either the Chi-Square (χ2) test or Fisher's exact test. Continuous variables were expressed as mean±standard deviation and two independent samples t-tests were used to compare the two groups. As statistically significant, p<0.05 was considered.
Results and Discussion
Study participants included 55 patients, with 19 suffering from sepsis-induced ARDS and 36 suffering from non-induced sepsis-induced ARDS. There was no statistical difference in age, gender, SOFA score, history of smoking and drinking, comorbidity, infection sites, hospitalization time and medications. However, the APACHE II score, the rate of tracheotomy and mechanical ventilation, and mortality within 28 d or 90 d were higher in the sepsis-induced ARDS group compared with the sepsis non-induced ARDS group as shown in Table 1.
| Characteristics | Sepsis-induced ARDS (n=19) | Sepsis non-induced ARDS (n=36) | p |
|---|---|---|---|
| Age (years) | 67.00±15.04 | 69.02±15.67 | 0.64 |
| Males | 14 | 19 | 0.13 |
| APACHE II score | 30.31±7.75 | 24.14±6.37 | 0.002 |
| SOFA score | 10.42±3.04 | 10.63±3.51 | 0.82 |
| History of smoking | 9 | 11 | 0.21 |
| History of drinking | 5 | 6 | 0.39 |
| Tracheotomy | 10 | 5 | 0.002 |
| Mechanical ventilation | 17 | 16 | 0.001 |
| Duration of mechanical ventilation (day) | 11.42±6.03 | 7.80±5.61 | 0.02 |
| Comorbidity | |||
| Cancer | 3 | 4 | 0.62 |
| Diabetes | 3 | 9 | 0.51 |
| Cardiovascular disease | 9 | 21 | 0.56 |
| Respiratory disorders | 3 | 6 | |
| Liver cirrhosis | 1 | 3 | |
| Autoimmune diseases | 1 | 3 | |
| Others | 10 | 22 | |
| Infection sites | |||
| Pulmonary infection | 16 | 28 | 0.57 |
| Abdominal infection | 3 | 10 | |
| Blood infection | 4 | 5 | |
| Urinary tract infection | 0 | 3 | 0.27 |
| Clinical outcomes | |||
| Mortality within 28 d | 11 | 7 | 0.004 |
| Mortality within 90 d | 11 | 8 | 0.008 |
| Hospitalization time (days) | 22.66±19.89 | 19.41±15.21 | 0.51 |
| Medications | |||
| Glucocorticoids | 5 | 10 | 0.9 |
| Immunomodulation drugs | 3 | 1 | 0.07 |
Table 1: The Characteristics of Patients Enrolled in Our Study
Only 12 sepsis-induced ARDS patients and 15 sepsis non-induced ARDS patients have detected the peripheral T lymphocytes. We next compared the expression of peripheral T lymphocytes of sepsis patients with or without ARDS. As shown in Table 2, the expression of peripheral T lymphocytes of sepsis patients with ARDS differed from those of patients without ARDS. The percentage of CD3+, CD4+, CD3+, CD8+ and ThTs were much higher in the patients with ARDS.
