- *Corresponding Author:
- Jingwei Zhang
Department of Breast and Thyroid Surgery, Zhongnan Hospital, Wuhan University, Wuhan, Hubei 430071, China
E-mail: zn001213@whu.edu.cn
| This article was originally published in a special issue, “Advanced Targeted Therapies in Biomedical and Pharmaceutical Sciences” |
| Indian J Pharm Sci 2023:85(1) Spl Issue “182-191” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
In this study, it was reported that basement membrane-associated long non-coding ribonucleic acid predicts prognosis and effectively enhances the individualized treatment of breast cancer by effectively identifying hot and cold tumors. Breast cancer transcriptional sequencing data were downloaded from The Cancer Genome Atlas and searched for prognostic long non-coding ribonucleic acids associated with basement membrane by univariate Cox regression and co-expression analysis. The least absolute shrinkage and selection operator analysis was employed to investigate the long non-coding ribonucleic acid prognostic model that is related to the basement membrane. Then, the following analyses were used to validate and evaluate the model, including univariate Cox regression, Kaplan-Meier analysis, multivariate Cox regression, receiver operating characteristic curve, calibration curves and nomogram. Immunoassay, principal component analysis, immunocytometric analysis and half-maximal inhibitory concentration analysis were conducted on the risk groups. To distinguish cold and hot tumors in terms of drug immunotherapy sensitivity, all inflammation-related long non-coding ribonucleic acids were divided into two groups. A model containing 4 basement membranerelated long non-coding ribonucleic acids was developed in this study. The area under the receiver operating characteristic curve was 0.742, 0.759 and 0.840 for 1, 2 and 3 y, respectively. High-risk patients were associated with tumor invasion and immunity and had a high immune infiltration status. Immune cells and checkpoints were infiltrated and activated in the high-risk group. Hot and cold tumors could be effectively distinguished by tumor clusters. Among the two clusters, cluster 2 was identified as hot tumors, which indicated that they were more sensitive to immunotherapeutic agents. This study provides evidence to support the hypothesis that basement membrane-related long non-coding ribonucleic acids can accurately predict patient prognosis and differentiate between hot and cold tumors. As a result, individualized immunotherapy for breast cancer patients will be improved and patients will have access to new treatment options.
Keywords
Breast cancer, basement membrane, long non-coding ribonucleic acid, Kaplan-Meier analysis, immunotherapy
Breast Cancer (BC) is essentially a malignant tumor that can be attributed to the deterioration of epithelial tissue lesions in the breast. About 1.4 million people worldwide are diagnosed with BC, resulting in about 500 000 deaths each year[1]. As the greatest threat to women’s lives, BC has the highest incidence and second highest mortality rate among the female population[2,3].
Nowadays, BC has become the most widely observed cancer among women across the world and about 2.3 million new cases were estimated to be diagnosed by 2020[4]. Despite significant progress in treatment such as anti-cancer drugs, radiation, surgery and diagnostics have lowered the cancer-related mortality where clinicians still need to address the challenge of cancer recurrence, metastasis and death induced by treating resistance[5,6]. Therefore, there is a need to uncover the pathogenesis of BC and to discover effective new biomarkers for its diagnosis, treatment and prognosis. The Basement Membrane (BM) is a specific Extracellular Matrix (ECM) that inhibits the proliferation of cancer cells to distant sites[7]. Dynamic remodeling of ECM is often involved in cancer development[8]. The Tumor Microenvironment (TME) includes various soluble growth factors, ECM and Cellular Components (CC), and has a close relation with tumor progression. Additionally, the BM is a key histological boundary distinguishing the invasive tumors and non-invasive (carcinoma in situ) tumors. BM damage leads to exacerbated local metastasis, as well as invasion of tumor cells. Cell migration could be put under strict regulation by means of BMs. Herein, the breakdown is an important step of tumor progression.
Long non-coding Ribonucleic Acids (lncRNAs) are a specific group of RNA molecules with a size of 200 nucleotides (nt), which regulate gene expression[9]. In addition to the gene regulation, lncRNAs are also involved in different Biological Processes (BP) related to the occurrence, development and metastasis of tumors[10]. Currently, there are few studies on BM-associated lncRNAs. Increasing evidence supports that lncRNAs play a critical role in occurrence and development of cancer. It has been shown that, Plasmacytoma Variant Translocation 1 (PVT1) lncRNAs are involved in the BC progression by promoting proliferation and metastasis of Breast Cancer Cells (BCCs)[11]. A recent study showed that lncRNA Growth Arrest‑Specific Transcript 5 (GAS5) has a stimulation effect on the apoptosis of BCC through multiple pathways, including mitochondrial signaling pathways and cell apoptosis receptors, and that GAS5 also serves as a regulator of several key signaling pathways (example: Winglessrelated integration site (Wnt)/beta (β)-catenin, Phosphoinositide 3 Kinase (PI3K)/Protein Kinase B (Akt)/mammalian Target of Rapamycin (mTOR) and Nuclear Factor kappa B (NF-κB) signaling) in BC[12]. In this study, a prognostic profile of differentially expressed BM-associated lncRNAs was developed based on The Cancer Genome Atlas (TCGA) data. Additionally, the roles of BM-associated messenger RNA (mRNA) and immune response in the prognosis of BC were investigated.