| Characteristics | Sepsis-induced ARDS (n=12) | Sepsis non- induced ARDS (n=15) | p |
|---|---|---|---|
| CD3+ CD4+ (%) | 62.72±5.81 | 50.65±16.28 | 0.01 |
| CD3+ CD8+ (%) | 42.62±11.54 | 31.72±14.07 | 0.005 |
| ThTs (%) | 2.05±1.39 | 2.29±1.54 | 0.02 |
Table 2: The Characteristics of Patients Enrolled in Our Study
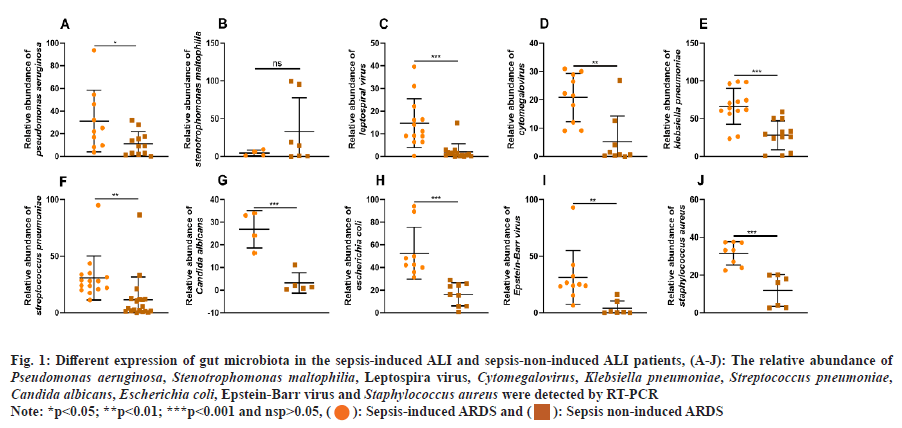
Then, the differences in lung microbiota between the sepsis-induced ARDS group and the sepsis-non- induced ARDS group were investigated. Based on the abundance of Stenotrophomonas maltophilia, there were no significant differences between the two groups (fig. 1A). As shown in fig. 1B-fig. 1J, the relative abundance of Pseudomonas aeruginosa, leptospiral virus, Cytomegalovirus, Klebsiella pneumoniae, Streptococcus pneumoniae, Candida albicans, Escherichia coli, Epstein-Barr virus and Staphylococcus aureus were significantly higher in the sepsis-induced ARDS group than in the sepsis-non-induced ARDS group.
Fig. 1: Different expression of gut microbiota in the sepsis-induced ALI and sepsis-non-induced ALI patients, (A-J): The relative abundance of Pseudomonas aeruginosa, Stenotrophomonas maltophilia, Leptospira virus, Cytomegalovirus, Klebsiella pneumoniae, Streptococcus pneumoniae, Candida albicans, Escherichia coli, Epstein-Barr virus and Staphylococcus aureus were detected by RT-PCR
Note: *p<0.05; **p<0.01; ***p<0.001 and nsp>0.05, (): Sepsis-induced ARDS and (
): Sepsis non-induced ARDS
This case-control study analyzed factors related to sepsis-induced ARDS based on the clinical characteristics and abundance of lung microflora. There were a total of 19 cases with sepsis-induced ARDS and 36 cases with sepsis-non-induced ARDS. The results of this research have revealed that there were several factors independently associated with an increased risk of sepsis-induced ARDS, including lung microorganism, peripheral T lymphocytes, APACHE II scores, duration of mechanical ventilation and mortality with 28 d and 90 d.
In a seminal study by Johanson et al., decades before the dawn of high-throughput sequencing, they showed that critical illness alters the ecosystem of the body's microbiota, confirming what was shown in previous studies[17]. As a result of their illness, patients staying in hospitals will have a different microbiota than those who are not in hospitals. In the event of a critical illness, the physiology of the host is substantially altered, which alters the conditions in the environment, as well as the structure of the communities of microbes that reside there.
Previous culture-dependent studies suggest a relationship between respiratory tract bacteria and ARDS pathophysiology. Pneumonia occurs far more frequently in ARDS patients than in other mechanically ventilated patients[18]. There is evidence that pulmonary exposure to bacterial factors (e.g. Lipopolysaccharide (LPS) and flagellin) contributes to ARDS-type inflammation and injury. In studies conducted on mechanically ventilated patients without culture, the previously unappreciated complexity of the respiratory microbiome was revealed. Animal experiments recently showed that direct lung injury alters the lung microbiome composition in a similar way to culture-independent studies[19]. Earlier studies suggested that disorder of the lung microbiome, as identified by the presence of Proteobacteria phylum (commonly occurring in the microbiome of patients with inflammatory lung conditions), was significantly associated with elevated alveolar concentrations of Tumour Necrosis Factor-Alpha (TNF-α)[20]. By contrast, decreased alveolar concentrations of TNF-α were associated with increased levels of Bacteroidota in the lung microbiomes of healthy subjects, which is the most abundant phylum in the lung microbiomes of healthy individuals[21].