Materials and Methods
Data collection:
The RNA sequencing (RNA-seq) dataset was extracted from the TCGA database[13], including 344 BC specimens and 41 normal breast specimens and clinical data from 1097 cases of BC that were retrieved from the TCGA database. In a study that was recently conducted, 224 Basement Membrane Genes (BMGs) were identified[14]. Pearson’s correlation analysis was conducted to assess the correlation of BM lncRNAs and BC, which was considered significant if p<0.001 when the correlation coefficient was |r2|>0.4. Clinicopathological data including age, gender, grade, stage, Tumor, Nodes and Metastases (TNM) and survival status and time were collected from BC patients, False Discovery Rate (FDR)<0.05, |log2 Fold Change (FC)|≥1 for significantly differential expression of BMassociated lncRNAs was considered. We explored the functions of up-regulation and down-regulation of BM-associated Differentially Expressed Genes (DEGs) and then, we used the Gene Ontology (GO) to evaluate the DEG-associated biological pathways. On the basis of the Kyoto Encyclopedia of Genes and Genomes (KEGG) data, the R software grammar of graphics plot 2 (ggplot2) package was employed for functional analysis of CC, Molecular Functions (MF) and BP, which were under regulation by differentially expressed BM genes.
Development of the BM-related lncRNAs prognostic signature:
Univariate Cox (uniCox) regression analyses and Least Absolute Shrinkage and Selection Operator (LASSO)‐penalized Cox regression were employed to develop the BM-related lncRNAs signature. The formula for the risk score of each sample has the following formula: Risk score= , where "expr" denotes the levels of gene expression determined by using the prognostic risk score model and "coef" represents the non-zero regression coefficients that are determined by using LASSO Cox regression analysis[15]. The corresponding risk score for each BC patient was assessed as well. The RNAs were classified in both High-Risk (HR) (≥median number) group and Low-Risk (LR) (<median number) group according to the specific median score.
Correlation of risk scores and clinical characteristics:
In the TCGA cohort, the clinical features were incorporated with the risk score of each sample based on the sample ID. The correlation of clinical features and risk score was investigated with the application of limma R package. The comparison of clinical features between different groups was carried out after Kruskal-Wallis and Wilcoxon Rank-Sum (WRS) tests, where p<0.05 showed a considerable difference.
Prediction of potential compounds for BC treatment:
Half-Maximal Inhibitory Concentration (IC50) of conventional chemotherapeutic medicine was determined by using the pRRophetic R package[16]. IC50 represents the efficacy of a substance in inhibition of particular biological and biochemical functions. Wilcoxon signed-rank test was performed to evaluate the inter-group differences. The limma, pRRophetic, ggpubr and ggplot2 R packages were used in an attempt to predict which compounds could be used for BC treatment.
TME estimation:
The single sample Gene Set Enrichment Analysis (ssGSEA) was carried out with the "GSEABase" and "Gene Set Variation Analysis (GSVA)" R packages for the purpose of evaluating the immune-related infiltration in each sample. In addition, the gene sets acquired from previous studies were used for the assessment of the characteristics related to immune system in TME, which included various functions related to human immune and subtypes of immune cells. Examples included Cluster of Differentiation 8 (CD8+) T cells, Natural Killer (NK) T cells and regulatory T cells (Tregs)[17]. The correlation of risk scores and immune cells was investigated by means of Tumor Immune Estimation Resource (TIMER), Quantifying Tumor-Infiltrating Immune Cells from RNA sequence (quanTIseq), Cell-type Identification by Estimating Relative Subsets of RNA Transcripts (CIBERSORT), Estimate the Proportion of Immune and Cancer Cells (EPIC), Microenvironment Cell Population counter method (MCPcounter) and xCell in order to evaluate the status of immune infiltration, where p<0.05, it indicates statistical significance.
Consensus clustering using four BM-related prognostic lncRNAs:
To study their responses to immunotherapy, we divided BC patients into different subtypes using the R package ConsensesClusterPlus and the potential molecular subgroup immunotherapy response was explored according to the lncRNA expression level. The R package Rtsne was used to execute the Principal Component Analysis (PCA), t-distributed Stochastic Neighbor Embedding (t-SNE) and Kaplan-Meier (KM) survival curve. Immunoassay and drug sensitivity analyses were completed with R packages Limma, scales and pRRophetic.
Results and Discussion
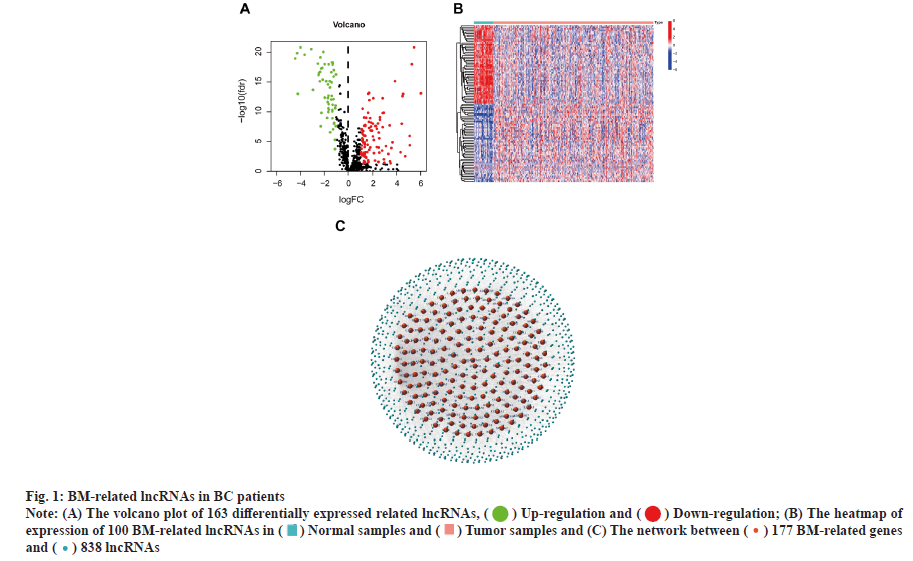
Identification of the BM-related lncRNAs in BC is shown in fig. 1. RNA-seq data from 385 samples, including 41 normal samples and 344 tumor samples, and their corresponding 1097 clinical samples were analyzed. Correlation analysis of expression of 16 773 lncRNAs with mRNA expression of 224 BM genes identified 838 BM-related lncRNAs. 163 lncRNAs associated with differentially expressed BM were reported. Fig. 1A-fig. 1C shows the correlation of 177 BM genes and 838 lncRNAs.
Fig. 1: BM-related lncRNAs in BC patients
Note: (A) The volcano plot of 163 differentially expressed related lncRNAs, (![]() ) Up-regulation and (
) Up-regulation and ( ) Down-regulation; (B) The heatmap of
expression of 100 BM-related lncRNAs in (
) Down-regulation; (B) The heatmap of
expression of 100 BM-related lncRNAs in (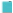 ) Normal samples and (
) Normal samples and ( ) Tumor samples and (C) The network between (
) Tumor samples and (C) The network between ( ) 177 BM-related genes
and (
) 177 BM-related genes
and ( ) 838 lncRNAs
) 838 lncRNAs
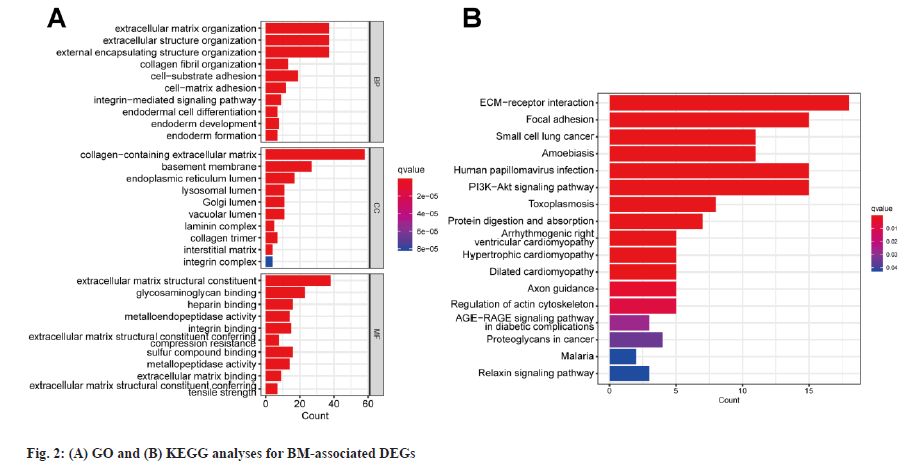
Enrichment analysis was shown in fig. 2. The comparison of expression levels of BMGs between different cancerous samples and healthy samples was carried out. 87 differentially expressed BMGs in BC tissue samples were acquired and they consisted of 33 upregulated and 54 downregulated differentially expressed BMGs. The GO enrichment analysis showed that structural components of ECM, ECM containing collagen and ECM tissues were exposed to high enrichment for GO terms (fig. 2A). The outcomes of the KEGG pathway enrichment assay showed the highly enriched ECM receptor interaction terms (fig. 2B). Overall, BMGs play a key role in the progression of BC.
Development and verification of the prognostic signature was shown in fig. 3. 7 out of 163 BMrelated lncRNAs were significantly correlated with Overall Survival (OS) as determined by uniCox regression analysis (p<0.05 in all cases) (fig. 3A). To prevent overfitting of prognostic signatures, we identified 4 lncRNAs associated with BM of BC through LASSO regression analysis. Fig. 3B is a heat map of the 4 constructed model lncRNAs. The following formula was employed to determine the risk score: AL691482.3×(- 0 . 3 1 8 4 7 8 6 6 7 0 6 3 3 0 8 ) + A C 0 9 7 7 1 3 . 1 × ( - 3 . 2 7 4 8 0 4 4 2 2 5 9 0 2 ) + L I N C 0 1 6 1 4 × Z ( 0 . 0 3 6 4 7 4 0 1 3 6 9 8 0 5 1 3 ) + S I A H 2 - AS1×(-1.24510230264465). Furthermore, survival probability, the distribution of survival time and risk scores, and the relevant expression of these lncRNAs between two risk groups were compared, which revealed that the HR group exhibited a low OS rate and worse prognosis (fig. 3C and fig. 3D).
Survival results and multivariate examination was shown in fig. 4. Univariate and multivariate Cox (multi-Cox) analyses indicated that age (Hazard Ratio (HR): 1.036, Confidence Interval (CI): 1.010- 1.062), lncRNAs characteristics (HR: 1.201, CI: 1.123-1.285) and tumor stage (HR: 1.916, 95% CI: 1.226-2.994) were independent prognostic factors for OS of patients with BC (fig. 4A and fig. 4B). The KM analyses demonstrated that the expression of HR lncRNAs signatures was correlated with degraded OS (p<0.001 and fig. 4C). According to the status plot of risk survival, the risk score was inversely proportional to the OS of patients with BC. The Area Under the Curve (AUC) predictions of the novel lncRNAs signature on 1 y, 2 y and 3 y survival were 0.742, 0.759 and 0.840, respectively (fig. 4D).