According to a study published in 2016, 10.4 % of all patients who were admitted to an ICU and 23.4 % of all patients who were ventilated in an ICU suffered from ARDS[22]. Although there is still no consensus as to what a healthy lung microbiome is, there does seem to be an increased mortality rate linked to ARDS, with as many as 43 % of patients dying due to the illness[23]. Patients with healthy lungs are often found to have specific microorganisms residing in their lungs[24]. Nonetheless, in clinical practice, it seems to be difficult for microorganisms of this type to be detected. There were almost 40 % of bronchoalveolar lavage cultures that were negative in critically ill patients who underwent mechanical ventilation. However, NGS showed a positive result in patients undergoing mechanical ventilation[25]. A lower alpha diversity not only indicates poor lung health, but it also signals that the patient will eventually need to be ventilated invasively at some point in their lives[26]. A significant difference was found between patients with ventilated ARDS and patients without ventilated ARDS when the alpha diversity of their Bronchoalveolar Lavage (BAL) cultures was also positive, as well as a high dominance of one bacterial species (>50 %)[27]. In a study by Kyo et al.,[26] it was demonstrated that in ARDS patients, there is a decrease in alpha diversity of the lung microbiome, which is associated with increased mortality in the hospital. A study also revealed that patients with ARDS frequently had Betaproteobacteria, Staphylococcus, Streptococcus and Enterobacteriaceae in abundance, which may play a crucial role in the pathophysiology of patients with ARDS[26]. An ARDS is more likely to occur in patients who have suffered a trauma caused by smoking cigarettes. As compared to non-smokers, smokers have a microbiome that contains a higher abundance of bacteria such as Streptococcus, Fusobacterium, Prevotella, Haemophilus and Treponema[28]. The microbiome of the lungs has been significantly altered by smoking. An experiment carried out on mice showed that mice exposed to smoke for 2 h every day for 90 d were more likely to have higher levels of Staphylococcus, Acinetobacter and Bacillus, all of which are considered pathogenic microorganisms[29]. Last but not least, it is extremely important to distinguish between microorganisms that cause diseases and those that are only a part of the infection process. It has been shown that in pneumonia patients with a positive BAL culture for Methicillin-Resistant Staphylococcus aureus (MRSA), often this is just colonization without disease value, as it was shown in the case of pneumonia patients. In a clinical setting, it is much harder to interpret microbiome data due to the complexity of the interpretation process[30]. However, our study found that no relationship between smoking and sepsis induced ARDS.
As part of the host, the gut microbiome coexists harmoniously with the host and contributes to a wide range of beneficial functions, such as influencing immunity, maintaining homeostasis and many others[31]. It has become increasingly evident that the gut microbiome plays an important role in the occurrence, development and outcome of septic[32]. The gut barrier integrity is significantly compromised during sepsis[10], As a result, intact microbes and microbiota products are able to translocate, contributing to exacerbated systemic inflammation and multi-organ failure[33]. By deteriorating the systemic inflammatory response and by potentiating ALI/ARDS, this imbalanced interaction between the gut barrier, immune system, endogenous microorganisms, and lung may result in a systemic inflammatory response. This means that a better understanding of gut microbiome in sepsis-related ALI/ARDS is likely to contribute to the development of new therapeutic approaches. There has been evidence published that patients with sepsis have a greater abundance of intestinal microbiota, which is closely related to inflammation caused by Parabacteroides, Clostridium, Bilophila and other organisms. Additionally, researchers have found an increased number of Enterococcus and other pathogenic bacteria in sepsis patients who have died, suggesting that these bacteria may be useful biomarkers for ICU management[34]. Moreover, septic patients have a reduced abundance of Faecalibacterium which is associated with a reduced degree of intestinal inflammation[35]. According to a case control study conducted at a single center, the abundance of the following 13 bacteria was significantly higher among septic children than among healthy control children. A positive correlation was observed between C-Reactive Protein (CRP) and White Blood Cell (WBC) levels and the abundance of several bacteria, including Enterococcaceae, Enterococcus and Enterococcus durans[36]. Besides, an earlier study found that sepsis-induced cardiomyopathy was associated with higher levels of Cronobacter and Cronobacter phage abundance, and both Cronobacter and Cronobacter phage had a good predictive ability for sepsis-induced cardiomyopathy, according to the receiver operating characteristic curve[37]. In this study, we revealed the different species of gut bacteria between the sepsis-induced ARDS group and the sepsis non-induced ARDS group and proposed that the abundance of Escherichia coli associated with an increased risk of sepsis-induced ARDS.