Additionally, the heat map indicating the relationship between prognostic features of BM-related lncRNA and clinicopathological presentation was analyzed (fig. 4E). The combination of clinicopathological features and BM-associated lncRNA prognostic features can be used for the clinical management of BC.
Fig. 4: uniCox and multi-Cox analysis for the expression of BM-related lncRNAs and BM-related lncRNAs prognostic signature and clinicopathological
manifestations
Note: (A) uniCox regression analysis; (B) Multi-Cox regression analysis; (C) KM curves results, ( ) HR; (
) HR; ( ) LR; (D) The AUC for the prediction
of (
) LR; (D) The AUC for the prediction
of ( ) 1 y, (
) 1 y, (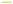 ) 2 y and (
) 2 y and (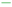 ) 3 y survival rate of BC; (E) Heatmap for BM-related lncRNAs prognostic signature and clinicopathological
manifestations
) 3 y survival rate of BC; (E) Heatmap for BM-related lncRNAs prognostic signature and clinicopathological
manifestations
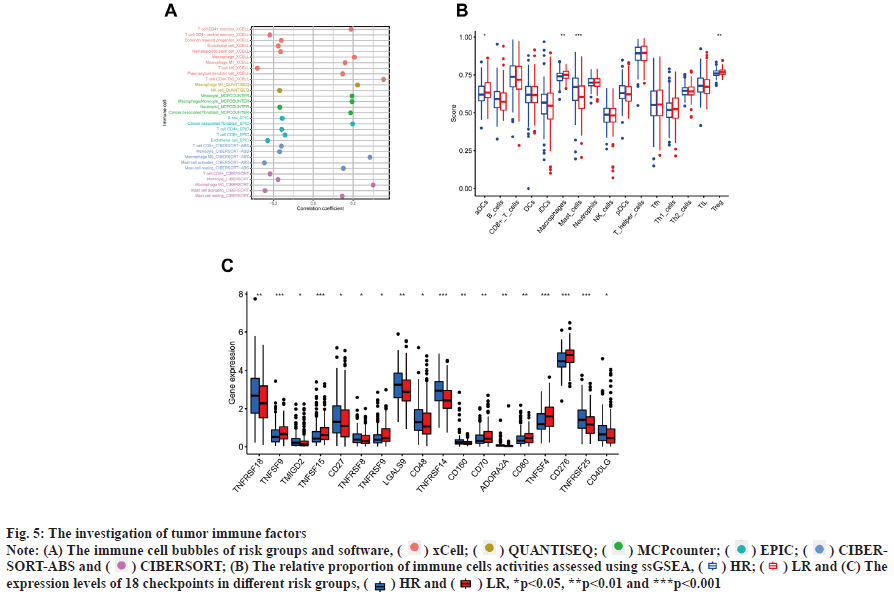
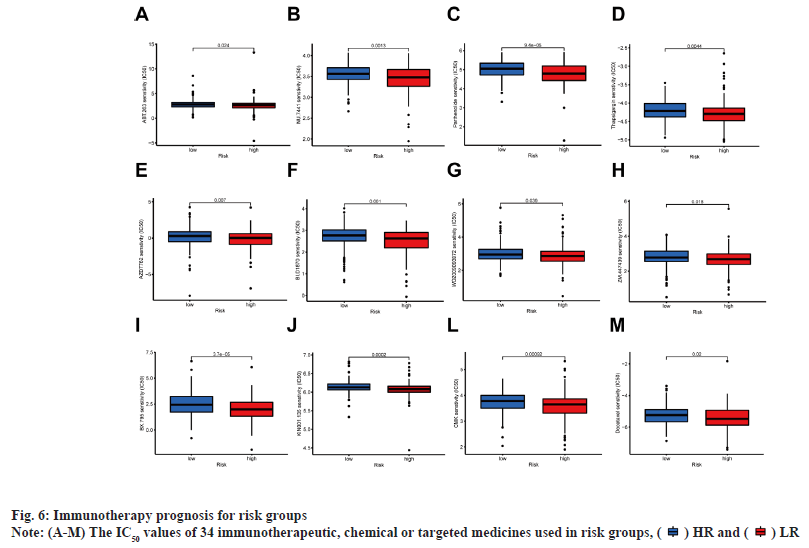
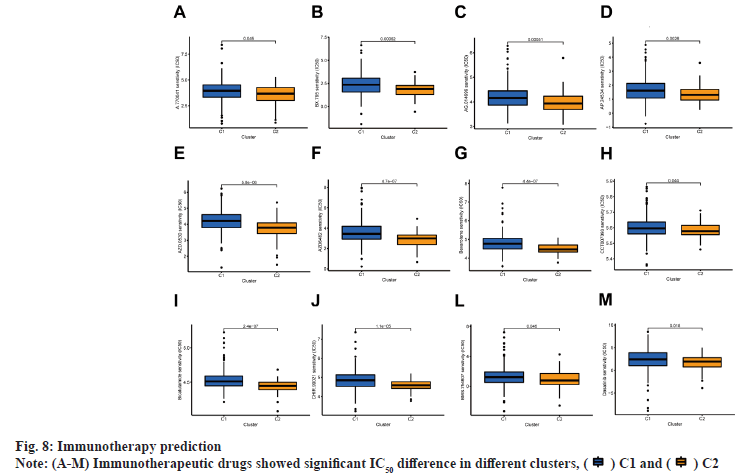
Immunoassay and drug sensitivity analysis of the risk population was shown in fig. 5. The correlation of immune cells calculated on different platforms revealed that the risk score was proportional to T cell Cluster of Differentiation 4 (CD4+) T-helper 2 cells (Th2)_xCell, Macrophage M0_CIBERSORT, Macrophage M0_CIBERSORT-ABS, Macrophage M1_QUANTISEQ, Macrophage_xCell, cancer associated fibroblast_EPIC and Monocyte_ MCPcounter. The risk score was inversely proportional to T cell NK_ xCell, mast cell activated_CIBERSORT-ABS, mast cell activated_CIBERSORT and endothelial cell_EPIC (p<0.05) (fig. 5A). The relative percentages of immune function as well as immune cells were higher in the HR group according to the results of ssGSEA analysis (fig. 5B). On the basis of the proposed risk model, we analyzed the expression levels of appropriate immune checkpoint inhibitors and found that most immune cells and immune checkpoints were infiltrated, followed by activation in the HR group (fig. 5C). IC50 of twelve immunotherapeutic, chemical and targeted medicines, such as Navitoclax (ABT-263) and NU- 7441 were low for the HR group, indicating that the HR group responded better to medications (fig. 6). The semi-inhibitory concentration values of 63 immunotherapeutic, chemotherapeutic or targeted drugs used in patients with BC were different in cluster 1 and cluster 2, including A-770041 and AG- 014699 which responded better to drug therapy in the HR group (fig. 6A-fig. 6M).
Fig. 5: The investigation of tumor immune factors
Note: (A) The immune cell bubbles of risk groups and software, ( ) xCell; (
) xCell; ( ) QUANTISEQ; (
) QUANTISEQ; ( ) MCPcounter; (
) MCPcounter; ( ) EPIC; (
) EPIC; ( ) CIBERSORT-
ABS and (
) CIBERSORT-
ABS and ( ) CIBERSORT; (B) The relative proportion of immune cells activities assessed using ssGSEA, (
) CIBERSORT; (B) The relative proportion of immune cells activities assessed using ssGSEA, ( ) HR; (
) HR; ( ) LR and (C) The
expression levels of 18 checkpoints in different risk groups, (
) LR and (C) The
expression levels of 18 checkpoints in different risk groups, ( ) HR and (
) HR and ( ) LR, *p<0.05, **p<0.01 and ***p<0.001
) LR, *p<0.05, **p<0.01 and ***p<0.001
The patients in this study were divided into two clusters on the basis of the expression level of BMrelated lncRNAs (fig. 7A)[18]. These two groups could be clearly distinguished among two clusters by PCA (fig. 7B) and t-SNE analysis (fig. 7C). According to the Sankey diagram, most patients in cluster 1 belonged to the LR group, while most patients in cluster 2 belonged to the HR group (fig. 7D). In KM analysis, the OS rate of cluster 1 was lower compared with that of other clusters (p<0.01) (fig. 7E). The TME was different in different clusters. Based on the results conducted on several platforms, cluster 2 exhibited a greater degree of immune cell infiltration (fig. 7F). The expression level of most immunological checkpoints in the cluster, including Cluster of Differentiation 276 (CD276), Hepatitis A Virus Cellular Receptor 2 (HAVCR2) (fig. 7G), which are highly effective for the treatment of hot tumors, decreased in descending order (cluster 2>cluster 1). Depending on various TMEs and immunotherapeutic responses, cluster 2 were classified as "hot" tumors, whereas cluster 1 was classified as "cold" tumor[19]. Based on cold and hot tumors, cluster 2 was more associated with cell infiltration and more sensitive for the immunotherapy. By comparing drug sensitivity, we discovered that 63 immunotherapeutic, chemical or targeted medicines, such as A-770041 had IC50 values that were lower in cluster 2 (fig. 8A -fig. 8M).
Fig. 7: Difference between cold and hot tumors
Note: (A) Patients were divided into two clusters by ConsensusClusterPlus, (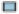 ) 1, (
) 1, ( ) 2 ; (B) The PCA of clusters, (
) 2 ; (B) The PCA of clusters, ( ) HR and (
) HR and ( ) LR; (C)
The t-SNE of two clusters, (
) LR; (C)
The t-SNE of two clusters, ( ) C1 and (
) C1 and ( ) C2; (D) The Sankey diagram of risk groups; (E) KM analysis, (
) C2; (D) The Sankey diagram of risk groups; (E) KM analysis, ( ) cluster C1 and (
) cluster C1 and ( ) C2; (F)
The heatmap of immune cells in clusters and (G) The difference of 18 checkpoints expression in clusters, (
) C2; (F)
The heatmap of immune cells in clusters and (G) The difference of 18 checkpoints expression in clusters, ( ) C1 and (
) C1 and ( ) C2, *p<0.05, **p<0.01
and ***p<0.001
) C2, *p<0.05, **p<0.01
and ***p<0.001
87 DEGs associated with BM were identified in this study. KEGG analysis further revealed that these genes are mainly related to ECM-receptor interaction signaling pathway, small cell lung cancer signaling pathway, amoebiasis, focal adhesion and Human Papillomavirus (HPV) infection signaling pathways.