It is important to note, however, that this study has some limitations. The study was conducted at a single center with a small sample size and a large multicenter cohort study is needed to confirm our findings in more detail. Due to the dynamic and temporally heterogeneous clinical and biological profiles of sepsis and ARDS, our study of BAL specimens from patients with established ARDS may not be able to accurately predict the microbiology of the ARDS early stages as well as developing stages based on the microbiology of the specimens from patients with established ARDS. Additionally, the role of microbiota and its metabolites on the sepsis need to be investigated in the future. It remains unknown whether ubiquitous antibiotic exposure will affect the lung microbiota in humans with ARDS. The dysregulation of the host’s response to critical illness can only be understood in its complex context. There will need to be further studies conducted to determine whether lung microbiota play a role in the onset, augmentation and perpetuation of ARDS.
In conclusion, we have found that there were several factors independently associated with sepsis- induced ARDS, including a higher expression of peripheral T lymphocytes, as well as the relative abundance of microbiota dysbiosis.
Funding:
This work was supported by Science and Technology Project of Panyu District (2020-Z04-085).
Conflict of interests:
The authors declared no conflict of interests.
References
- Meyer NJ, Gattinoni L, Calfee CS. Acute respiratory distress syndrome. Lancet 2021;398(10300):622-37.
[Crossref] [Google Scholar] [PubMed]
- Butt Y, Kurdowska A, Allen TC. Acute lung injury: A clinical and molecular review. Arch Pathol Lab Med 2016;140(4):345-50.
[Crossref] [Google Scholar] [PubMed]
- Brower RG, Ware LB, Berthiaume Y, Matthay MA. Treatment of ARDS. Chest 2001;120(4):1347-67.
- Schneider J, Sweberg T. Acute respiratory failure. Crit Care Clin 2013;29(2):167-83.
[Crossref] [Google Scholar] [PubMed]
- Zhou X, Liao Y. Gut-lung crosstalk in sepsis-induced acute lung injury. Front Microb 2021;12:779620.
[Crossref] [Google Scholar] [PubMed]
- Li W, Li D, Chen Y, Abudou H, Wang H, Cai J, et al. Classic signaling pathways in alveolar injury and repair involved in sepsis-induced ALI/ARDS: New research progress and prospect. Dis Markers 2022;2022:6362344.
- Vincent JL, Sakr Y, Groeneveld J, Zandstra DF, Hoste E, Malledant Y, et al. ARDS of early or late onset: Does it make a difference? Chest 2010;137(1):81-7.
[Crossref] [Google Scholar] [PubMed]
- Cresci GA, Bawden E. Gut microbiome: What we do and don't know. Nutr Clin Pract 2015;30(6):734-46.
[Crossref] [Google Scholar] [PubMed]
- Patterson E, Ryan PM, Cryan JF, Dinan TG, Ross RP, Fitzgerald GF, et al. Gut microbiota, obesity and diabetes. Postgrad Med J 2016;92(1087):286-300.
[Crossref] [Google Scholar] [PubMed]
- Adelman MW, Woodworth MH, Langelier C, Busch LM, Kempker JA, Kraft CS, et al. The gut microbiome’s role in the development, maintenance and outcomes of sepsis. Crit Care 2020;24(1):278.
[Crossref] [Google Scholar] [PubMed]
- Kullberg RF, Wiersinga WJ, Haak BW. Gut microbiota and sepsis: From pathogenesis to novel treatments. Curr Opin Gastroenterol 2021;37(6):578-85.
[Crossref] [Google Scholar] [PubMed]
- Wei Y, Yang J, Wang J, Yang Y, Huang J, Gong H, et al. Successful treatment with fecal microbiota transplantation in patients with multiple organ dysfunction syndrome and diarrhea following severe sepsis. Crit Care 2016;20(1):322.
[Crossref] [Google Scholar] [PubMed]
- Dickson RP, Singer BH, Newstead MW, Falkowski NR, Erb-Downward JR, Standiford TJ, et al. Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat Microbiol 2016;10(1):16113.
[Crossref] [Google Scholar] [PubMed]
- Truwit JD, Bernard GR, Steingrub J, Matthay MA, Liu KD, Albertson TE, et al. Rosuvastatin for sepsis-associated acute respiratory distress syndrome. N Engl J Med 2014;370(23):2191-200.
[Crossref] [Google Scholar] [PubMed]
- Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, et al. The third international consensus definitions for sepsis and septic shock (sepsis-3). JAMA 2016;315(8):801-10.
- Bernard GR, Artigas A, Brigham KL, Carlet J, Falke K, Hudson L, et al. Report of the American-European consensus conference on ARDS: Definitions, mechanisms, relevant outcomes and clinical trial coordination. Intensive Care Med 1994;20:225-32.
[Crossref] [Google Scholar] [PubMed]
- Johanson WG, Pierce AK, Sanford JP. Changing pharyngeal bacterial flora of hospitalized patients: Emergence of gram-negative bacilli. New Engl J Med 1969;281(21):1137-40.
[Crossref] [Google Scholar] [PubMed]
- Alverdy JC, Aoys E, Moss GS. Total parenteral nutrition promotes bacterial translocation from the gut. Surgery 1988;104(2):185-90.
[Google Scholar] [PubMed]
- Zaborina O, Lepine F, Xiao G, Valuckaite V, Chen Y, Li T, et al. Dynorphin activates quorum sensing quinolone signaling in Pseudomonas aeruginosa. PLoS Pathog 2007;3(3):e35.
[Crossref] [Google Scholar] [PubMed]
- Albenberg L, Esipova TV, Judge CP, Bittinger K, Chen J, Laughlin A, et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology 2014;147(5):1055-63.
[Crossref] [Google Scholar] [PubMed]
- Dickson RP. The microbiome and critical illness. Lancet Respir Med 2016;4(1):59-72.
[Crossref] [Google Scholar] [PubMed]
- Bellani G, Laffey JG, Pham T, Fan E, Brochard L, Esteban A, et al. Epidemiology, patterns of care and mortality for patients with acute respiratory distress syndrome in intensive care units in 50 countries. JAMA 2016;315(8):788-800.
[Crossref] [Google Scholar] [PubMed]
- Bos LD, Ware LB. Acute respiratory distress syndrome: Causes, pathophysiology and phenotypes. Lancet 2022;400:11245-56.
- Smith AD, Zhang Y, Barber RC, Minshall CT, Huebinger RM, Allen MS. Common lung microbiome identified among mechanically ventilated surgical patients. PLoS One 2016;11(11):e0166313.
[Crossref] [Google Scholar] [PubMed]
- Schmitt FC, Lipinski A, Hofer S, Uhle F, Nusshag C, Hackert T, et al. Pulmonary microbiome patterns correlate with the course of disease in patients with sepsis-induced ARDS following major abdominal surgery. J Hosp Infect 2020;105(3):438-46.