It has been shown that overexpression of Long Intergenic Non-Protein Coding RNA 1615 (LINC01615) promotes BCC metastasis and reduces the OS of BC patients. The transcriptional activator, Signal-Induced Proliferation-Associated Protein 1 (SIPA1) can regulate the expression of LINC01615 and thus promote breast cell carcinogenesis[20].
In addition, LINC01614 proves to be highly expressed in BC and is related to poor Disease-Free Survival (DFS). Bioinformatics and pathway analyses were conducted on LINC01614 high vs. LINC01614 low in BC tissues, and the results demonstrated that ECM and Transforming Growth Factor beta 1 (TGF-β1) were the most activated networks in LINC01614 high tumors. Indeed, exogenous TGF-β1 may induce expression of LINC01614 in the human Breast Tumor cell line (BT474) triple positive BC model, while Focal Adhesion Kinase (FAK, PF-573228) or small-molecule inhibition of TGF-β (4-[4-(2H-1,3- Benzodioxol-5-yl)-5-(pyridin-2-yl)-1H-imidazol-2- yl]Benzamide (SB-431542)) abrogated LINC01614 expression. It was shown that LINC01614 is regulated by TGF-β and FAK signaling and can serve as a prognostic marker for BC[21]. LncRNA Forkhead Box D3 Antisense RNA 1 (FOXD3-AS1) plays a key role in BC. The expression of FOXD3-AS1 increased greatly in BC tissues and correlates with prognostic survival of patients. FOXD3-AS1 can function as a competitive endogenous RNA (ceRNA) and FOXD3-AS1 can affect cell proliferation, migration, ADP Ribosylation Factor 6 (ARF6) axis by targeting microRNA (miR)-127-3p/ARF6 invasion and tumor growth[22].
In addition, 4 lncRNA models associated with the BM were developed. The new models also divided patients into HR and LR groups, which were then analyzed using KM analysis, ROC analysis and semi-inhibitory concentration prediction. However, risk groups can be used as guidance for clinical evaluation and prognostic prediction, but cannot effectively differentiate between hot and cold tumors. Patients with BC are classified into different subtypes and these subtypes are also referred to as clusters. The TME also varies among subtypes. Different subtypes have different prognosis and immune response[19]. By dividing these lncRNAs into two groups by consensus clustering methods, we can see that cluster 1 has reduced CD8+ T cell infiltration and immunosuppressive TME, while cluster 2 has more CD8+ T infiltration and increased immune score and activity of Indoleamine 2,3-Dioxygenase 2 (IDO2) and T cell Immunoglobulin and Mucin-Domain Containing-3 (TIM3), thus cluster 2 is identified as a hot tumor and more sensitive to immunotherapeutic agents[18,23]. As we know, immunotherapy is more effective against hot tumors, such as anti-Programmed Death Ligand-1 (PD-L1)/Programmed Cell Death Protein 1 (PD-1) therapy. In order to obtain a better response of immunotherapy to tumors, various studies have focused on converting cold tumors into hot tumors[24,25].
From these results, it can be seen that BM-associated lncRNA can be used not only as a prognostic indicator, but also as a theoretical basis for individualized treatment of clinical patients. There is no reported role of BM-associated lncRNA in BC. In this study, an association between BM genes and lncRNA was identified. Therefore, the role of BM biomarkers in prediction of BC prognosis was explored, which could inform the treatment modalities for this disease. These BM-associated lncRNAs, may not only serve as a predictor of BC prognosis, but may also provide guidance for clinical drug use and contribute to further experimental studies and better understanding of BC pathogenesis. However, this study exhibits several limitations. First of all, the data for risk assessment of lncRNAs were acquired from databases that are available online and more external transcriptomic information for the validation of the role of inflammation-associated lncRNAs in BC was unavailable. The exact molecular mechanism of BM-associated lncRNAs in BC is not known and therefore additional molecular experiments are required. Additionally, 344 tumor samples and 41 normal samples were involved in the BC book and the sample heterogeneity may compromise the accuracy and precision of the data analysis. Overall, the prognostic prediction model proposed here needs further validation.
BM-associated lncRNAs can be used for prognosis prediction and improvement of personalized treatment of BC by effectively identifying hot and cold tumors, and provide new therapeutic targets for tumor patients. Immunotherapy and targeted therapy with checkpoint inhibition have completely changed the original treatment of BC. Targeting BM and lncRNAs will provide more immunotherapeutic options for BC patients, elucidate and identify the immune mechanisms of BM-associated lncRNAs in patients with BC, and offer new therapeutic approaches for patients with BC.
Author’s contributions:
Jiangyao Zhou contributed equally to this work. Jingwei Zhang designed the project and supervised the study; Jiangyao Zhou and Lei Wei contributed to the data analysis and original draft preparation; Jiangyao Zhou and Fangfang Chen reviewed and edited the manuscript; Yiqing Xi and Meishun Hu provided valuable suggestions for study and Jun Li and Qianqian Tang edited the language. All authors have read and approved the version of the manuscript.
Funding:
This study was supported by the National Natural Science Foundation of China (8217113494).