[Crossref] [Google Scholar] [PubMed]
- Kyo M, Nishioka K, Nakaya T, Kida Y, Tanabe Y, Ohshimo S, et al. Unique patterns of lower respiratory tract microbiota are associated with inflammation and hospital mortality in acute respiratory distress syndrome. Respir Res 2019;20(1):246.
[Crossref] [Google Scholar] [PubMed]
- Kitsios GD, Fitch A, Manatakis DV, Rapport SF, Li K, Qin S, et al. Respiratory microbiome profiling for etiologic diagnosis of pneumonia in mechanically ventilated patients. Front Microbiol 2018;9:1413.
[Crossref] [Google Scholar] [PubMed]
- Panzer AR, Lynch SV, Langelier C, Christie JD, McCauley K, Nelson M, et al. Lung microbiota is related to smoking status and to development of acute respiratory distress syndrome in critically ill trauma patients. Am J Respir Crit Care Med 2018;197(5):621-31.
[Crossref] [Google Scholar] [PubMed]
- Li KJ, Chen ZL, Huang Y, Zhang R, Luan XQ, Lei TT, et al. Dysbiosis of lower respiratory tract microbiome are associated with inflammation and microbial function variety. Respir Res 2019;20(1):1-6.
- Kawanami T, Yatera K, Yamasaki K, Noguchi S, Fukuda K, Akata K, et al. Clinical impact of methicillin-resistant Staphylococcus aureus on bacterial pneumonia: Cultivation and 16S ribosomal RNA gene analysis of bronchoalveolar lavage fluid. BMC Infect Dis 2016;16(1):155.
[Crossref] [Google Scholar] [PubMed]
- Morais LH, Schreiber IV HL, Mazmanian SK. The gut microbiota–brain axis in behaviour and brain disorders. Nat Rev Microbiol 2021;19(4):241-55.
[Crossref] [Google Scholar] [PubMed]
- Sun J, Zhang J, Wang X, Ji F, Ronco C, Tian J, et al. Gut-liver crosstalk in sepsis-induced liver injury. Crit Care 2020;24(1):1-8.
[Crossref] [Google Scholar] [PubMed]
- Dickson RP, Schultz MJ, van der Poll T, Schouten LR, Falkowski NR, Luth JE, et al. Lung microbiota predict clinical outcomes in critically ill patients. Am J Respir Crit Care Med 2020;201(5):555-63.
[Crossref] [Google Scholar] [PubMed]
- Agudelo-Ochoa GM, Valdés-Duque BE, Giraldo-Giraldo NA, Jaillier-Ramírez AM, Giraldo-Villa A, Acevedo-Castaño I, et al. Gut microbiota profiles in critically ill patients, potential biomarkers and risk variables for sepsis. Gut Microbes 2020;12(1):1707610.
[Crossref] [Google Scholar] [PubMed]
- Lankelma JM, van Vught LA, Belzer C, Schultz MJ, van der Poll T, de Vos WM, et al. Critically ill patients demonstrate large interpersonal variation in intestinal microbiota dysregulation: A pilot study. Intensive Care Med 2017;43(1):59-68.
[Crossref] [Google Scholar] [PubMed]
- Liu J, Wang M, Chen W, Ma J, Peng Y, Zhang M, et al. Altered gut microbiota taxonomic compositions of patients with sepsis in a pediatric intensive care unit. Front Pediatr 2021;9:645060.
[Crossref] [Google Scholar] [PubMed]
- Deng F, Chen Y, Sun QS, Lin ZB, Min Y, Zhao BC, et al. Gut microbiota dysbiosis is associated with sepsis-induced cardiomyopathy in patients: A case‒control study. J Med Virol 2023;95(1):e28267.
[Crossref] [Google Scholar] [PubMed]