Conflict of interests:
The authors declare that there are no conflicts of interest.
References
- Fan L, Strasser-Weippl K, Li JJ, St Louis J, Finkelstein DM, Yu KD, et al. Breast cancer in China. Lancet Oncol 2014;15(7):e279-89.
[Crossref] [Google scholar] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69(1):7-34.
[Crossref] [Google scholar] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70(1):7-30.
[Crossref] [Google scholar] [PubMed]
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71(3):209-49.
[Crossref] [Google scholar] [PubMed]
- Houthuijzen JM, Jonkers J. Cancer-associated fibroblasts as key regulators of the breast cancer tumor microenvironment. Cancer Metastasis Rev 2018;37:577-97.
[Crossref] [Google scholar] [PubMed]
- Mao Y, Keller ET, Garfield DH, Shen K, Wang J. Stromal cells in tumor microenvironment and breast cancer. Cancer Metastasis Rev 2013;32:303-15.
[Crossref] [Google scholar] [PubMed]
- Reuten R, Zendehroud S, Nicolau M, Fleischhauer L, Laitala A, Kiderlen S, et al. Basement membrane stiffness determines metastases formation. Nat Mater 2021;20(6):892-903.
[Crossref] [Google scholar] [PubMed]
- Mohan V, Das A, Sagi I. Emerging roles of ECM remodeling processes in cancer. Semin Cancer Biol 2020;62:192-200.
[Crossref] [Google scholar] [PubMed]
- Zhang T, Li K, Zhang ZL, Gao K, Lv CL. LncRNA Airsci increases the inflammatory response after spinal cord injury in rats through the nuclear factor kappa B signaling pathway. Neural Regen Res 2021;16(4):772-7.
[Crossref] [Google scholar] [PubMed]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010;464(7291):1071-6.
[Crossref] [Google scholar] [PubMed]
- Liu S, Chen W, Hu H, Zhang T, Wu T, Li X, et al. Long noncoding RNA PVT1 promotes breast cancer proliferation and metastasis by binding miR-128-3p and UPF1. Breast Cancer Res 2021;23(1):1-13.
[Crossref] [Google scholar] [PubMed]
- Filippova EA, Fridman MV, Burdennyy AM, Loginov VI, Pronina IV, Lukina SS, et al. Long noncoding RNA GAS5 in breast cancer: Epigenetic mechanisms and biological functions. Int J Mol Sci 2021;22(13):1-17.
[Crossref] [Google scholar] [PubMed]
- Tomczak K, Czerwińska P, Wiznerowicz M. Review The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp Oncol 2015;19(1):A68-77.
[Crossref] [Google scholar] [PubMed]
- Jayadev R, Morais MR, Ellingford JM, Srinivasan S, Naylor RW, Lawless C, et al. A basement membrane discovery pipeline uncovers network complexity, regulators and human disease associations. Sci Adv 2022;8(20):1-19.
[Crossref] [Google scholar] [PubMed]
- Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 2010;33(1):1-22.
[Crossref] [Google scholar] [PubMed]
- Geeleher P, Cox N, Huang RS. pRRophetic: An R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One 2014;9(9):1-3.
[Crossref] [Google scholar] [PubMed]
- Zhao Z, Liu H, Zhou X, Fang D, Ou X, Ye J, et al. Necroptosis-related lncRNAs: Predicting prognosis and the distinction between the cold and hot tumors in gastric cancer. J Oncol 2021;2021:1-16.
[Crossref] [Google scholar] [PubMed]
- Wilkerson MD, Hayes DN. ConsensusClusterPlus: A class discovery tool with confidence assessments and item tracking. Bioinformatics 2010;26(12):1572-3.
[Crossref] [Google scholar] [PubMed]
- Todenhöfer T, Seiler R. Molecular subtypes and response to immunotherapy in bladder cancer patients. Transl Androl Urol 2019;8(3):S293-S5.
[Crossref] [Google scholar] [PubMed]
- Xiang Y, Feng L, Liu H, Liu Y, Li J, Su L, et al. SIPA1 Regulates LINC01615 to Promote Metastasis in Triple-Negative Breast Cancer. Cancers 2022;14(19):1-13.
[Crossref] [Google scholar] [PubMed]
- Vishnubalaji R, Shaath H, Elkord E, Alajez NM. Long non-coding RNA (lncRNA) transcriptional landscape in breast cancer identifies LINC01614 as non-favorable prognostic biomarker regulated by TGFβ and Focal Adhesion Kinase (FAK) signaling. Cell Death Discov 2019;5(1):1-15.
[Crossref] [Google scholar] [PubMed]
- Zhang X, Zhao X, Chang L, Liu F, Li C, Ge P. LncRNA FOXD3-AS1 promotes breast cancer progression by mediating ARF6. Breast Cancer 2022;29(5):908-20.
[Crossref] [Google scholar] [PubMed]
- Galon J, Bruni D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat Rev Drug Discov 2019;18(3):197-218.
[Crossref] [Google scholar] [PubMed]
- Gajewski TF. The next hurdle in cancer immunotherapy: Overcoming the non-T-cell-inflamed tumor microenvironment. Semin Oncol 2015:42(4):663-671.
[Crossref] [Google scholar] [PubMed]
- Zemek RM, De Jong E, Chin WL, Schuster IS, Fear VS, Casey TH, et al. Sensitization to immune checkpoint blockade through activation of a STAT1/NK axis in the tumor microenvironment. Sci Transl Med 2019;11(501):eaav7816.
[Crossref] [Google scholar] [PubMed]
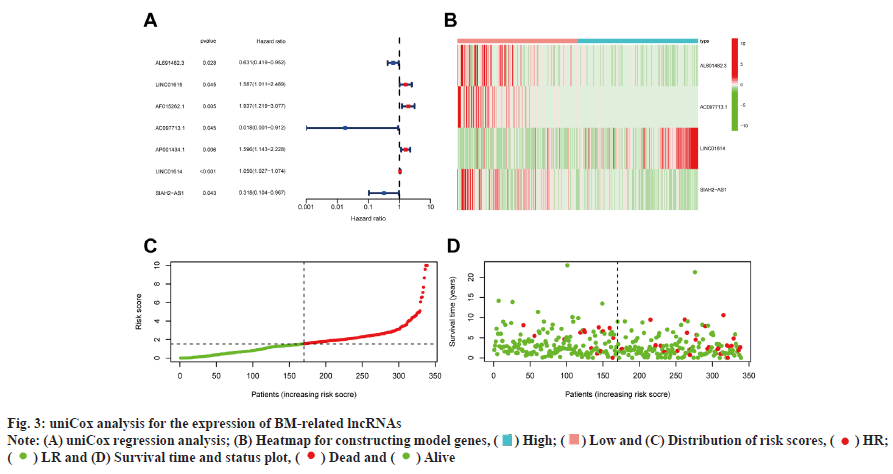
 ) Low and (C) Distribution of risk scores, (
) Low and (C) Distribution of risk scores, ( ) HR;
(
) HR;
( ) LR and (D) Survival time and status plot, (
) LR and (D) Survival time and status plot, (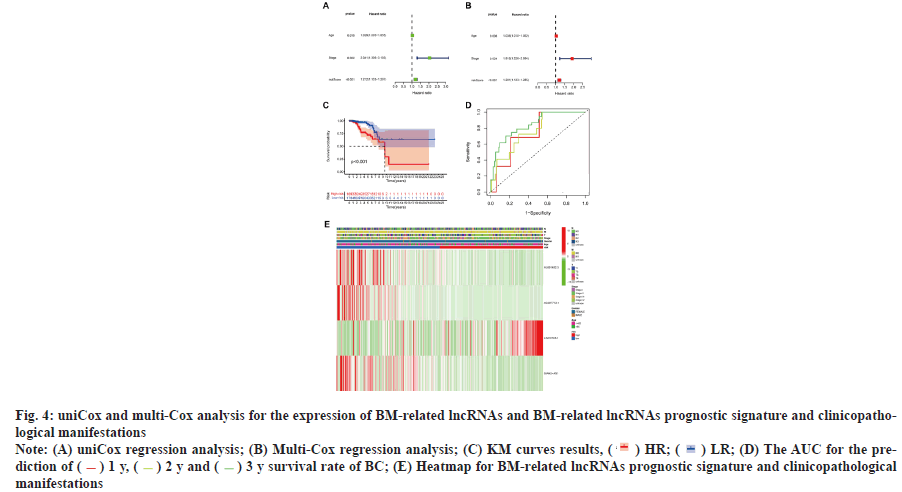
 ) HR and (
) HR and ( ) LR
) LR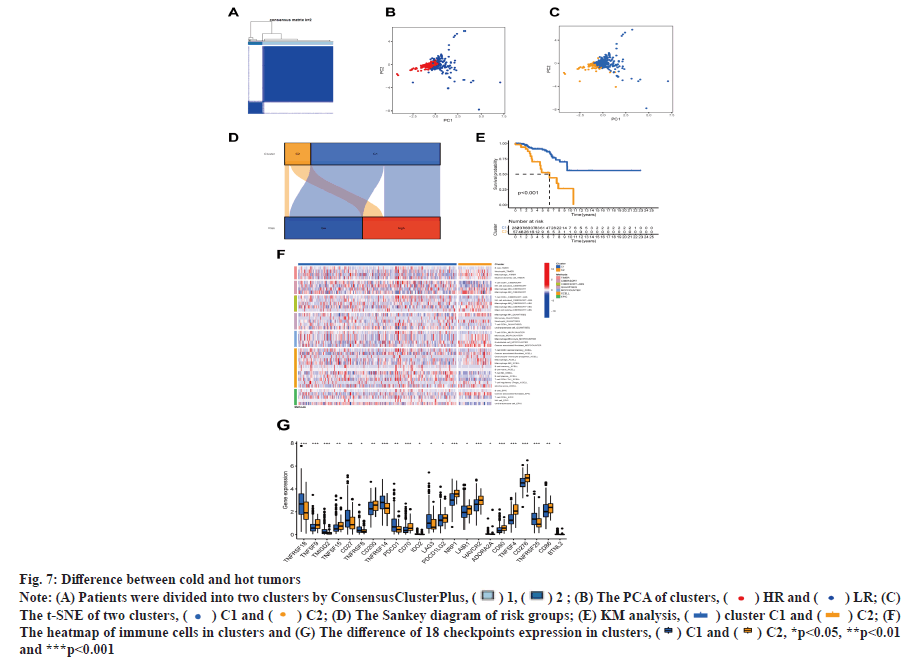
 ) C1 and (
) C1 and ( ) C2
) C2



